Genome-Wide Identification of DREB Gene Family in Kiwifruit and Functional Characterization of Exogenous 5-ALA-Mediated Cold Tolerance via ROS Scavenging and Hormonal Signaling
Abstract
1. Introduction
2. Results
2.1. Physicochemical Properties of Kiwifruit DREB Family Genes
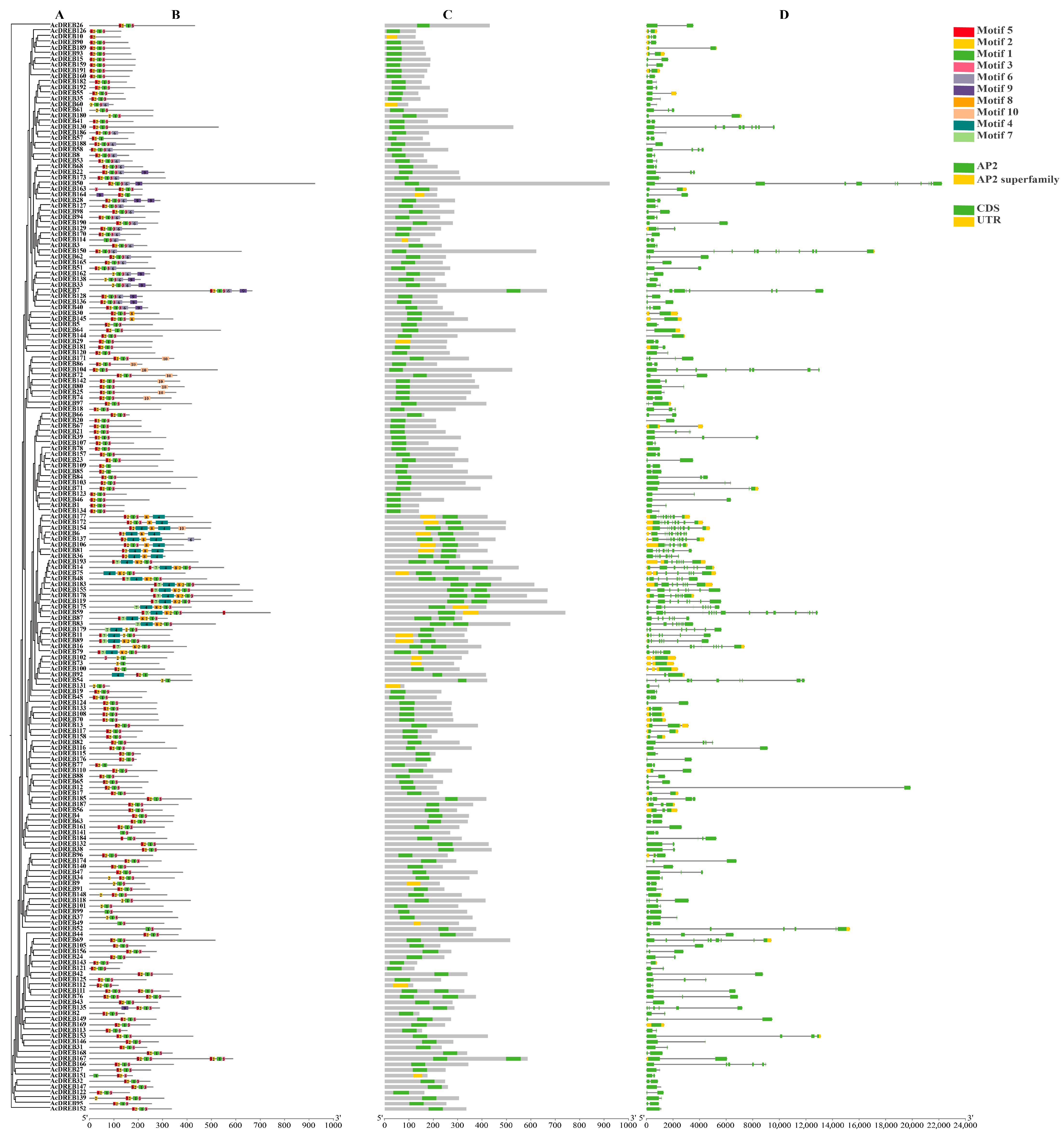
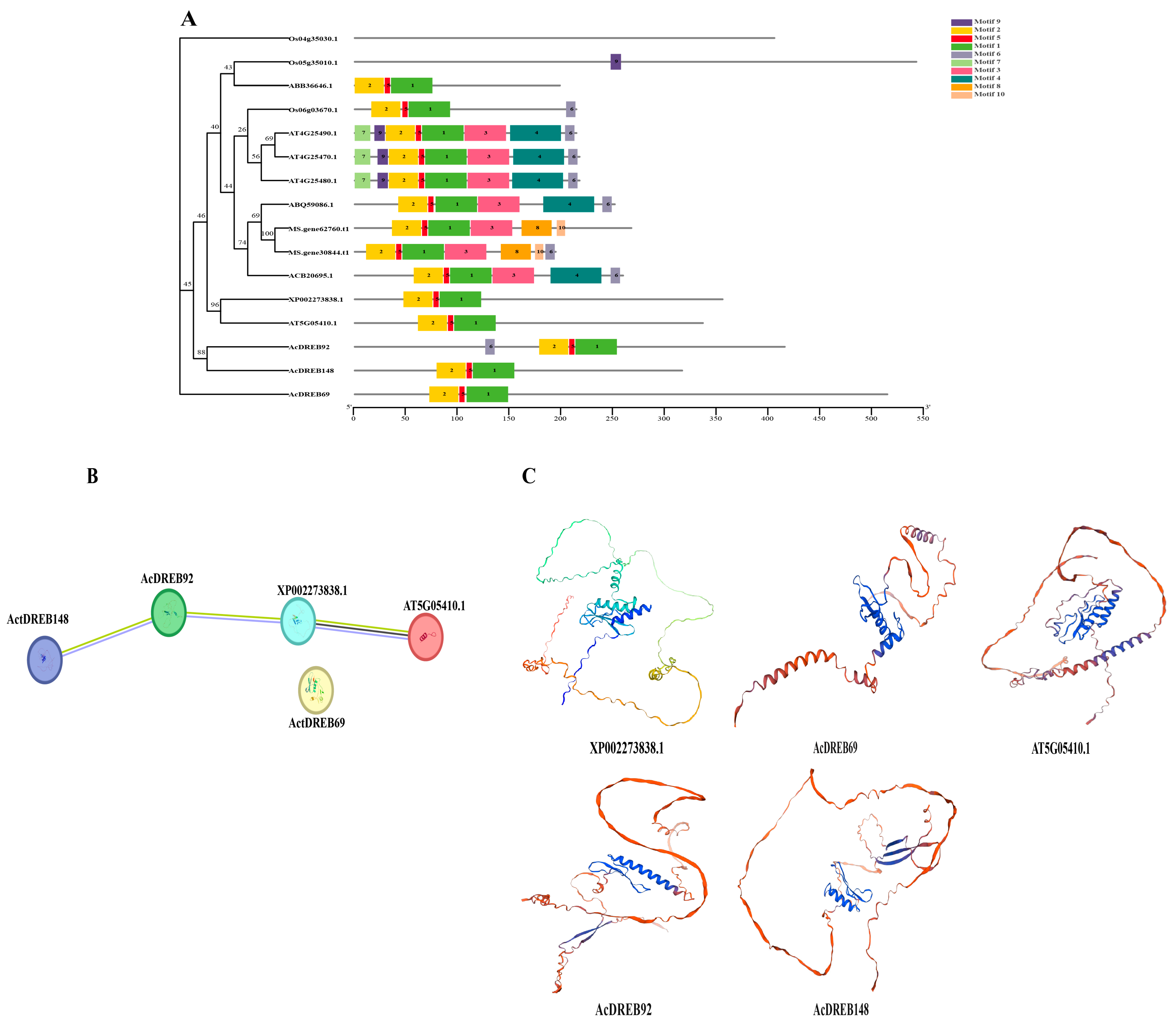
2.2. Phylogenetic Analysis of the Kiwifruit DREB Family Genes
2.3. Structure and Conserved Domain Analysis of the Kiwifruit DREB Gene
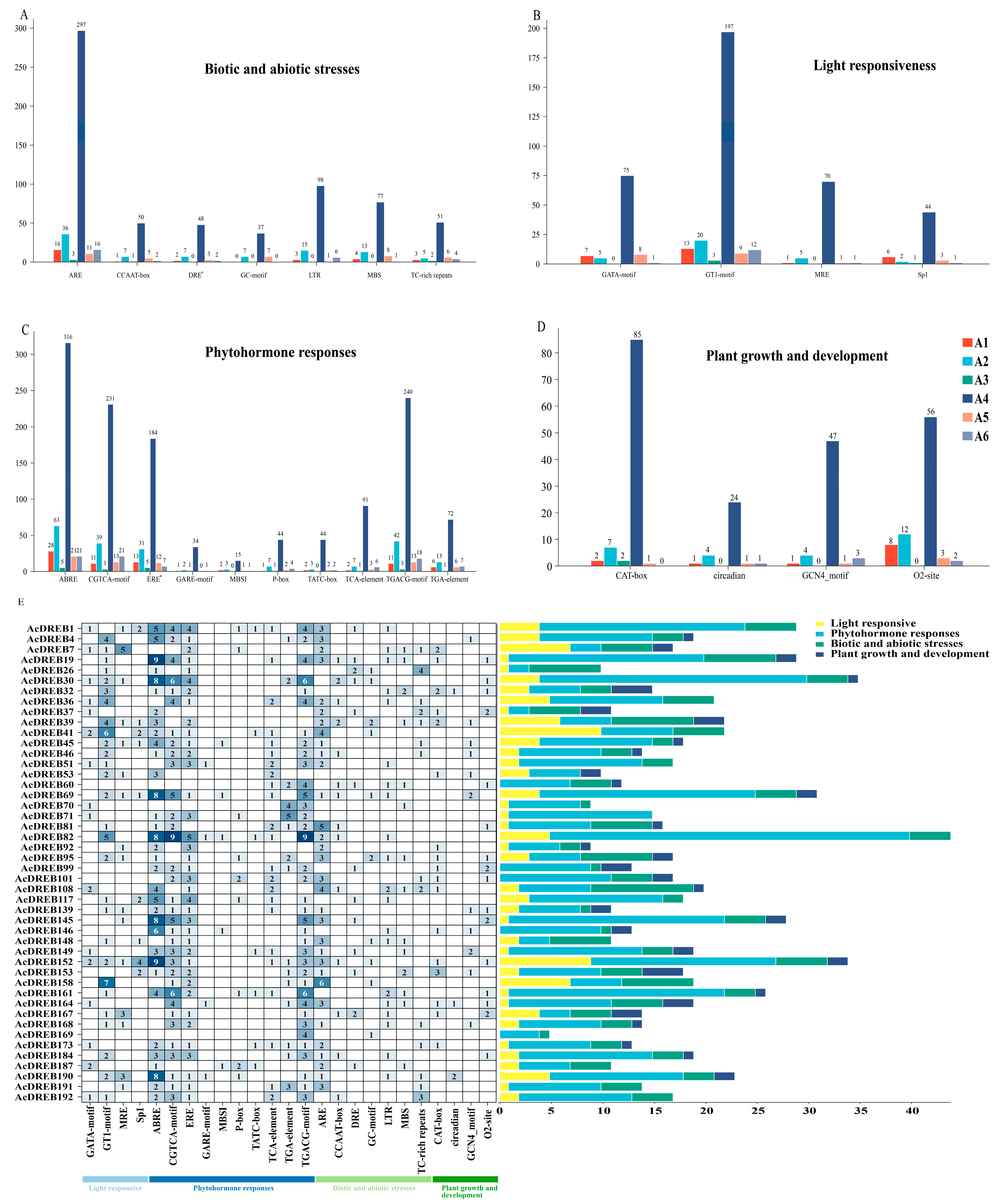
2.4. Analysis of Cis-Acting Elements in Gene Promoters
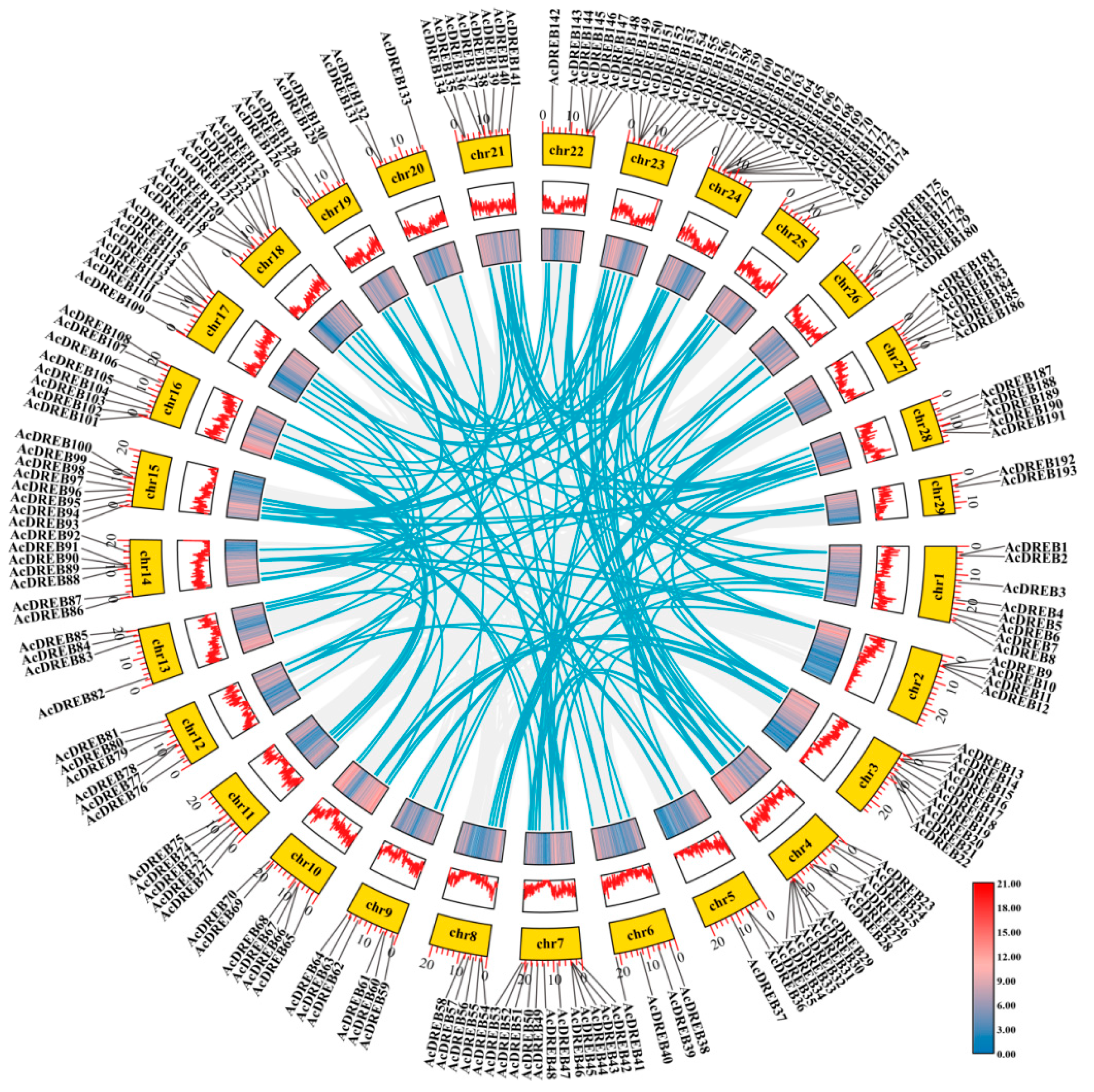
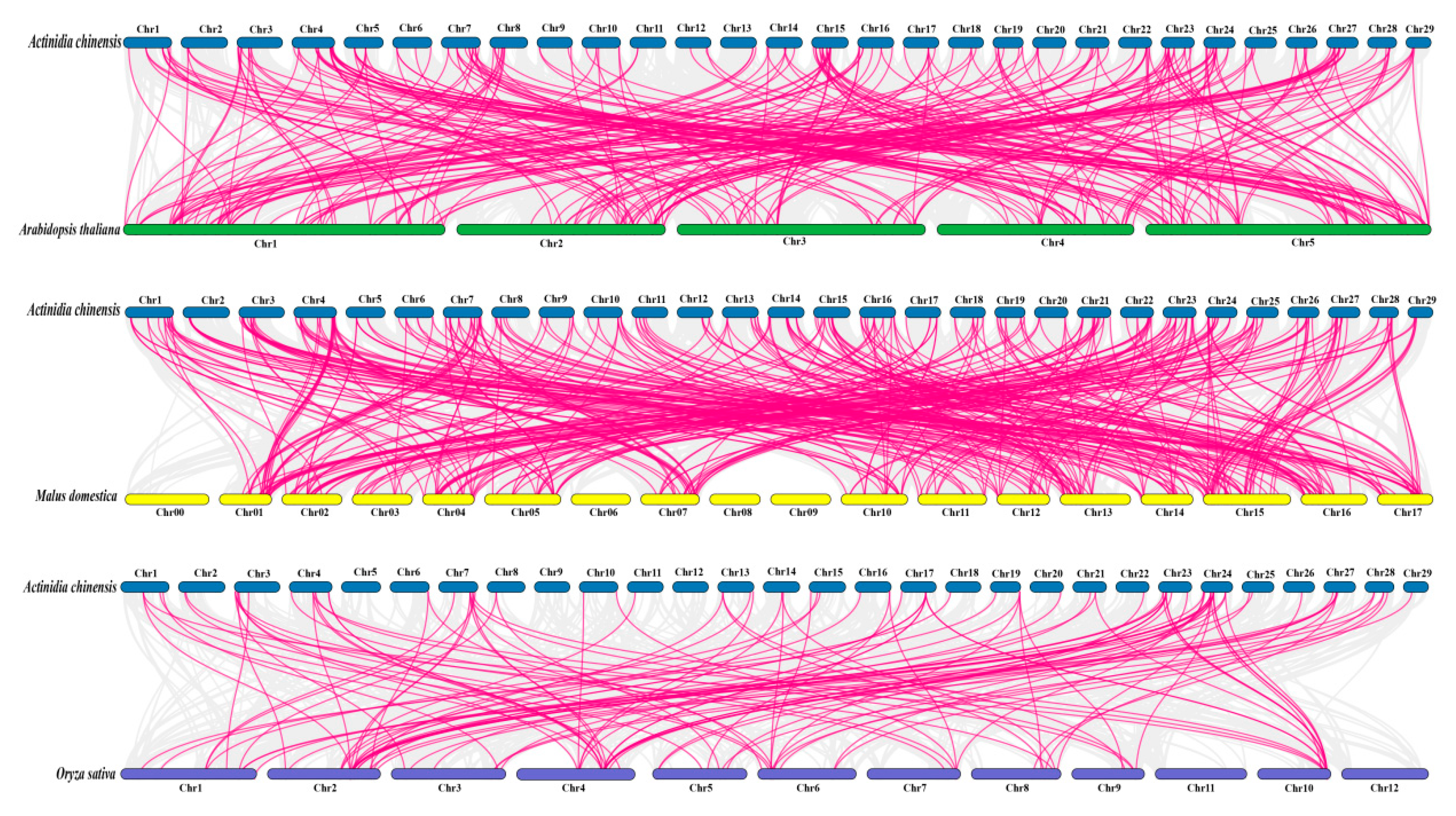
2.5. Chromosomal Mapping and Collinearity Analysis of the Kiwifruit DREB Gene Family
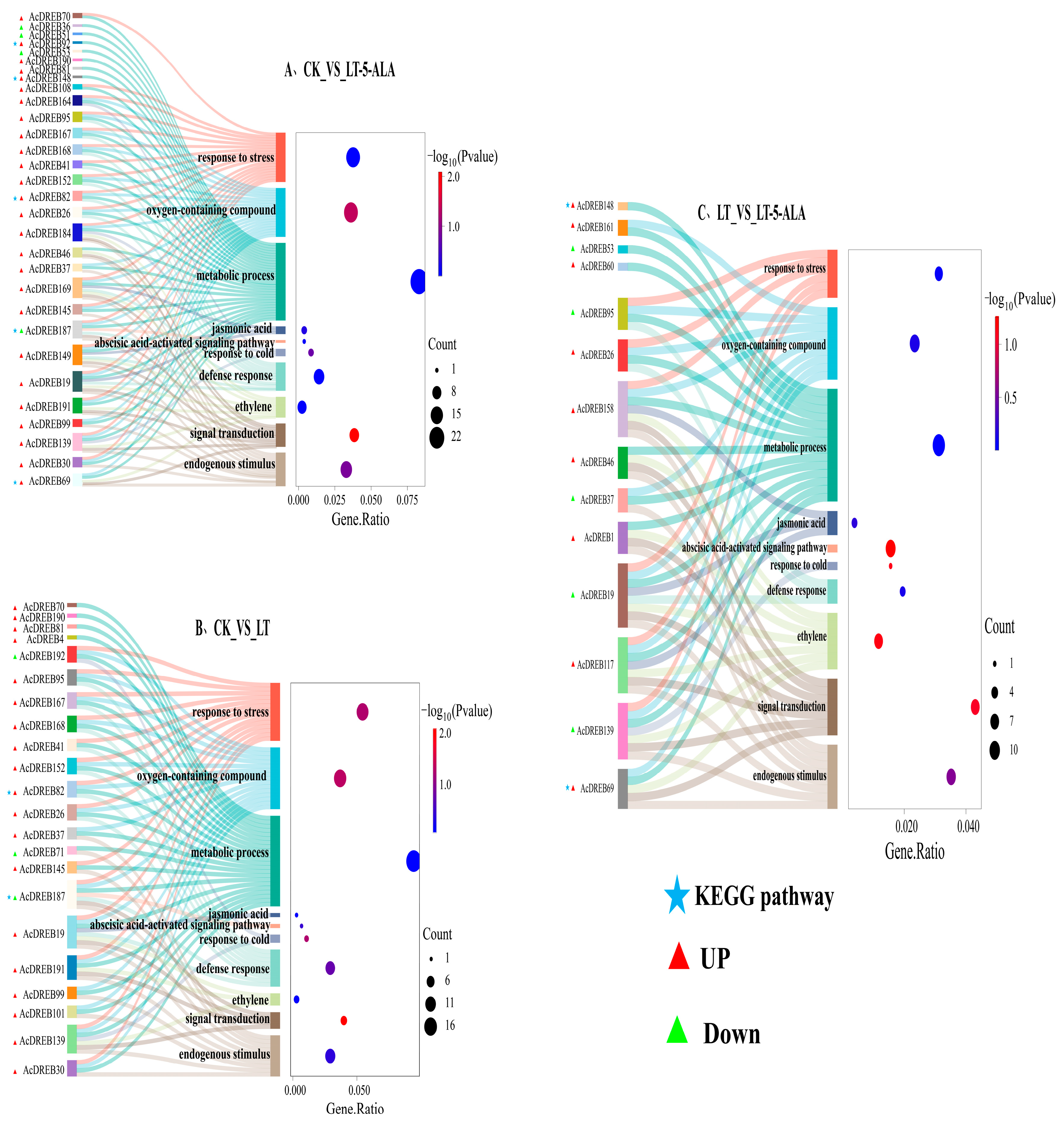
2.6. GO and KEGG Enrichment Analyses of the Kiwifruit DREB Gene Family
2.7. Expression Pattern of the AcDREB Gene Family in Kiwifruit Under Exogenous 5-ALA-Regulated Low-Temperature Stress
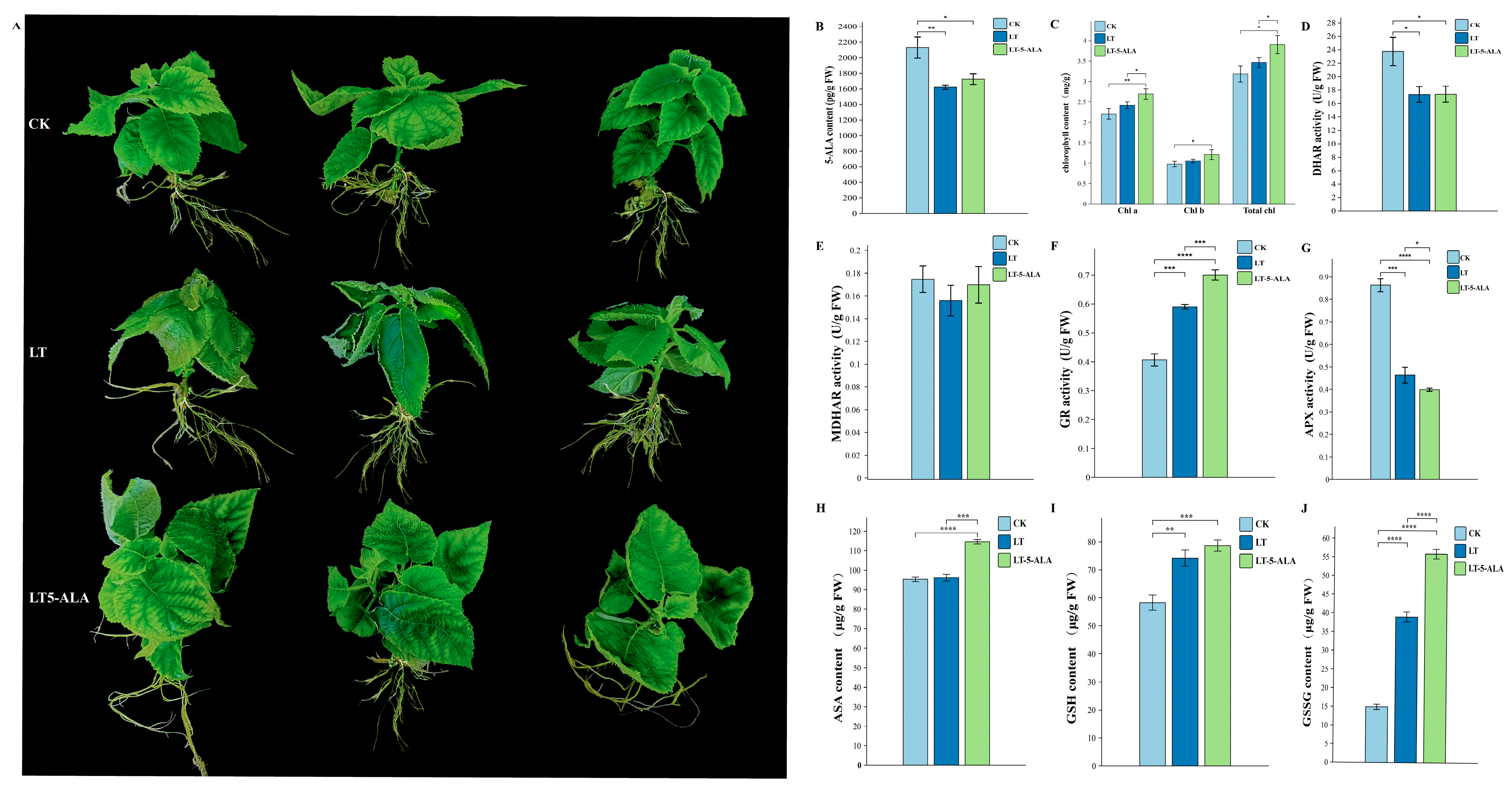
2.7.1. Effects of Exogenous 5-ALA on the Physiological Indicators of Kiwifruit Seedlings Under Low-Temperature Stress
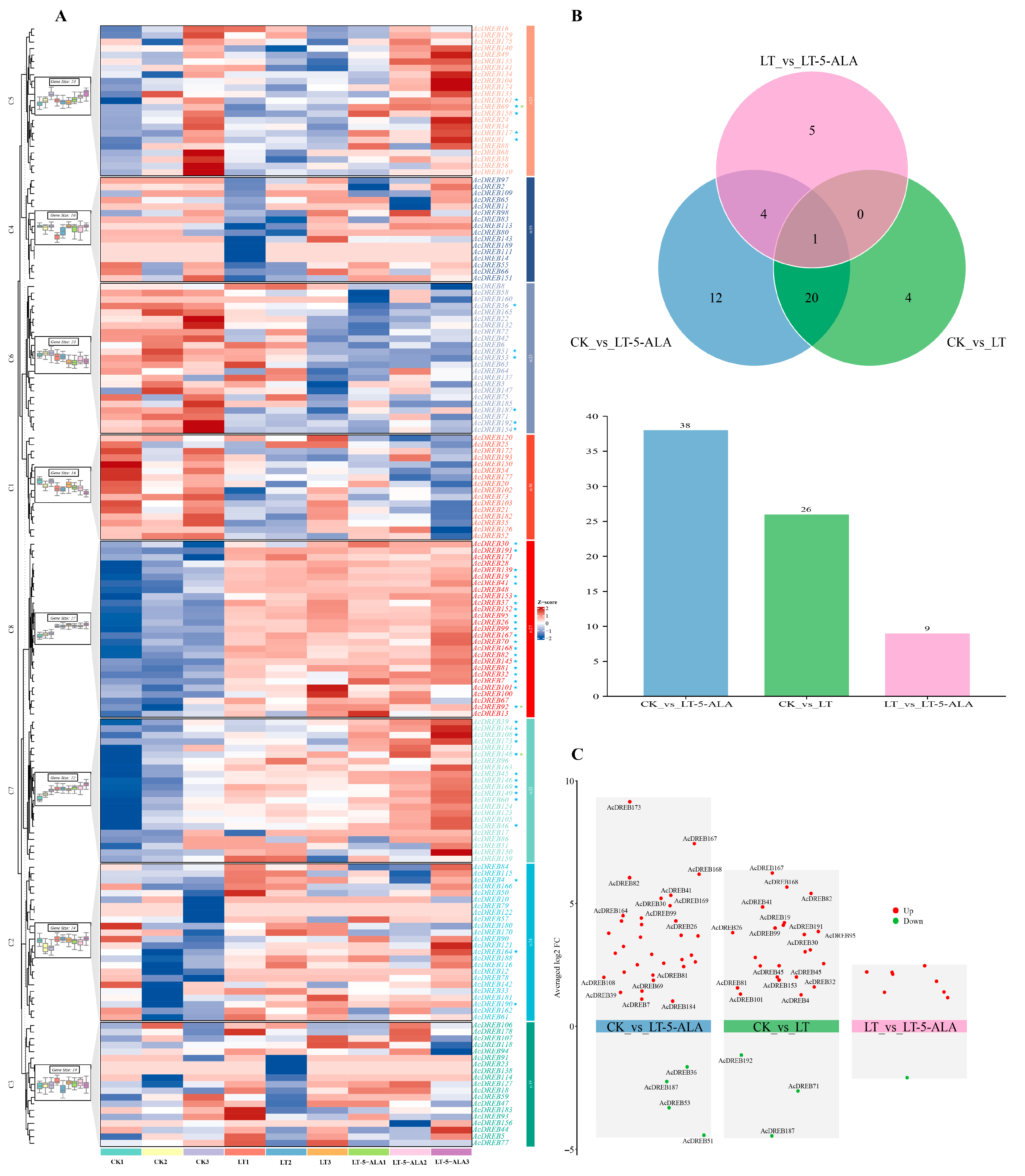
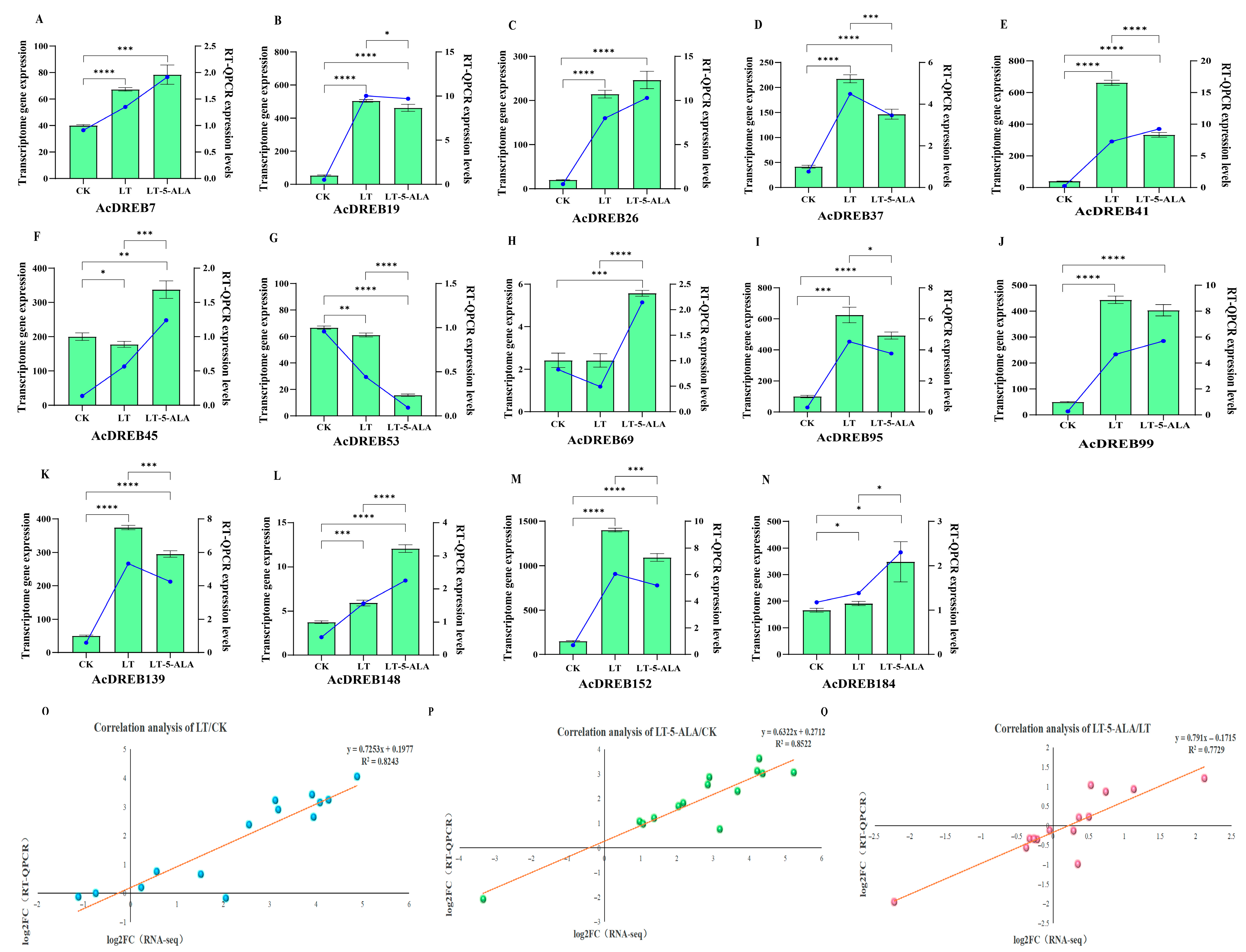
2.7.2. Expression Profiling of the AcDREB Gene Family in Kiwifruit Under Exogenous 5-ALA-Regulated Low-Temperature Stress
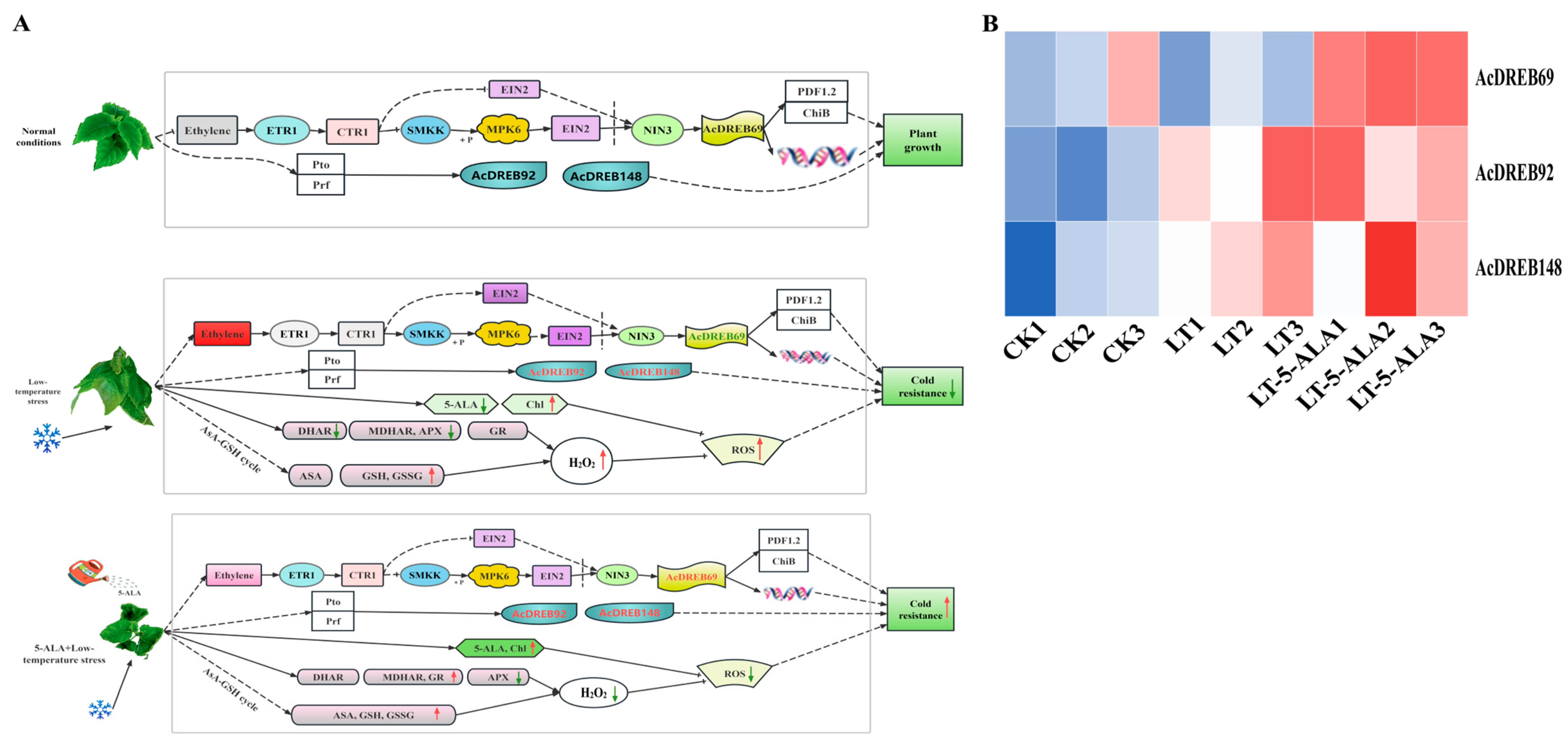
2.7.3. Functional Analysis of the AcDREB Gene Family in Kiwifruit Under Exogenous 5-ALA-Regulated Low-Temperature Stress
2.8. RT–qPCR Expression Analysis
3. Discussions
4. Materials and Methods
4.1. Data Acquisition
4.2. Identification and Classification of the AcDREB Gene Family
4.3. Gene Structure and Conserved Motif Analysis of the AcDREB Gene Family
4.4. Chromosomal Positioning and Collinearity Analysis
4.5. Analysis of Cis-Acting Elements in Kiwifruit DREB Subgroups
4.6. GO and KEGG Enrichment Analyses of the Kiwifruit DREB Gene
4.7. Analysis of the Expression Pattern of the AcDREB Gene in Kiwifruit Under Low-Temperature Stress Regulated by Exogenous 5-ALA
4.7.1. Plant Materials and Experimental Design
4.7.2. Measurement of Physiological Indicators
Determination of the 5-ALA Content and Chlorophyll Content
Determination of Antioxidant Enzyme Activities
Ascorbic Acid–Glutathione Cycle Analysis
4.7.3. RNA Extraction, cDNA Library Construction, and Sequencing
4.7.4. Expression Pattern Analysis of the Kiwifruit DREB Gene Family
4.7.5. qRT-PCR Analysis
4.8. Phylogenetic, Conserved Motif, Protein Structure, and Interaction Analyses of the Cold Resistance DREB Gene Family
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, B.C.; Shi, Y.T.; Zhang, X.Y.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, W.; Wang, M.; Khan, M.; Liu, J.H. A C2H2-type zinc finger protein ZAT12 of Poncirus trifoliata acts downstream of CBF1 to regulate cold tolerance. Plant J. 2023, 117, 1317–1329. [Google Scholar] [CrossRef]
