Molecular Decoding of Phytohormone Crosstalk: JA-Mediated Key Regulatory Nodes and Signal Integration
Abstract
1. Introduction
2. Molecular Mechanisms of Jasmonate Interactions with Other Hormones
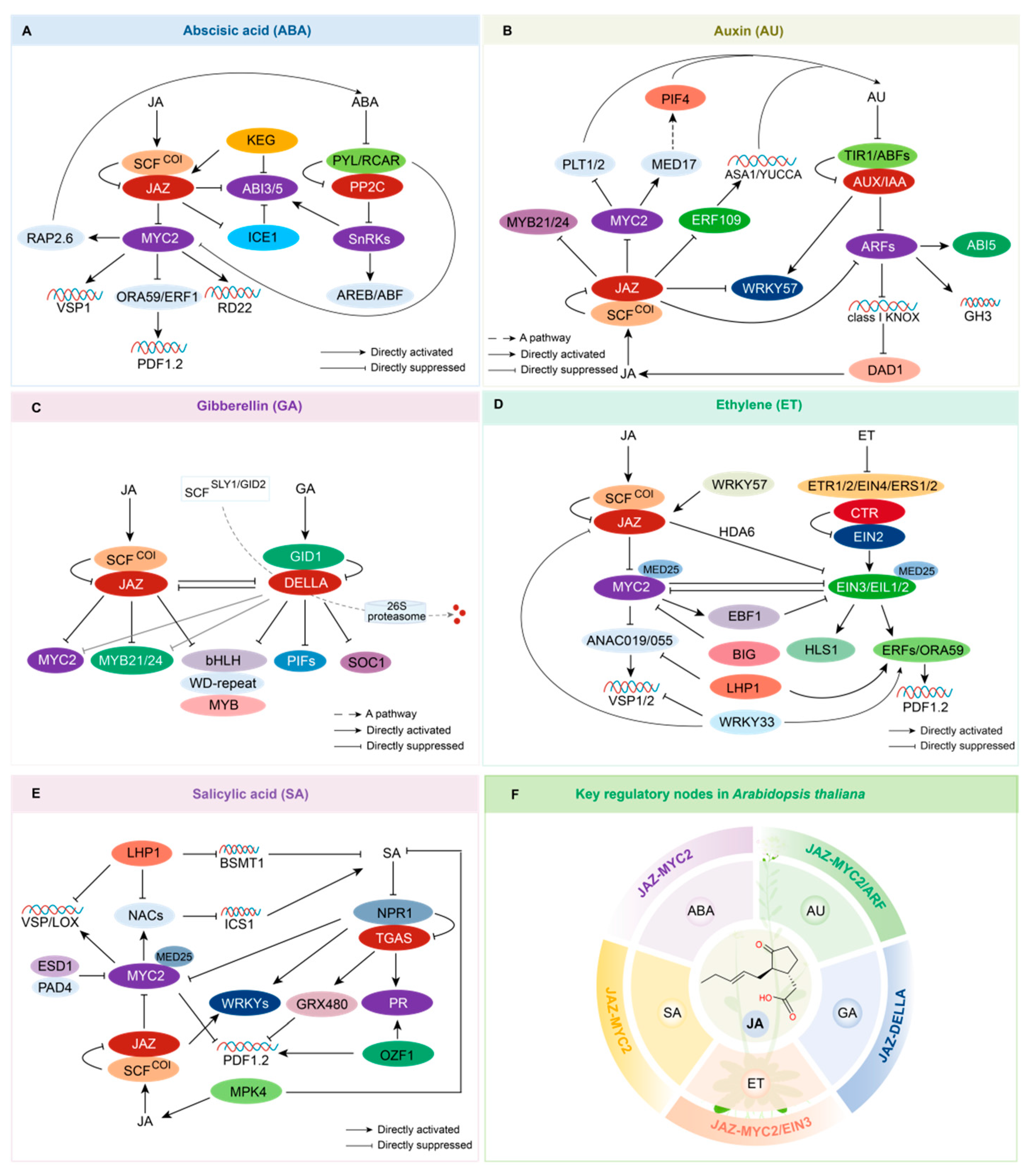
2.1. JA and ABA Crosstalk: JAZ-MYC2
2.2. JA and Auxin (AU) Crosstalk: JAZ, MYC2, and ARF
2.3. JA and GA Crosstalk: JAZ-DELLA
2.4. JA and Ethylene Crosstalk: JAZ, MYC2, and EIN3
2.5. JA and SA Crosstalk: MYC2, NPR1
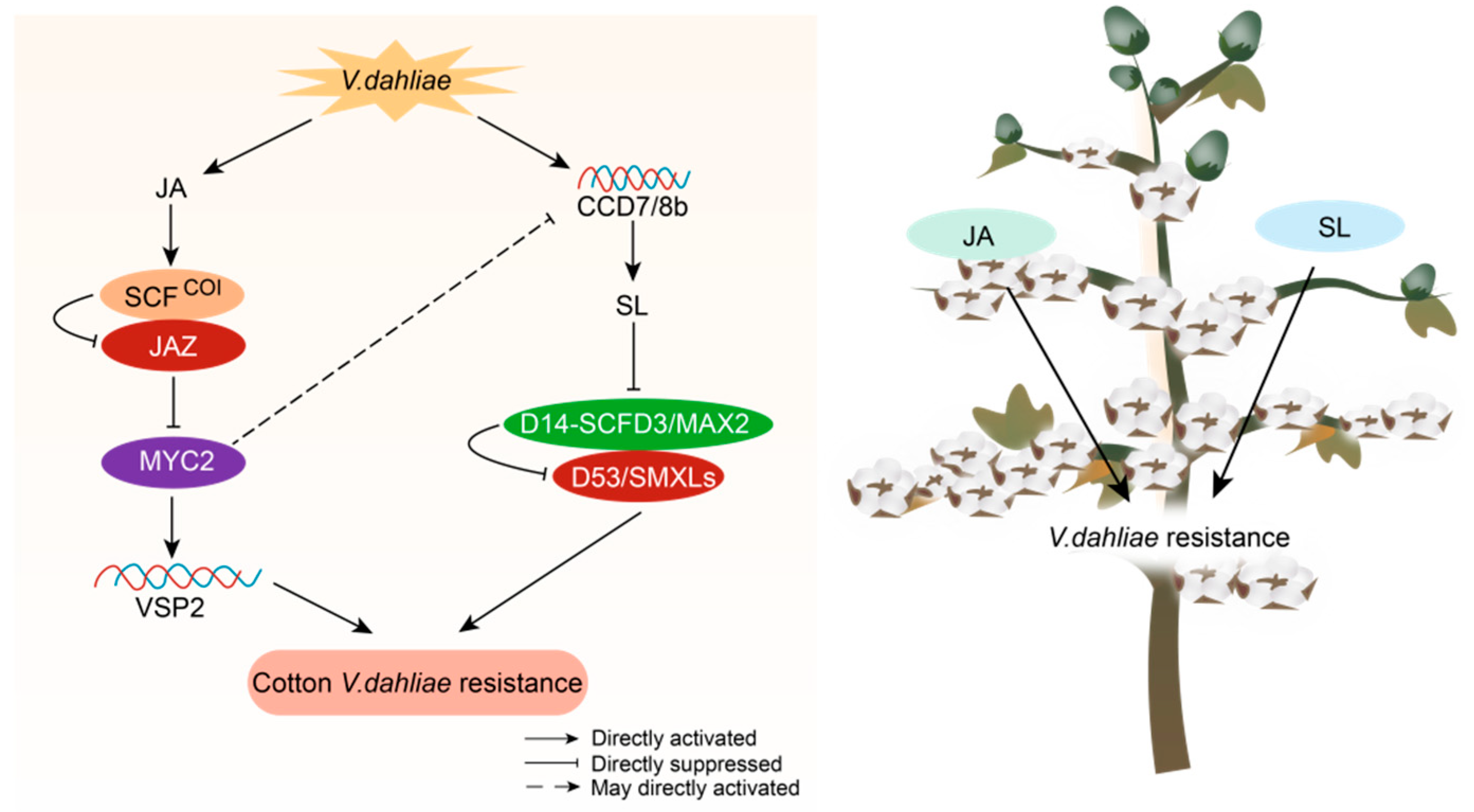
2.6. JA and SL Crosstalk
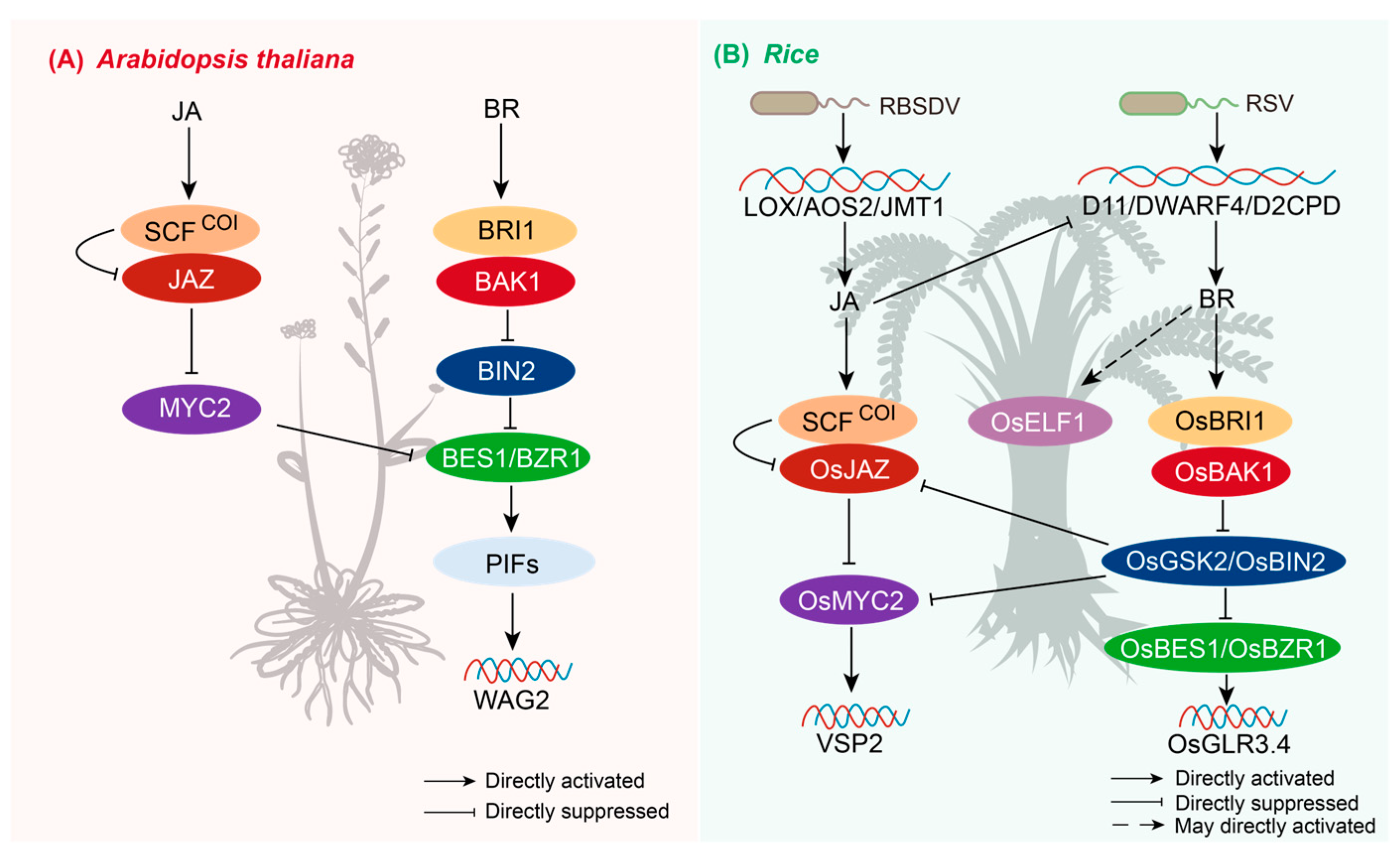
2.7. JA and BR Crosstalk
2.8. JA in Multi-Stress and Symbiotic Contexts
2.9. JA-Mediated Chromatin Remodeling
3. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lyons, R.; Manners, J.M.; Kazan, K. Jasmonate biosynthesis and signaling in monocots: A comparative overview. Plant Cell Rep. 2013, 32, 815–827. [Google Scholar] [CrossRef]
- Widemann, E.; Smirnova, E.; Aubert, Y.; Miesch, L.; Heitz, T. Dynamics of Jasmonate Metabolism upon Flowering and across Leaf Stress Responses in Arabidopsis thaliana. Plants 2016, 5, 4. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Liu, H.; Timko, M.P. Jasmonic Acid Signaling and Molecular Crosstalk with Other Phytohormones. Int. J. Mol. Sci. 2021, 22, 2914. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Chung, H.S.; Koo, A.J.K.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef]
- Reinbothe, C.; Springer, A.; Samol, I.; Reinbothe, S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009, 276, 4666–4681. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Devoto, A.; Nieto-Rostro, M.; Xie, D.; Ellis, C.; Harmston, R.; Patrick, E.; Davis, J.; Sherratt, L.; Coleman, M.; Turner, J.G. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002, 32, 457–466. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernández-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular Mechanism Underlying the Synergetic Effect of Jasmonate on Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489. [Google Scholar] [CrossRef] [PubMed]
- Herud-Sikimić, O.; Stiel, A.C.; Kolb, M.; Shanmugaratnam, S.; Berendzen, K.W.; Feldhaus, C.; Höcker, B.; Jürgens, G. A biosensor for the direct visualization of auxin. Nature 2021, 592, 768–772. [Google Scholar] [CrossRef]
- Zhai, Q.; Deng, L.; Li, C. Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86. [Google Scholar] [CrossRef]
