Necrosis-Suppressing Effector Protein ChEC88 Adopts a Novel Structural Motif Conserved Among Genus-Spanning Hemibiotrophic Phytopathogens
Abstract
1. Introduction
2. Results
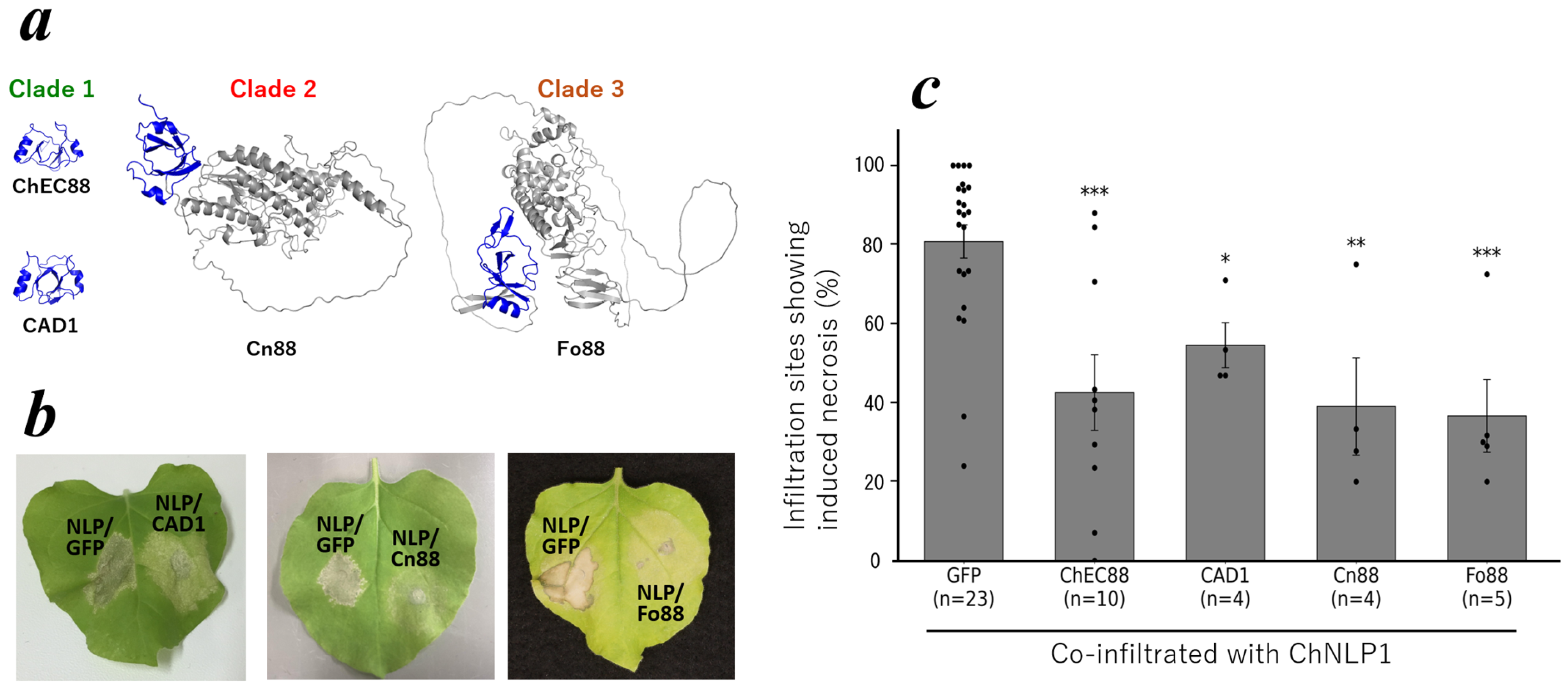
2.1. ChEC88 Suppresses NLP1-Induced Cell Death
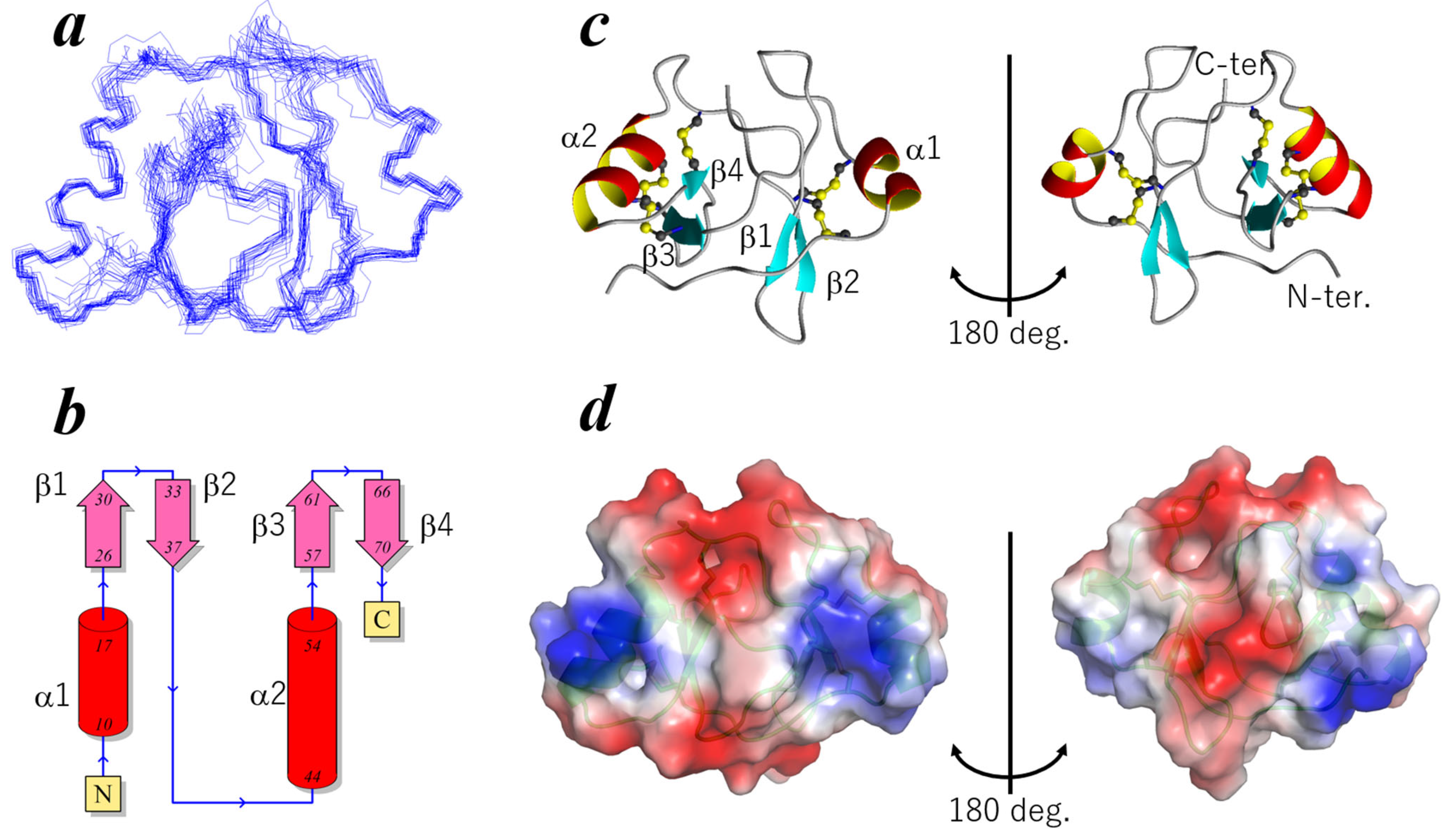
2.2. Three-Dimensional Structure of ChEC88
2.3. Dynamics and Thermal Stability of ChEC88
2.4. Conservation of the ChEC88-like Motif Across Hemibiotrophic Phytopathogens
2.5. Functional Conservation of ChEC88 Homologs
2.6. Identification of ChEC88 Functional Residues
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Activity Measurements of the ChEC88 Family and Its Mutants
5.2. RT-PCR Analysis to Assess Expression Level
5.3. Preparation of Stable Isotope-Labeled Samples for NMR Analysis
5.4. Multidimensional NMR and Structure Determination of ChEC88
5.5. NMR Relaxation Experiments of ChEC88
5.6. Bioinformatic Analysis
5.7. Plasmid Construction for Plant Expression
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [PubMed]
- Mine, A.; Seyfferth, C.; Kracher, B.; Berens, M.L.; Becker, D.; Tsuda, K. The defense phytohormone signaling network enables rapid and robust immune responses. Plant Cell 2018, 30, 1199–1219. [Google Scholar] [CrossRef] [PubMed]
- Nobori, T.; Velásquezb, A.; Wua, J.; Kvitkob, B.H.; Kremer, J.M.; Wanga, Y.; He, S.Y.; Tsuda, K. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E3055–E3064. [Google Scholar] [CrossRef]
- Zhou, D.; Chen, X.; Chen, X.; Xia, Y.; Liu, J.; Zhou, G. Plant immune receptors interact with hemibiotrophic pathogens to activate plant immunity. Front. Microbiol. 2023, 14, 1252039. [Google Scholar] [CrossRef]
- McCombe, C.L.; Greenwood, J.R.; Solomon, P.S.; Williams, S.J. Molecular plant immunity against biotrophic, hemibiotrophic, and necrotrophic fungi. Essays Biochem. 2022, 66, 581–593. [Google Scholar] [CrossRef]
- Pradhan, A.; Ghosh, S.; Sahoo, D.; Jha, G. Fungal effectors, the double edge sword of phytopathogens. Curr. Genet. 2021, 67, 27–40. [Google Scholar] [CrossRef]