- Li, Y.H.; Lin, M.M.; Zhang, Q.N.; Zhang, P.; Zhang, Z.Z.; Li, Y.K.; Sun, L.M.; Li, S.M.; Li, C.C.; Chen, D.X.; et al. Overexpression of the Kiwifruit Transcription Factor AaMYB44 Decreases the Cold Tolerance in Arabidopsis thaliana. Plants 2024, 13, 3126. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Liang, G.P.; Lu, S.X.; Wang, H.; Zeng, F.W.; Nai, G.J.; Mao, J.; Chen, B.H. Over expression of VaSS4 negatively regulates cold tolerance by disturbing ROS balance and decreasing soluble sugar content. Sci. Hortic. 2024, 337, 113590. [Google Scholar] [CrossRef]
- Zhao, J.J.; Feng, N.F.; Wang, X.X.; Cai, G.R.; Cao, M.Y.; Zheng, D.F.; Zhu, H.D. Uniconazole confers chilling stress tolerance in soybean (Glycine max L.) by modulating photosynthesis, photoinhibition, and activating oxygen metabolism system. Photosynthetica 2019, 57, 446–457. [Google Scholar] [CrossRef]
- Cakmak, I.; Marschner, H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 1992, 98, 1222–1227. [Google Scholar] [CrossRef]
- Choudhary, S.P.; Yu, J.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012, 17, 594–605. [Google Scholar] [CrossRef]
- Cui, X.A.; Wang, Y.W.; Wu, J.X.; Han, X.; Gu, X.; Lu, T.; Zhang, Z. The RNA editing factor DUA 1 is crucial to chloroplast development at low temperature in rice. New Phytol. 2019, 221, 834–849. [Google Scholar] [CrossRef]
- Munro, D.; Treberg, J.R. A radical shift in perspective: Mitochondria as regulators of reactive oxygen species. J. Exp. Biol. 2017, 220 Pt 7, 1170–1180. [Google Scholar] [CrossRef]
- Han, J.P.; Umer, M.J.; Yang, M.Y.; Hou, Y.; Gereziher Mehari, T.; Zheng, J.; Wang, H.; Liu, J.; Dong, W.; Xu, Y.; et al. Genome-wide identification and functional analysis of ICE genes reveal that Gossypium thurberi “GthICE2” is responsible for cold and drought stress tolerance. Plant Physiol. Biochem. 2023, 199, 107708. [Google Scholar] [CrossRef]
- Wu, J.C.; Cao, X.F.; Sun, X.Z.; Chen, Y.; Zhang, P.; Li, Y.; Ma, C.; Wu, L.; Liang, X.; Fu, Q.; et al. OsEL2 Regulates Rice Cold Tolerance by MAPK Signaling Pathway and Ethylene Signaling Pathway. Int. J. Mol. Sci. 2025, 26, 1633. [Google Scholar] [CrossRef]
- Ali, N.; Hadi, F. CBF/DREB transcription factor genes play role in cadmium tolerance and phytoaccumulation in Ricinus communis under molybdenum treatments. Chemosphere 2018, 208, 425–432. [Google Scholar] [CrossRef]
- Eckardt, N.A. DREB duo defines distinct drought and cold response pathways. Plant Cell 2019, 31, 1196–1197. [Google Scholar] [CrossRef]
- Niu, X.; Luo, T.; Zhao, H.; Su, Y.; Ji, W.; Li, H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Kazuo, S.; Kazuko, Y.S. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 2903, 998–1009. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 1210, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological responses to drought, salinity, and heat stress in plants: A review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Han, Q.H.; Chen, K.L.; Yan, D.; Hao, G.; Qi, J.; Wang, C.; Dirk, L.M.A.; Bruce Downie, A.; Gong, J.; Wang, J.; et al. ZmDREB2A regulates ZmGH3.2 and ZmRAFS, shifting metabolism towards seed aging tolerance over seedling growth. Plant J. 2020, 104, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Yang, Q.; Zhang, C.L.; Wei, L.L.; Yue, R.; Li, G.J.; Lin, X.F.; Wang, R.G. A CkDREB1 gene isolated from Caragana korshinskii Kom. Enhances Arabidopsis drought and cold tolerance. Braz. J. Bot. 2019, 42, 97–105. [Google Scholar] [CrossRef]