- Reynolds, M.; Atkin, O.K.; Bennett, M.; Cooper, M.; Dodd, I.C.; Foulkes, M.J.; Frohberg, C.; Hammer, G.; Henderson, I.R.; Huang, B.; et al. Addressing Research Bottlenecks to Crop Productivity. Trends Plant Sci. 2021, 26, 607–630. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ Proteins: A Crucial Interface in the Jasmonate Signaling Cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Nguyen, T.-N.; Tuan, P.A.; Ayele, B.T. Jasmonate regulates seed dormancy in wheat via modulating the balance between gibberellin and abscisic acid. J. Exp. Bot. 2022, 73, 2434–2453. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bhardwaj, M.; Tran, L.-S.P. JASMONATE ZIM-DOMAIN Family Proteins: Important Nodes in JasmonicAcid-Abscisic Acid Crosstalk for Regulating Plant Response toDrought. Curr. Protein Pept. Sci. 2021, 22, 759–766. [Google Scholar] [CrossRef]
- Vos, I.A.; Verhage, A.; Schuurink, R.C.; Watt, L.G.; Pieterse, C.M.J.; Van Wees, S.C.M. Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 2013, 4, 539. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Dong, C.; Wu, Y.; Fu, S.; Tauqeer, A.; Gu, X.; Li, Q.; Niu, X.; Liu, P.; Zhang, X.; et al. The JA-to-ABA signaling relay promotes lignin deposition for wound healing in Arabidopsis. Mol. Plant 2024, 17, 1594–1605. [Google Scholar] [CrossRef]
- Lijun, L.; Tang, C.; Zhang, Y.; Sha, X.; Tian, S.; Luo, Z.; Wei, G.; Zhu, L.; Li, Y.; Jingye, F.; et al. The SnRK2.2-ZmHsf28-JAZ14/17 module regulates drought tolerance in maize. New Phytol. 2024, 245, 1985–2003. [Google Scholar] [CrossRef]
- Wan, Q.; Yao, R.; Zhao, Y.; Xu, L. JA and ABA signaling pathways converge to protect plant regeneration in stress conditions. Cell Rep. 2025, 44, 115423. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72- AOC ’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Wei, X.; Wei, X.; Guan, W.; Nong, W.; Chen, R.; Tao, X.; Mao, L. ABA-responsive transcription factor ABF1-1 promotes JA biosynthesis to accelerate suberin polyphenolic formation in wounded kiwifruit (Actinidia chinensis). Postharvest Biol. Technol. 2022, 187, 111850. [Google Scholar] [CrossRef]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The Transcription Factor INDUCER OF CBF EXPRESSION1 Interacts with ABSCISIC ACID INSENSITIVE5 and DELLA Proteins to Fine-Tune Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef]
- Varshney, V.; Majee, M. JA Shakes Hands with ABA to Delay Seed Germination. Trends Plant Sci. 2021, 26, 764–766. [Google Scholar] [CrossRef]
- Liu, H.; Stone, S.L. Abscisic Acid Increases Arabidopsis ABI5 Transcription Factor Levels by Promoting KEG E3 Ligase Self-Ubiquitination and Proteasomal Degradation[W][OA]. Plant Cell 2010, 22, 2630–2641. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 Ligase Essential for Arabidopsis Growth and Development, Is Involved in Abscisic Acid Signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar] [CrossRef]