- Kleeman, J.; Rincon-Rivera, L.J.; Takahara, H.; Neumann, U.; van Themaat, E.V.L.; van der Does, H.C.; Hacquard, S.; Stüber, K.; Will, I.; Schmalenbach, W.; et al. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 2012, 8, e1002643. [Google Scholar] [CrossRef]
- Dodds, P.N.; Chen, J.; Outram, M. Pathogen perception and signaling in plant immunity. Plant Cell 2024, 36, 1465–1481. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Irieda, H.; Inoue, Y.; Mori, M.; Yamada, K.; Oshikawa, Y.; Saitoh, H.; Uemura, A.; Terauchi, R.; Kitakura, S.; Kosaka, A.; et al. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Di, R.; Zhu, L.; Huang, Z.; Lu, M.; Yin, L.; Wang, C.; Bao, Y.; Duan, Z.; Powell, C.A.; Zhang, J.; et al. Fusarium sacchari FsNis1 induces plant immunity. Gene 2024, 907, 148260. [Google Scholar] [CrossRef] [PubMed]
- Pirc, K.; Hodnik, V.; Snoj, T.; Lenarčič, T.; Simon, C.S.; Podobnik, M.; Böhm, H.; Albert, I.; Kotar, A.; Plavec, J.; et al. Nep1-like proteins as a target for plant pathogen control. PLoS Pathog. 2021, 17, e1009477. [Google Scholar] [CrossRef] [PubMed]
- Qutob, D.; Kemmerling, B.; Brunner, F.; Küfner, I.; Engelhardt, S.; Gust, A.A.; Luberacki, B.; Seitz, H.U.; Stahl, D.; Rauhut, T.; et al. Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 2006, 18, 3721–3744. [Google Scholar] [CrossRef]
- Xiang, J.; Cheng, J.; Wei, L.; Li, M.; Wu, J. Functional analysis of the Nep1-like proteins from Plasmopara viticola. Plant Signal. Behav. 2022, 17, e2000791. [Google Scholar] [CrossRef]
- Mosquera, G.; Giraldo, M.C.; Khang, C.H.; Coughlan, S.; Valent, B. Interaction transcriptome analysis identifies Magaporthe oryzae BAS1-4 as biotropy-associated secreted proteins in rice blast disease. Plant Cell 2009, 21, 1273–1290. [Google Scholar] [CrossRef]
- Sharpee, W.; Oh, Y.; Yi, M.; Franck, W.; Eyre, A.; Okagaki, L.H.; Valent, B.; Dean, R.A. Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 850–863. [Google Scholar] [CrossRef]
- Martin, O.A.; Villegas, M.E.; Vila, J.A.; Cheraga, H.A. Analysis of 13Ca and 13Cb chemical shifts of cysteine and cystine residues in proteins: A quantum chemical approach. J. Biomol. NMR 2010, 46, 217–225. [Google Scholar] [CrossRef]