- Sun, S.; Yu, J.P.; Chen, F.; Zhao, T.J.; Fang, X.H.; Li, Y.Q.; Sui, S.F. Tiny, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 2008, 283, 6261–6271. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 108, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.T.; Ding, Y.L.; Yang, S.H. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Song, Q.Q.; Zhu, P.J.; He, J.; Ye, W.Y.; Ou, J.L.; Tang, X.G.; Li, J.H. Effect of low temperature on photosynthetic physiological characteristics of different Artocarpus heterophyllus Lam. Germplasm resources. Agricul. Res Appl. 2021, 34, 7. [Google Scholar]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Chen, M.; Xu, Z.; Xia, L.; Li, L.; Cheng, X.; Dong, J.; Wang, Q.; Ma, Y. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot. 2009, 60, 121–135. [Google Scholar] [CrossRef]
- Hotta, Y.; Tanaka, T.; Takaoka, H.; Takeuchi, Y.; Konnai, M. New physiological effects of 5-Aminolevulinic acid in plants: The increase of photosynthesis, chlorophyll content, and plant growth. Biosci. Biotechnol. Biochem. 1997, 61, 2025–2028. [Google Scholar] [CrossRef]
- Czarnecki, O.; Hedtke, B.; Melzer, M.; Rothbart, M.; Richter, A.; Schröter, Y.; Pfannschmidt, T.; Grimm, B. An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 2011, 23, 4476–4491. [Google Scholar] [CrossRef]
- Zhang, L.Z.; Tao, H.H.; Zhang, J.N.T.; An, Y.; Wang, L. 5-Aminolevulinic acid activates the MdWRKY71-MdMADS1 module to enhance anthocyanin biosynthesis in apple. Mol. Hortic. 2025, 5, 10. [Google Scholar] [CrossRef]
- Wang, Y.X.; Tan, C.L.; Li, Y.H.; Meng, F.; Du, Y.; Zhang, S.; Jiang, W.; Feng, N.; Zhao, L.; Zheng, D. 5-ALA, DTA-6, and nitrogen mitigate NaCl stress by promoting photosynthesis and carbon metabolism in rice seedlings. Metabolites 2024, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.L.; Song, L.L.; Cao, L.F.; Meng, L. Alleviation of shade stress in Chinese Yew (Taxus chinensis) seedlings with 5-aminolevulinic acid (ALA). Plants 2023, 12, 2333. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Qu, D.; Chen, X.; Zeng, H.; Li, X.; Hu, C.Y. Metabolomics reveals 5-aminolevulinic acid improved the ability of tea leaves (Camellia sinensis L.) against cold stress. Metabolites 2022, 12, 392. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Zhang, J.; Lai, G.T.; He, L.; Xu, H.; Li, S.; Che, J.; Wang, Q.; Guan, X.; Huang, J.; et al. Targeted regulation of 5-aminolevulinic acid enhances flavonoids, anthocyanins and proanthocyanidins accumulation in Vitis davidii callus. BMC Plant Biol. 2024, 24, 944. [Google Scholar] [CrossRef]
- Li, Y.Y.; Guo, Q.; Zhang, K.Y.; Wang, H.; Jia, C.; Guo, D.; Guo, L.; Hou, X. Dormancy-release, germination and seedling growth of Paeonia ostii ‘Fengdan’ seeds under measures of physical and chemical treatment and sowing. PLoS ONE 2022, 17, e027076. [Google Scholar] [CrossRef]
- Liu, R.D.; Huang, R.H.; Wu, S.Q.; Yu, Z.S.; Li, X.X. Changes in anthocyanin content of ‘Hongyang’ kiwifruit and the effects of ring stripping and ABA on its formation. J. Hortic. 2009, 3606, 793–798. [Google Scholar]