- Pauwels, L.; Ritter, A.; Goossens, J.; Durand, A.N.; Liu, H.; Gu, Y.; Geerinck, J.; Boter, M.; Vanden Bossche, R.; De Clercq, R.; et al. The RING E3 Ligase KEEP ON GOING Modulates JASMONATE ZIM-DOMAIN12 Stability. Plant Physiol. 2015, 169, 1405–1417. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, D.; Yan, X.; Wu, Z.; Kayani, S.I.; Shen, Q.; Fu, X.; Xie, L.; Hao, X.; Hassani, D.; et al. Jasmonate- and abscisic acid-activated AaGSW1-AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 2021, 19, 1412–1428. [Google Scholar] [CrossRef]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K. The AP2/ERF transcription factor PtoERF15 confers drought tolerance via JA -mediated signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.J.; Spatola Rossi, T.; Kriechbaumer, V. Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Vanneste, S.; Pei, Y.; Friml, J. Mechanisms of auxin action in plant growth and development. Nat. Rev. Mol. Cell Biol. 2025. ahead of print. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef]
- Cai, X.T.; Xu, P.; Zhao, P.-X.; Liu, R.; Yu, L.-H.; Xiang, C.-B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 Is Important for Jasmonate-Mediated Regulation of Auxin Biosynthesis and Transport during Lateral Root Formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Păcurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Păcurar, M.; Demailly, H.; Geiss, G.; et al. Auxin Controls Arabidopsis Adventitious Root Initiation by Regulating Jasmonic Acid Homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Zhang, M.; Ye, J.; Du, J.; Jiang, Y.; Hu, Y. Auxin contributes to jasmonate-mediated regulation of abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2023, 35, 1110–1133. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.-F.; Yadav, V.; Tholl, D.; Chételat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A Regulatory Network for Coordinated Flower Maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef]
- Qiao, Z.W.; Ji, X.L.; Li, H.L.; Wang, X.; Zhang, C.L.; Wang, X.F.; You, C.X. MdARF8: An Auxin Response Factor Involved in Jasmonate Signaling Pathway in Malus domestica. J. Plant Growth Regul. 2023, 42, 1738–1749. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 Functions as a Node of Convergence for Jasmonic Acid– and Auxin-Mediated Signaling in Jasmonic Acid–Induced Leaf Senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef]