- Inagaki, A.; Takano, Y.; Kubo, Y.; Mise, K.; Furusawa, I. Construction of an equalized cDNA library from Colletotrichum lagenarium and its application to the isolation of differentially expressed genes. Can. J. Microbiol. 2000, 46, 150–158. [Google Scholar] [CrossRef]
- Ohki, S.; Takeuchi, M.; Mori, M. NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2011, 2, 512. [Google Scholar] [CrossRef]
- Salomão, S.S.; Oliveira, S.; Cherene, M.B.; Taveira, G.B.; Mello, É.O.; Carvalho, A.O.; Gomes, V.M. Plant antimicrobial peptides and their main families and roles: A review of the literature. Curr. Issues Mol. Biol. 2025, 47, 1. [Google Scholar] [CrossRef]
- Baxter, A.A.; Richter, V.; Lay, F.T.; Poon, I.K.H.; Adda, C.G.; Veneer, P.K.; Phan, T.K.; Bleackley, M.R.; Anderson, M.A.; Kvansakul, M.; et al. The tomato defensin TPP3 binds phosphatidylinositol (4,5)-bisphosphate via conserved dimeric cationic grip conformation to mediate cell lysis. Mol. Cell. Biol. 2015, 35, 1964–1978. [Google Scholar] [CrossRef] [PubMed]
- Wai, A.H.; Waseen, M.; Khan, A.B.M.M.M.; Nath, U.K.; Lee, D.J.; Kim, S.T.; Kim, C.K.; Chung, M.Y. Genome-wide identification and expression profiling of the PDI gene family reveals their probable involvement in abiotic stress tolerance in tomato (Solanum lycopersicum L.). Genes 2021, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Zhang, Q.; Huang, G.; Liang, X.; Wang, C.; Wang, L.; Lu, D. Two protein disulfide isomerase subgroups work synergistically in catalyzing oxidative protein folding. Plant Physiol. 2021, 188, 241–254. [Google Scholar] [CrossRef]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N.; et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genetics 2012, 44, 1060–1067. [Google Scholar] [CrossRef]
- Hacuard, S.; Kracher, B.; Hiura, K.; Münch, P.C.; Garrido-Oter, R.; Thon, M.R.; Weimann, A.; Damm, U.; Dallery, J.-F.; Hainaut, M.; et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 2016, 7, 11362. [Google Scholar] [CrossRef]