- Jiang, L.L.; Gong, X.; Ji, M.Y.; Wang, C.C.; Wang, J.H.; Li, M.H. Bioactive compounds from plant-based functional foods: A promising choice for the prevention and management of hyperuricemia. Foods 2020, 9, 973. [Google Scholar] [CrossRef]
- Gong, X.C.; Lin, M.F.; Song, J.; Mao, J.P.; Yao, D.L.; Gao, Z.; Wang, X.L. Genome-wide identification of the AcBAM family in kiwifruit (Actinidia chinensis cv. Hongyang) and the expression profiling analysis of AcBAMs reveal their role in starch metabolism. BMC Plant Biol. 2025, 25, 415. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Bai, Y.; Dai, Q.Y.; He, Y.H.; Yan, L.; Niu, J.; Wang, X.; Xie, Y.; Yu, X.; Tang, W.; Li, H.; et al. Exogenous diethyl aminoethyl hexanoate alleviates the damage caused by low-temperature stress in Phaseolus vulgaris L. seedlings through photosynthetic and antioxidant systems. BMC Plant Biol. 2025, 25, 75. [Google Scholar] [CrossRef]
- Liu, T.; Hu, X.H.; Zhang, J.; Zhang, J.H.; Du, Q.J.; Li, J.M. H2O2 mediates ALA-induced glutathione and ascorbate accumulation in the perception and resistance to oxidative stress in Solanum lycopersicum at low temperatures. BMC Plant Biol. 2018, 18, 34. [Google Scholar] [CrossRef]
- Fu, J.J.; Sun, Y.F.; Chu, X.T.; Xu, Y.; Hu, T. Exogenous 5-aminolevulenic acid promotes seed germination in Elymus nutans against oxidative damage induced by cold stress. PLoS ONE 2014, 99, e107152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Yuan, L.; Zhou, F.; Gao, Y.; Kang, Z.; Li, T.; Hu, X. Exogenous 5-aminolevulinic acid alleviates low-temperature injury by regulating glutathione metabolism and β-5-ALAnine metabolism in tomato seedling roots. Ecotoxicol. Environ. Saf. 2022, 245, 114112. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Jiang, W.B.; Zhang, Z.; Yao, Q.H.; Hiroyuki, M.; Jun, K. Biosynthesis and physiological activity of 5-aminolevulinic acid and its potential application in agriculture. Plant Physiol. Lett. 2003, 393, 185–192. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 281, 27–30. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Chory, J.; Wang, X. The role of ethylene in plant temperature stress response. Trends Plant Sci. 2023, 28, 808–824. [Google Scholar] [CrossRef]
- Chai, M.; Cheng, H.; Yan, M.; Priyadarshani, S.; Zhang, M.; He, Q.; Huang, Y.; Chen, F.; Liu, L.; Huang, X.; et al. Identification and expression analysis of the DREB transcription factor family in pineapple (Ananas comosus (L.) Merr.). PeerJ 2020, 8, e9006. [Google Scholar] [CrossRef]
- Sun, N.; Sun, X.; Zhou, J.; Zhou, X.; Gao, Z.; Zhu, X.; Xu, X.; Liu, Y.; Li, D.; Zhan, R.; et al. Genome-wide characterization of pepper DREB family members and biological function of CaDREB32 in response to salt and osmotic stresses. Plant Physiol. Biochem. 2025, 222, 109736. [Google Scholar] [CrossRef]
- Ma, X.W.; Zhu, P.J.; Du, Y.J.; Song, Q.Q.; Ye, W.Y.; Tang, X.G.; He, J.; Zhong, Y.J.; Ou, J.L.; Pang, X.H. Transcriptome analysis and genome-wide identification of the dehydration-responsive element binding gene family in jackfruit under cold stress. BMC Genom. 2024, 25, 833. [Google Scholar] [CrossRef]