- Lahey, K.A.; Yuan, R.; Burns, J.K.; Ueng, P.P.; Timmer, L.W.; Kuang-Ren, C. Induction of phytohormones and differential gene expression in citrus flowers infected by the fungus Colletotrichum acutatum. Mol. Plant Microbe Interact. 2004, 17, 1394–1401. [Google Scholar] [CrossRef]
- Shinde, R.; Ayyanath, M.-M.; Shukla, M.; El Kayal, W.; Saxena, P.K.; Subramanian, J. Salicylic and Jasmonic Acid Synergism during Black Knot Disease Progression in Plums. Plants 2024, 13, 292. [Google Scholar] [CrossRef]
- Vinutha, T.; Vanchinathan, S.; Bansal, N.; Kumar, G.; Permar, V.; Watts, A.; Ramesh, S.V.; Praveen, S. Tomato auxin biosynthesis/signaling is reprogrammed by the geminivirus to enhance its pathogenicity. Planta 2020, 252, 51. [Google Scholar] [CrossRef]
- Agrawal, R.; Sharma, M.; Dwivedi, N.; Maji, S.; Thakur, P.; Junaid, A.; Fajkus, J.; Laxmi, A.; Thakur, J.K. MEDIATOR SUBUNIT17 integrates jasmonate and auxin signaling pathways to regulate thermomorphogenesis. Plant Physiol. 2022, 189, 2259–2280. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Jones, R.L. Gibberellin signaling. Curr. Opin. Plant Biol. 1998, 1, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Sun, T. Gibberellin-GID1-DELLA: A Pivotal Regulatory Module for Plant Growth and Development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Um, T.Y.; Lee, H.Y.; Lee, S.; Chang, S.H.; Chung, P.J.; Oh, K.-B.; Kim, J.-K.; Jang, G.; Choi, Y.D. Jasmonate Zim-Domain Protein 9 Interacts With Slender Rice 1 to Mediate the Antagonistic Interaction Between Jasmonic and Gibberellic Acid Signals in Rice. Front. Plant Sci. 2018, 9, 1866. [Google Scholar] [CrossRef]
- Li, C.; He, X.; Luo, X.; Xu, L.; Liu, L.; Min, L.; Jin, L.; Zhu, L.; Zhang, X. Cotton WRKY1 Mediates the Plant Defense-to-Development Transition during Infection of Cotton by Verticillium dahliae by Activating JASMONATE ZIM-DOMAIN1 Expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 Interacts with DELLA Proteins in Regulating Sesquiterpene Synthase Gene Expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed]
- Jin, G. Jasmonate-mediated gibberellin catabolism constrains growth during herbivore attack in rice. Plant Cell 2023, 35, 3828–3844. [Google Scholar] [CrossRef]
- Panda, S.; Jozwiak, A.; Sonawane, P.D.; Szymanski, J.; Kazachkova, Y.; Vainer, A.; Kilambi, H.V.; Almekias-Siegl, E.; Dikaya, V.; Bocobza, S.; et al. Steroidal alkaloids defence metabolism and plant growth are modulated by the joint action of gibberellin and jasmonate signalling. New Phytol. 2022, 233, 1220–1237. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, R.-R.; Guo, X.-L.; Liu, Y.-J.; You, C.; Han, Y.; An, J.-P. The MdNAC72-MdABI5 module acts as an interface integrating jasmonic acid and gibberellin signals and undergoes ubiquitination-dependent degradation regulated by MdSINA2 in apple. New Phytol. 2024, 243, 997–1016. [Google Scholar] [CrossRef]