- Yu, D.S.; Outram, M.A.; Smith, A.; McCombe, C.L.; Khambalkar, P.B.; Jones, D.A.; Williams, S.J. The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies. eLife 2024, 12, RP89280. [Google Scholar] [CrossRef]
- Le Naour-Vernet, M.L.; Lahfa, M.; Maidment, J.H.R.; Padilla, A.; Roumestand, C.; de Guillen, K.; Kroj, T.; Césari, S. Structure-guided insights into the biology of fungal effectors. New Phytol. 2025, 246, 1460–1477. [Google Scholar] [CrossRef]
- Takahara, H.; Yamaguchi, S.; Omura, N.; Nakajima, S.; Otoku, K.; Tanaka, S.; Ogura, K.; Kleeman, J.; O’Connel, R. The Colletotrichum higginsianum secreted effector protein ChEC91 induces plant cell death. J. Geneal. Plant Pathol. 2021, 87, 344–353. [Google Scholar] [CrossRef]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyán, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol, J. 2009, 7, 682–693. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Hann, D.R.; Chang, J.H.; Segonzac, C.; Boller, T.; Rathjen, J.P. Differential suppression of Nicotiana benthamiana innate immune responses by transiently expressed Pseudomonas syringae type III effectors. Front. Plant Sci. 2018, 9, 2021. [Google Scholar] [CrossRef] [PubMed]
- Ikura, M.; Kay, L.E.; Bax, A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of larger proteins: Heteronuclear triple-resonance three-dimensional NMR spectroscopy. application to calmodulin. Biochemistry 1990, 29, 4659–4667. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G., III; Skelton, N.J. Protein NMR Spectroscopy: Principles and Practice; Academic Press: San Diego, CA, USA, 2018. [Google Scholar]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Rahimi, M.; Lee, Y.; Chiu, A. POKY: A software suite for multidimensional NMR and 3D structure calculation of biomolecules. Bioinformatics 2021, 37, 3041–3042. [Google Scholar] [CrossRef]
- Cornilescu, G.; Delaglio, F.; Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 1999, 13, 289–302. [Google Scholar] [CrossRef]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Clore, G.M. Using Xplor-NIH for NMR molecular structure determination. Progr. NMR Spectrosc. 2006, 48, 47–62. [Google Scholar] [CrossRef]