- Hu, Y.T.; Xu, Z.C.; Tian, Y.; Gao, R.R.; Ji, A.J.; Pu, X.D.; Wang, Y.; Liu, X.; Song, J.Y. Genome-wide identification and analysis of AP2/ERF transcription factors related to camptothecin biosynthesis in Camptotheca acuminata. Chin. J. Nat. Med. 2020, 18, 582–593. [Google Scholar] [CrossRef]
- Sheng, S.; Guo, X.; Wu, C.; Xiang, Y.; Duan, S.; Yang, W.; Li, W.; Cao, F.; Liu, L. Genome-wide identification and expression analysis of DREB genes in alfalfa (Medicago sativa) in response to cold stress. Plant Signal Behav. 2022, 17, 2081420. [Google Scholar] [CrossRef]
- Su, J.C.; Song, S.L.; Wang, Y.T.; Zeng, Y.; Dong, T.; Ge, X.; Duan, H. Genome-wide identification and expression analysis of DREB family genes in cotton. BMC Plant Biol. 2023, 23, 169. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, L.; Nie, L.; Zheng, Y.; Zhu, S.; Hou, J.; Li, R.; Chen, G.; Tang, X.; Wang, C.; et al. Genome-wide analysis of the DREB family genes and functional identification of the involvement of BrDREB2B in abiotic stress in wucai (Brassica campestris L.). BMC Genom. 2022, 23, 598. [Google Scholar] [CrossRef]
- Xu, B.; Chen, B.; Qi, X.; Liu, S.; Zhao, Y.; Tang, C.; Meng, X. Genome-wide identification and expression analysis of RcMYB genes in Rhodiola Grenulata. Front. Genet. 2022, 13, 831611. [Google Scholar]
- Mushtaq, N.; Munir, F.; Gul, A.; Amir, R.; Zafar Paracha, R. Genome-wide analysis, identification, evolution and genomic organization of dehydration responsive element-binding (DREB) gene family in Solanum tuberosum. PeerJ 2021, 9, e1164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.D.; Bai, B.X.; Xu, M.; Liu, Y.L.; Wu, Y.F.; Zhao, L. Advances in and prospects for Actinidia viruses. Plant Dis. 2022, 106, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Y.; Ma, F.; Zhao, J.; Yang, H.; Song, S.; Zhang, S. Identification of DREB family genes in banana and their function under drought and cold stress. Plants 2024, 13, 2119. [Google Scholar] [CrossRef] [PubMed]
- Baxter, L.; Jironkin, A.; Hickman, R.; Moore, J.; Barrington, C.; Krusche, P.; Dyer, N.P.; Buchanan-Wollaston, V.; Tiskin, A.; Beynon, J.; et al. Conserved noncoding sequences highlight shared components of regulatory networks in dicotyledonous plants. Plant Cell 2012, 24, 3949–3965. [Google Scholar] [CrossRef]
- Liu, X.Q.; Zhu, J.J.; Wei, C.J.; Guo, Q.; Bian, C.K.; Xiang, Z.H.; Zhao, A.C. Genome-wide identification and characterization of the DREB transcription factor gene family in mulberry. Bio Plant. 2015, 59, 253–265. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Gupta, A.; Soni, D.; Garg, R.; Pathre, U.V.; Nath, P.; Sane, A.P. Ectopic expression of a tomato DREB gene affects several ABA processes and influences plant growth and root architecture in an age-dependent manner. J. Plant Physiol. 2017, 214, 97–107. [Google Scholar] [CrossRef]
- Xue, X.; Xie, M.; Zhu, L.; Wang, D.; Xu, Z.; Liang, L.; Zhang, J.; Xu, L.; Zhou, P.; Ran, J.; et al. 5-ALA Improves the low temperature tolerance of common Bean seedlings through a combination of hormone transduction pathways and chlorophyll metabolism. Int. J. Mol. Sci. 2023, 24, 13189. [Google Scholar] [CrossRef] [PubMed]