- Ji, X.L.; Zhao, L.L.; Liu, B.; Yuan, Y.B.; Han, Y.; You, C.X.; An, J.-P. MdZFP7 integrates JA and GA signals via interaction with MdJAZ2 and MdRGL3a in regulating anthocyanin biosynthesis and undergoes degradation by the E3 ubiquitin ligase MdBRG3. J. Integr. Plant Biol. 2025, 67, 1339–1363. [Google Scholar] [CrossRef]
- Wen, J.; Li, Y.; Qi, T.; Gao, H.; Liu, B.; Zhang, M.; Huang, H.; Song, S. The C-terminal domains of Arabidopsis GL3/EGL3/TT8 interact with JAZ proteins and mediate dimeric interactions. Plant Signal. Behav. 2018, 13, e1422460. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM Domain Proteins Interact with the R2R3-MYB Transcription Factors MYB21 and MYB24 to Affect Jasmonate-Regulated Stamen Development in Arabidopsis[C][W]. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef]
- Huang, H.; Gong, Y.; Liu, B.; Wu, D.; Zhang, M.; Xie, D.; Song, S. The DELLA proteins interact with MYB21 and MYB24 to regulate filament elongation in Arabidopsis. BMC Plant Biol. 2020, 20, 64. [Google Scholar] [CrossRef]
- Cheng, H.; Song, S.; Xiao, L.; Soo, H.M.; Cheng, Z.; Xie, D.; Peng, J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009, 5, e1000440. [Google Scholar] [CrossRef]
- Wang, Z.; Wong, D.C.J.; Wang, Y.; Xu, G.; Ren, C.; Liu, Y.; Kuang, Y.; Fan, P.; Li, S.; Xin, H.; et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021, 186, 1660–1678. [Google Scholar] [CrossRef]
- Virág, E.; Hegedűs, G.; Nagy, Á.; Pallos, J.P.; Kutasy, B. Temporal Shifts in Hormone Signaling Networks Orchestrate Soybean Floral Development Under Field Conditions: An RNA-Seq Study. Int. J. Mol. Sci. 2025, 26, 6455. [Google Scholar] [CrossRef]
- Zhu, Z. Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis. J. Exp. Bot. 2014, 65, 5743–5748. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N. The Molecular Mechanism of the BZR1-EIN3 Transcription Complex Integrating Brassinosteroid and Ethylene Signals to Regulate Plant Growth and Development. Available online: https://kns.cnki.net/kcms2/article/abstract?v=CdHX_LbaUYwxRJCJVfRaJYfQUVeIGmfEj3mcw319VW7dsuEYHjx-CoHeOBJWIGibp1MRNlRcuhvr5LKO_wqXU2lm0Nyi3s0LUH95DYt1oWDFUxxOnGVe07TtflTCfKUO5s3L1oPA6HldyX9kYG-WRGkNrXMYaSo4pCpltx6p-Kd_S1SNCRZ8IazG8FuDDYc_dRMh-57us94=&uniplatform=NZKPT&language=CHS (accessed on 9 November 2021).