- Willard, L.; Rajan, A.; Zhang, H.; Monzavi, H.; Boyko, R.F.; Sykes, B.D.; Wishart, D.S. VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acid. Res. 2003, 31, 3316–3319. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of protein as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using Modeller. Cur. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef]
- Imamura, T.; Takagi, H.; Miyazato, A.; Ohki, S.; Mizukoshi, H.; Mori, M. Isolation and characterization of the betalain biosynthesis gene involved in hypocotyl pigmentation of the allotetraploid Chenopodium quinoa. Biochem. Biophys. Res. Commun. 2018, 496, 280–286. [Google Scholar] [CrossRef]
- Wise, A.A.; Liu, Z.; Binns, A.N. Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol. Biol. 2006, 343, 43–54. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohki, S.; Takahara, H.; Imamura, T.; Sakane, K.; Bai, A.; Sasaki, K.; Nishiuchi, T.; Mori, M. Necrosis-Suppressing Effector Protein ChEC88 Adopts a Novel Structural Motif Conserved Among Genus-Spanning Hemibiotrophic Phytopathogens. Plants 2025, 14, 2562. https://doi.org/10.3390/plants14162562
Ohki S, Takahara H, Imamura T, Sakane K, Bai A, Sasaki K, Nishiuchi T, Mori M. Necrosis-Suppressing Effector Protein ChEC88 Adopts a Novel Structural Motif Conserved Among Genus-Spanning Hemibiotrophic Phytopathogens. Plants. 2025; 14(16):2562. https://doi.org/10.3390/plants14162562
Chicago/Turabian StyleOhki, Shinya, Hiroyuki Takahara, Tomohiro Imamura, Kosei Sakane, Asihan Bai, Kazunori Sasaki, Takumi Nishiuchi, and Masashi Mori. 2025. "Necrosis-Suppressing Effector Protein ChEC88 Adopts a Novel Structural Motif Conserved Among Genus-Spanning Hemibiotrophic Phytopathogens" Plants 14, no. 16: 2562. https://doi.org/10.3390/plants14162562
APA StyleOhki, S., Takahara, H., Imamura, T., Sakane, K., Bai, A., Sasaki, K., Nishiuchi, T., & Mori, M. (2025). Necrosis-Suppressing Effector Protein ChEC88 Adopts a Novel Structural Motif Conserved Among Genus-Spanning Hemibiotrophic Phytopathogens. Plants, 14(16), 2562. https://doi.org/10.3390/plants14162562