- Balestrasse, K.B.; Tomaro, M.L.; Batlle, A.; Noriega, G.O. The role of 5-aminolevulinic acid in the response to cold stress in soybean plants. Phytochemistry 2010, 71, 2038–2045. [Google Scholar] [CrossRef]
- Korkmaz, A.; Korkmaz, Y.; Demirkiran, A.R. Enhancing chilling stress tolerance of pepper seedlings by exogenous application of 5-aminolevulinic acid. Environ. Exp. Bot. 2010, 67, 495–501. [Google Scholar] [CrossRef]
- Anwar, A.; Wang, J.; Yu, X.; He, C.; Li, Y. Substrate application of 5-aminolevulinic acid enhanced low-temperature and weak-light stress tolerance in cucumber (Cucumis sativus L.). Agronomy 2020, 10, 472. [Google Scholar] [CrossRef]
- Xu, F.; Cheng, S.Y.; Zhu, J.; Zhang, W.W. Effects of 5-aminolevulinic acid on chlorophyll, photosynthesis, soluble sugar and flavonoids of Ginkgo biloba. Not. Bot. Horti Agrobot. 2011, 39, 41–47. [Google Scholar] [CrossRef]
- Xu, F.; Chang, J.; Cheng, S.Y.; Zhu, J.; Li, L.L.; Wang, Y.; Chen, H. Promotive effect of 5-aminolevulinic acid on the antioxidant system in Ginkgo biloba leaves. Afr. J. Biotechnol. 2009, 8, 3769–3776. [Google Scholar]
- Liu, T.; Xu, G.; Gao, W.R.; Guo, S.R.; Li, D.C.; Sun, Y.J. Effects of ALA on AsA-GSH cycling in leaves of chili plants under low temperature stress. J. Agric. Sci. 2011, 4, 830–835. [Google Scholar]
- Zhao, J.J.; Zhou, N.; Feng, N.J.; Zheng, D.F. Effects of 5-aminolevulinic acid on osmotic adjustment and antioxidant system in mung bean under chilling stress. Biol. Plant. 2020, 64, 736–743. [Google Scholar] [CrossRef]
- Li, D.M.; Zhang, J.; Sun, W.J.; Li, Q.; Dai, A.H.; Bai, J.G. 5-Aminolevulinic acid pretreatment mitigates drought stress of cucumber leaves through altering antioxidant enzyme activity. Sci. Hortic. 2011, 30, 820–828. [Google Scholar] [CrossRef]
- Luo, X.H. Study on the mechanism of exogenous ALA alleviating pepper seedling growth under low temperature and weak light. Hunan Agricult. Univ. 2022. [Google Scholar] [CrossRef]
- Lv, R.; Gao, Y.; Yang, X.; Li, X.; Zhu, C.; Mo, F.; Li, K. Identification of Cysteine synthase (Cys) gene family in tomato (Solanum lycopersicum) and functional of SlCys5 in cold stress tolerance. Int. J. Mol. Sci. 2025, 26, 2801. [Google Scholar] [CrossRef]
- Li, S.J.; Zhang, W.P.; Si, C.; Chen, J.; Huang, Y.; Li, M.; Liang, H.; Duan, J.; He, C. Genome-wide identification and functional characterization of the Dof family in Dendrobium officinale. Int. J. Mol. Sci. 2025, 26, 2671. [Google Scholar] [CrossRef]
- Sabir, I.A.; Hu, X.; Khan, I.; Qin, Y. Regulatory mechanisms of bud dormancy: Environmental, hormonal, and genetic perspectives. Int. J. Mol. Sci. 2025, 26, 2517. [Google Scholar] [CrossRef]
- Yue, J.; Yuan, S.; Liu, L.; Niu, Z.; Ma, L.; Pu, Y.; Wu, J.; Fang, Y.; Sun, W. Genome-wide identification of the SWEET gene family and functional analysis of BraSWEET10 in winter B. rapa (Brassica rapa L.) under low-temperature stress. Int. J. Mol. Sci. 2025, 266, 2398. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Kopka, J.; Sung, D.Y.; Zhao, W.; Popp, M.; Porat, R.; Guy, C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007, 50, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cissé, N.; et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef]