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.-M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 Modulates Antagonism between Jasmonate and Ethylene Signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wei, C.; Ma, Q.; Dong, H.; Shi, K.; Zhou, Y.; Foyer, C.H.; Yu, J. Ethylene response factors 15 and 16 trigger jasmonate biosynthesis in tomato during herbivore resistance. Plant Physiol. 2021, 185, 1182–1197. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Liu, X.-F.; Fu, B.-L.; Zhang, Q.-Y.; Tong, Y.; Wang, J.; Wang, W.-Q.; Grierson, D.; Yin, X.-R. Methyl Jasmonate Enhances Ethylene Synthesis in Kiwifruit by Inducing NAC Genes That Activate ACS1. J. Agric. Food Chem. 2020, 68, 3267–3276. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef]
- Liu, R.; Niimi, H.; Ueda, M.; Takaoka, Y. Coordinately regulated transcription factors EIN3/EIL1 and MYCs in ethylene and jasmonate signaling interact with the same domain of MED25. Biosci. Biotechnol. Biochem. 2022, 86, 1405–1412. [Google Scholar] [CrossRef]
- Li, C.; Xu, M.; Cai, X.; Han, Z.; Si, J.; Chen, D. Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs. Int. J. Mol. Sci. 2022, 23, 3945. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xu, H.; Huang, J.; Kong, Y.; AbuQamar, S.; Yu, D.; Liu, S.; Zhou, G.; Chai, G. The Arabidopsis CCCH protein C3H14 contributes to basal defense against Botrytis cinerea mainly through the WRKY33-dependent pathway. Plant Cell Environ. 2020, 43, 1792–1806. [Google Scholar] [CrossRef]
- Zhou, J.; Mu, Q.; Wang, X.; Zhang, J.; Yu, H.; Huang, T.; He, Y.; Dai, S.; Meng, X. Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell 2022, 34, 3066–3087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ge, S.; He, J.; Li, S.; Hao, Y.; Du, H.; Liu, Z.; Cheng, R.; Feng, Y.; Xiong, L.; et al. BIG regulates stomatal immunity and jasmonate production in Arabidopsis. New Phytol. 2019, 222, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, T.; Sun, Y.; Zhang, Y.; Radojičić, A.; Ding, Y.; Tian, H.; Huang, X.; Lan, J.; Chen, S.; et al. Diverse Roles of the Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Plant Immunity. Plant Cell 2020, 32, 4002–4016. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef]
- Moreau, M.; Tian, M.; Klessig, D.F. Salicylic acid binds NPR3 and NPR4 to regulate NPR1-dependent defense responses. Cell Res. 2012, 22, 1631–1633. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Zheng, X.; Spivey, N.W.; Zeng, W.; Liu, P.-P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine Promotes Pseudomonas syringae Virulence in Plants by Activating a Signaling Cascade that Inhibits Salicylic Acid Accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Ortigosa, A.; García-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.R.; Ntoukakis, V.; Solano, R. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017, 213, 1378–1392. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, M.; Skelly, M.J.; Itaya, T.; Mori, T.; Suzuki, T.; Matsushita, T.; Tokizawa, M.; Kuwata, K.; Mori, H.; Yamamoto, Y.Y.; et al. Suppression of MYC transcription activators by the immune cofactor NPR1 fine-tunes plant immune responses. Cell Rep. 2021, 37, 110125. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent Defense Pathways through a Novel Function in the Cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef]
- Meng, F.; Yang, C.; Cao, J.; Chen, H.; Pang, J.; Zhao, Q.; Wang, Z.; Qing Fu, Z.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Verk, M.C.V.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic Acid Suppresses Jasmonic Acid Signaling Downstream of SCFCOI1-JAZ by Targeting GCC Promoter Motifs via Transcription Factor ORA59. Plant Cell 2013, 25, 744. [Google Scholar] [CrossRef]
- Balbi, V.; Devoto, A. Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 2008, 177, 301–318. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. Jasmonate Signaling: Toward an Integrated View. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of Transcription Factor MYC2 by EDS1/PAD4 Complexes Bolsters Salicylic Acid Defense in Arabidopsis Effector-Triggered Immunity. Mol. Plant 2018, 11, 1053–1066. [Google Scholar] [CrossRef]
- Singh, N.; Nandi, A.K. AtOZF1 positively regulates JA signaling and SA-JA cross-talk in Arabidopsis thaliana. J. Biosci. 2022, 47, 8. [Google Scholar] [CrossRef]
- Schwarzenbacher, R.E.; Wardell, G.; Stassen, J.; Guest, E.; Zhang, P.; Luna, E.; Ton, J. The IBI1 Receptor of β-Aminobutyric Acid Interacts with VOZ Transcription Factors to Regulate Abscisic Acid Signaling and Callose-Associated Defense. Mol. Plant 2020, 13, 1455–1469. [Google Scholar] [CrossRef]
- Huang, Z.; Li, B.; Wang, J. Physiological Response and Its Principal Component Analysis of Two Barley Varieties with Different Resistances to Bipolaris sorokiniana. Available online: https://kns.cnki.net/kcms2/article/abstract?v=rGdVK2OACdeQ4Gt0zB_I2uvTqgUohmYmOFTn-Gjf2Vec3meWfMu5lLhDHdWRgJ6QXVyqRmyj8P0Js6Q6CzBubLZlNeFO0cnbQ26Et6qTY9kyjFTCkUfHAg07wJl_RBaG4OX1JX2LIPex3n4p0Qwr2lfdoDIKhhsoXmN2tPK9i3PZJd1S-6qhEw==&uniplatform=NZKPT&language=CHS (accessed on 14 August 2025).