- Zandkarimi, H.; Ebadi, A.; Salami, S.A.; Alizade, H.; Baisakh, N. Analyzing the expression profile of AREB/ABF and DREB/CBF genes under drought and salinity stresses in grape (Vitis vinifera L.). PLoS ONE 2015, 10, e0134288. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.P. Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef]
- Yang, W.; Liu, X.D.; Chi, X.J.; Wu, C.A.; Li, Y.Z.; Song, L.L.; Liu, X.M.; Wang, Y.F.; Wang, F.W.; Zhang, C.; et al. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 2011, 233, 219–229. [Google Scholar] [CrossRef]
- Hou, L.X.; Wu, Q.Q.; Zhu, X.M.; Li, X.Y.; Fan, X.; Hui, M.; Ye, Q.; Liu, G.; Liu, X. Transcription factor VvDREB2A from Vitis vinifera improves cold tolerance. Int. J. Mol. Sci. 2023, 24, 9381. [Google Scholar] [CrossRef]
- Mao, D.H.; Chen, C.Y. Colinearity and similar expression pattern of Rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS ONE 2012, 7, e47275. [Google Scholar] [CrossRef]
- Hou, Y.J.; Wong, D.C.J.; Li, Q.Y.; Zhou, H.M.; Zhu, Z.F.; Gong, L.Z.; Liang, J.; Ren, H.S.; Liang, Z.C.; Wang, Q.F.; et al. Dissecting the effect of ethylene in the transcriptional regulation of chilling treatment in grapevine leaves. Plant Physiol Biochem. 2023, 196, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.; Liu, X.; Zhu, Z.; Chen, M. Genome-wide identification of the AP2/ERF gene family from Limonium bicolor and functional characterization of LbAP2/ERF32 under salt stress. Plant Physiol. Biochem. 2024, 215, 109035. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Hu, F.; Zhang, Y.; Guo, J. Identification and Characterization of AP2/ERF Transcription Factors in Yellow Horn. Int. J. Mol. Sci. 2022, 23, 14991. [Google Scholar] [CrossRef]
- Yue, J.; Liu, J.; Tang, W.; Wu, Y.Q.; Tang, X.; Li, W.; Yang, Y.; Wang, L.; Huang, S.; Fang, C.; et al. Kiwifruit Genome Database (KGD): A comprehensive resource for kiwifruit genomics. Hortic. Res. 2020, 7, 117. [Google Scholar] [CrossRef]
- Zi, Y.Q.; Zhang, Z.M.; Zhao, K.; Yang, X.Y.; Zhu, L.; Yin, T.; Chen, C.Y.; Wen, K.; Li, X.L.; Zhang, H.Y.; et al. Genome-wide identification of kiwifruit K channel Shaker family members and their response to low-K stress. BMC Plant Biol. 2024, 24, 833. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Chang, X. Preliminary Study on the Regulatory Pathway of CsCBF1 and Its In Vitro Functional Characterization in Tea Tree Under Low Temperature Stress. Master’s Thesis, Anhui Agricultural University, Anhui, China, 2012. [Google Scholar]
- Letunic, I.; Tobias, D.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.X.; Yin, T.; Han, P.C.; Yang, X.Y.; Zhang, M.J.; Du, C.J.; Zhang, H.Y.; Liu, X.Z. Genome-wide identification of sweet orange WRKY transcription factors and analysis of their expression in response to infection by Penicillium digitatum. Curr. Issues Mol. Biol. 2023, 45, 1250–1271. [Google Scholar] [CrossRef]
- Liu, F.; Li, H.; Wu, J.; Wang, B.; Tian, N.; Liu, J.; Sun, X.; Wu, H.; Huang, Y.; Lü, P.; et al. Genome-wide identification and expression pattern analysis of lipoxygenase gene family in banana. Sci. Rep. 2021, 11, 9948. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Salkind, N.J. Encyclopedia of Measurement and Statistics; SAGE Publications: Thousand Oaks, CA, USA, 2006. [Google Scholar]
- Zhu, L.; Zhang, M.J.; Yang, X.Y.; Zi, Y.Q.; Yin, T.; Li, X.L.; Wen, K.; Zhao, K.; Wan, J.Q.; Zhang, H.Y.; et al. Genome-wide identification of bZIP transcription factors in 12 Rosaceae species and modeling of novel mechanisms of EjbZIPs response to salt stress. Plant Genome 2024, 17, e20468. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Duricki, D.A.; Soleman, S.; Moon, L.D. Analysis of longitudinal data from animals with missing values using SPSS. Nat. Prot. 2016, 11, 1112–1129. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.J.; Yang, X.M.; Wu, J.X.; Cai, H.B.; Yang, N.; Li, X.L.; Wen, K.; Chen, D.M.; et al. Genome-wide identification and expression pattern analysis of the kiwifruit GRAS transcription factor family in response to salt stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef]
- Wen, K.; Li, X.L.; Yin, T.; Chen, C.Y.; Zhu, L.; Zi, Y.Q.; Zhao, K.; Zhou, X.Y.; Liu, X.Z.; Zhang, H.Y. Genome-wide identification of carotenoid cleavage oxygenase genes in Orah mandarin and the mechanism by which CrCCD4b1 affects peel color. Sci. Hortic. 2024, 338, 113652. [Google Scholar] [CrossRef]
- Seo, M.H.; Kim, P.M. The present and the future of motif-mediated protein-protein interactions. Curr. Opin. Struct. Biol. 2018, 50, 162–170. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, P.; Chen, D.; Wan, J.; Chen, C.; Zhao, K.; Zi, Y.; Liu, P.; Yang, C.; Zhang, H.; Liu, X. Genome-Wide Identification of DREB Gene Family in Kiwifruit and Functional Characterization of Exogenous 5-ALA-Mediated Cold Tolerance via ROS Scavenging and Hormonal Signaling. Plants 2025, 14, 2560. https://doi.org/10.3390/plants14162560
Tian P, Chen D, Wan J, Chen C, Zhao K, Zi Y, Liu P, Yang C, Zhang H, Liu X. Genome-Wide Identification of DREB Gene Family in Kiwifruit and Functional Characterization of Exogenous 5-ALA-Mediated Cold Tolerance via ROS Scavenging and Hormonal Signaling. Plants. 2025; 14(16):2560. https://doi.org/10.3390/plants14162560
Chicago/Turabian StyleTian, Ping, Daming Chen, Jiaqiong Wan, Chaoying Chen, Ke Zhao, Yinqiang Zi, Pu Liu, Chengquan Yang, Hanyao Zhang, and Xiaozhen Liu. 2025. "Genome-Wide Identification of DREB Gene Family in Kiwifruit and Functional Characterization of Exogenous 5-ALA-Mediated Cold Tolerance via ROS Scavenging and Hormonal Signaling" Plants 14, no. 16: 2560. https://doi.org/10.3390/plants14162560
APA StyleTian, P., Chen, D., Wan, J., Chen, C., Zhao, K., Zi, Y., Liu, P., Yang, C., Zhang, H., & Liu, X. (2025). Genome-Wide Identification of DREB Gene Family in Kiwifruit and Functional Characterization of Exogenous 5-ALA-Mediated Cold Tolerance via ROS Scavenging and Hormonal Signaling. Plants, 14(16), 2560. https://doi.org/10.3390/plants14162560