- Jia, H.; Hewitt, N.; Jordá, L.; Abramyan, T.M.; Tolliver, J.; Jones, J.L.; Nomura, K.; Yang, J.; He, S.-Y.; Tropsha, A.; et al. Phosphorylation-activated G protein signaling stabilizes TCP14 and JAZ3 to repress JA signaling and enhance plant immunity. Mol. Plant 2025, 18, 1171–1192. [Google Scholar] [CrossRef]
- Torres-Vera, R.; García, J.M.; Pozo, M.J.; López-Ráez, J.A. Do strigolactones contribute to plant defence? Mol. Plant Pathol. 2014, 15, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Nardini, M.; Dijkstra, B.W. α/β Hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999, 9, 732–737. [Google Scholar] [CrossRef]
- Zhao, L.-H.; Zhou, X.E.; Yi, W.; Wu, Z.; Liu, Y.; Kang, Y.; Hou, L.; de Waal, P.W.; Li, S.; Jiang, Y.; et al. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 2015, 25, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.; Lu, Z.; Meng, X.; Wang, Y.; Smith, S.M.; Li, J. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142. [Google Scholar] [CrossRef]
- Yi, F.; Song, A.; Cheng, K.; Liu, J.; Wang, C.; Shao, L.; Wu, S.; Wang, P.; Zhu, J.; Liang, Z.; et al. Strigolactones positively regulate Verticillium wilt resistance in cotton via crosstalk with other hormones. Plant Physiol. 2023, 192, 945–966. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wang, C.; Zhang, X.; Huang, W.; Xing, M.; Han, C.; Lei, C.; Zhang, Y.; Zhang, X.; Cheng, K.; et al. Histone deacetylase GhHDA5 negatively regulates Verticillium wilt resistance in cotton. Plant Physiol. 2024, 196, 2918–2935. [Google Scholar] [CrossRef]
- Chen, X.; Shi, X.; Ai, Q.; Han, J.; Wang, H.; Fu, Q. Transcriptomic and metabolomic analyses reveal that exogenous strigolactones alleviate the response of melon root to cadmium stress. Hortic. Plant J. 2022, 8, 637–649. [Google Scholar] [CrossRef]
- Liu, L.; Li, K.; Zhou, X.; Fang, C. Integrative Analysis of Metabolome and Transcriptome Reveals the Role of Strigolactones in Wounding-Induced Rice Metabolic Re-Programming. Metabolites 2022, 12, 789. [Google Scholar] [CrossRef] [PubMed]
- Lahari, Z.; Ullah, C.; Kyndt, T.; Gershenzon, J.; Gheysen, G. Strigolactones enhance root-knot nematode (Meloidogyne graminicola) infection in rice by antagonizing the jasmonate pathway. New Phytol. 2019, 224, 454–465. [Google Scholar] [CrossRef]
- Li, S.; Joo, Y.; Cao, D.; Li, R.; Lee, G.; Halitschke, R.; Baldwin, G.; Baldwin, I.T.; Wang, M. Strigolactone signaling regulates specialized metabolism in tobacco stems and interactions with stem-feeding herbivores. PLoS Biol. 2020, 18, e3000830. [Google Scholar] [CrossRef]
- Zhang, X.; Lai, C.; Liu, M.; Xue, X.; Zhang, S.; Chen, Y.; Xiao, X.; Zhang, Z.; Chen, Y.; Lai, Z.; et al. Whole Genome Analysis of SLs Pathway Genes and Functional Characterization of DlSMXL6 in Longan Early Somatic Embryo Development. Int. J. Mol. Sci. 2022, 23, 14047. [Google Scholar] [CrossRef] [PubMed]
- Sd, C. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 2011, 23, 1219–1230. [Google Scholar] [CrossRef]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.-W.; Zhou, H.-W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef]
- Zhang, J.J. Molecular Mechanisms of Antagonistic Regulation of Arabidopsis thaliana Apical Hook Development by Brassinosteroids and Jasmonates. Available online: https://kns.cnki.net/kcms2/article/abstract?v=LzEBRIJt2Q1EDpSunhVKZUkdtFd8oFh3QJsGDGKo87AOrDfbVRc7TEflqmUNLB905WfLCcHLxuaRdIEgyA08vWtUGaXM9HqknDAJKJrQZ7H-pREZm-6cMVLnJxRRT5YXdmjPLTYyZEkzHcMNddaYF0rBqmImiXSsK1Q9yHYft4tr-AH9WROBt6x9rPdCdd7m_WAltLS796w=&uniplatform=NZKPT&language=CHS (accessed on 25 February 2025).
- Ren, C.; Han, C.; Peng, W.; Huang, Y.; Peng, Z.; Xiong, X.; Zhu, Q.; Gao, B.; Xie, D. A Leaky Mutation in DWARF4 Reveals an Antagonistic Role of Brassinosteroid in the Inhibition of Root Growth by Jasmonate in Arabidopsis. Plant Physiol. 2009, 151, 1412–1420. [Google Scholar] [CrossRef]
- Yu, B.; Wu, Q.; Li, X.; Zeng, R.; Min, Q.; Huang, J. GLUTAMATE RECEPTOR-like gene OsGLR3.4 is required for plant growth and systemic wound signaling in rice (Oryza sativa). New Phytol. 2022, 233, 1238–1256. [Google Scholar] [CrossRef]
- Hu, J.; Huang, J.; Xu, H.; Wang, Y.; Li, C.; Wen, P.; You, X.; Zhang, X.; Pan, G.; Li, Q.; et al. Rice stripe virus suppresses jasmonic acid-mediated resistance by hijacking brassinosteroid signaling pathway in rice. PLoS Pathog. 2020, 16, e1008801. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hong, G.; Zhang, H.; Tan, X.; Li, L.; Kong, Y.; Sang, T.; Xie, K.; Wei, J.; Li, J.; et al. The OsGSK2 Kinase Integrates Brassinosteroid and Jasmonic Acid Signaling by Interacting with OsJAZ4. Plant Cell 2020, 32, 2806–2822. [Google Scholar] [CrossRef]
- He, Y.; Zhang, H.; Sun, Z.; Li, J.; Hong, G.; Zhu, Q.; Zhou, X.; MacFarlane, S.; Yan, F.; Chen, J. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2017, 214, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kitano, H.; Fujioka, S. ERECT LEAF1 suppresses jasmonic acid response in rice by decreasing OsWRKY4 stability. Plant Signal. Behav. 2019, 14, 1559578. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wei, J.; Wu, J.; Shi, B.; Wang, P.; Alabd, A.; Wang, D.; Gao, Y.; Ni, J.; Bai, S.; et al. Transcription factors BZR2/MYC2 modulate brassinosteroid and jasmonic acid crosstalk during pear dormancy. Plant Physiol. 2024, 194, 1794–1814. [Google Scholar] [CrossRef] [PubMed]
- Szentpéteri, V.; Virág, E.; Mayer, Z.; Duc, N.H.; Hegedűs, G.; Posta, K. First Peek into the Transcriptomic Response in Heat-Stressed Tomato Inoculated with Septoglomus constrictum. Plants 2024, 13, 2266. [Google Scholar] [CrossRef]
- Li, Z.; Luo, X.; Ou, Y.; Jiao, H.; Peng, L.; Fu, X.; Macho, A.P.; Liu, R.; He, Y. JASMONATE-ZIM DOMAIN proteins engage Polycomb chromatin modifiers to modulate Jasmonate signaling in Arabidopsis. Mol. Plant 2021, 14, 732–747. [Google Scholar] [CrossRef]
- Wang, H.; Li, S.; Li, Y.; Xu, Y.; Wang, Y.; Zhang, R.; Sun, W.; Chen, Q.; Wang, X.; Li, C.; et al. MED25 connects enhancer–promoter looping and MYC2-dependent activation of jasmonate signalling. Nat. Plants 2019, 5, 616–625. [Google Scholar] [CrossRef]
- Zander, M.; Lewsey, M.G.; Clark, N.M.; Yin, L.; Bartlett, A.; Saldierna Guzmán, J.P.; Hann, E.; Langford, A.E.; Jow, B.; Wise, A.; et al. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 2020, 6, 290–302. [Google Scholar] [CrossRef]
- Wu, F.; Sun, C.; Zhu, Z.; Deng, L.; Yu, F.; Xie, Q.; Li, C. A multiprotein regulatory module, MED16–MBR1&2, controls MED25 homeostasis during jasmonate signaling. Nat. Commun. 2025, 16, 772. [Google Scholar] [CrossRef]
- Zhang, C.; Xiong, A.-T.; Ren, M.-Y.; Zhao, Y.-Y.; Huang, M.-J.; Huang, L.-C.; Zhang, Z.; Wang, Y.; Zheng, Q.-Q.; Fan, J.; et al. An epigenetically mediated double negative cascade from EFD to HB21 regulates anther development. Nat. Commun. 2024, 15, 7796. [Google Scholar] [CrossRef]
- Lim, H.J.; Wang, Y.; Buzdin, A.; Li, X. A practical guide for choosing an optimal spatial transcriptomics technology from seven major commercially available options. BMC Genom. 2025, 26, 47. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, H.; Wang, S.; Yang, Z.; Ma, P. Molecular Decoding of Phytohormone Crosstalk: JA-Mediated Key Regulatory Nodes and Signal Integration. Plants 2025, 14, 2647. https://doi.org/10.3390/plants14172647
Gan H, Wang S, Yang Z, Ma P. Molecular Decoding of Phytohormone Crosstalk: JA-Mediated Key Regulatory Nodes and Signal Integration. Plants. 2025; 14(17):2647. https://doi.org/10.3390/plants14172647
Chicago/Turabian StyleGan, Hui, Shiying Wang, Zisong Yang, and Pengda Ma. 2025. "Molecular Decoding of Phytohormone Crosstalk: JA-Mediated Key Regulatory Nodes and Signal Integration" Plants 14, no. 17: 2647. https://doi.org/10.3390/plants14172647
APA StyleGan, H., Wang, S., Yang, Z., & Ma, P. (2025). Molecular Decoding of Phytohormone Crosstalk: JA-Mediated Key Regulatory Nodes and Signal Integration. Plants, 14(17), 2647. https://doi.org/10.3390/plants14172647





