1. Introduction
Boehmeria nivea (L.) Gaudich. (ramie) is a bast fiber crop used in textiles, medicine, feed, and environmental protection, making it a valuable alternative plant [
1,
2]. Lignins play an important role in plant defense and development and are a critical factor affecting the fiber and quality of ramie feed. Flavonoids are also important components of ramie feed and have medicinal value. Both lignins and flavonoids are formed via the phenylpropanoid pathway and share common precursors. Uridine diphosphate-glycosyltransferases (UGTs, EC 2.4.1.x) are a large family of enzymes that catalyze the transfer of sugar to a variety of plant secondary metabolites involved in lignan, flavonoid, salicylate, and phytohormone metabolism. This has potential implications for cell wall biosynthesis and biotic and abiotic stress responses [
3]. Plant UGTs play diverse roles at different developmental stages, and most of their substrates are derived from the phenylpropanoid pathway, whose precursors are the aromatic amino acids phenylalanine and tyrosine [
4,
5]. Therefore, this study focused on
UGT genes involved in monolignol and flavonoid biosynthesis and their abiotic stress responses in ramie (
Figure 1).
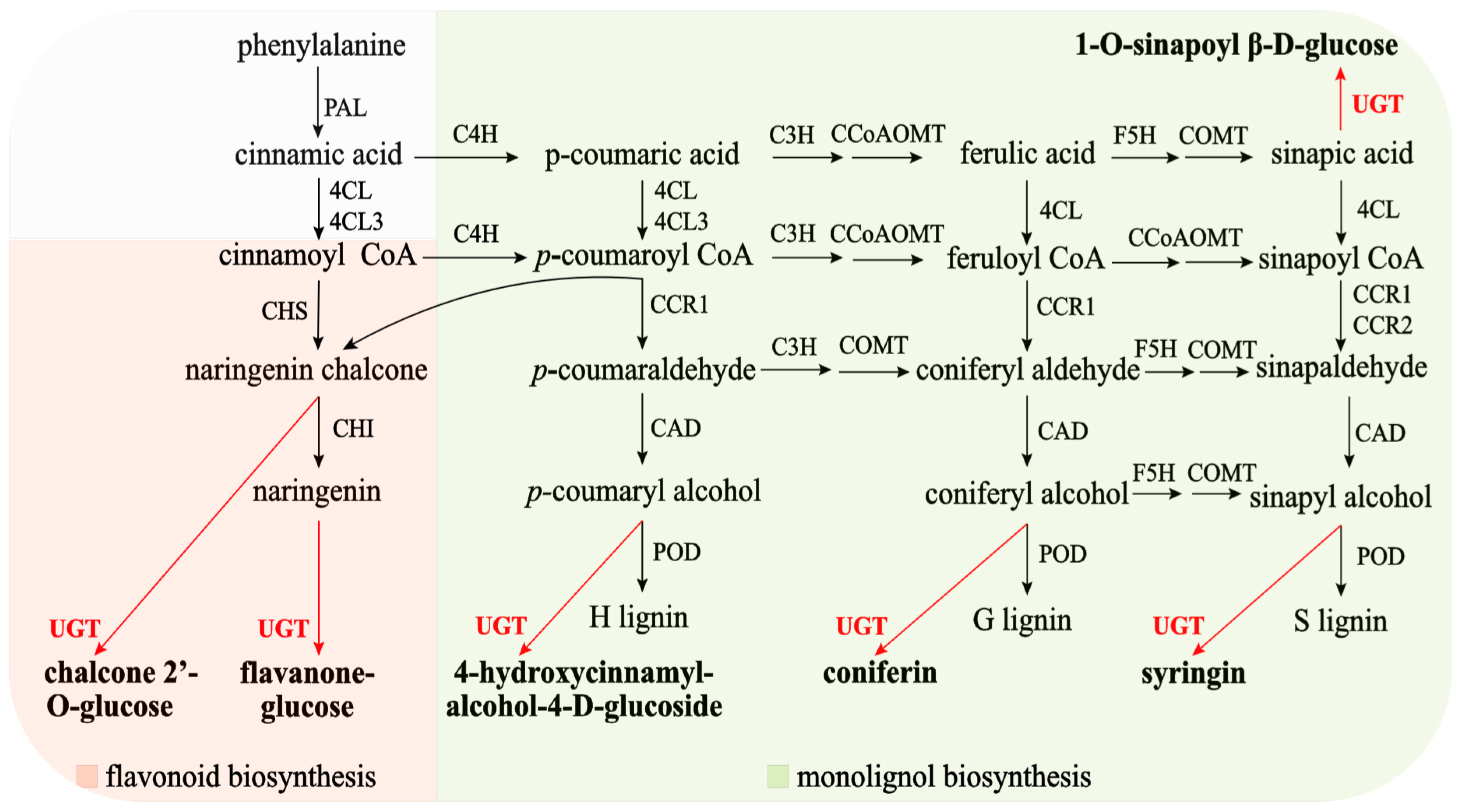
Monolignols (or (hydroxyl)cinnamyl alcohols) such as
p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol are the main building blocks of lignins. In plants, monolignol glycosylation is catalyzed by family 1 UGTs. When glycosylated by UGTs in the presence of uridine diphosphate (UDP)-glucose, these phenylpropanoids give rise to cinnamyl alcohol glucosides, coniferin, and syringin. In ramie, UGTs can also catalyze the glycosylation of sinapic acid (
Figure 1). Monolignol glucosides accumulate in vascular tissues, including the phloem, cambial tissue, and differentiating xylem, of both conifers and angiosperms and can be incorporated into lignin polymers [
7]. Monolignol glycosylation catalyzed by
UGT is essential for lignification; however, the role of
UGT genes in ramie lignin synthesis has not been well investigated. Most studies have focused on lignin biodegradation mechanisms, which are vital factors responsible for lignin modification and degradation [
8]. Monolignol glycosylation plays a key role in lignin metabolism in
Arabidopsis thaliana (L.) Heynh, poplar,
Ginkgo biloba L. [
9], and
Pyrus bretschneideri Rehder. This is carried out via
UGT72E [
10] and
UGT72B [
11] subfamilies in
Arabidopsis thaliana and
UGT72AZ1,
UGT72AZ2,
UGT72B37, and
UGT72B39 in poplar [
7] glycosylate monolignols.
Arabidopsis and poplar exhibit partial conservation of substrate recognition between
UGT72, and divergent functions exist between different groups of the
UGT72 family [
7].
PbUGT72AJ2 of
P. bretschneideri is considered a monolignol-glycosylation-related UGT [
12]. In
Arabidopsis, the guaiacyl (G) unit is 2–3 times more abundant than the syringyl (S) unit [
13]. The lignin in poplar mainly consists of G units, with a minor amount of H (hydroxyphenyl) units [
14]. The ratio of S, G, and H units in ramie lignin is approximately 6:3:1 [
15], which differs from that in the model plant. Therefore, research on
UGT genes in model plants has greatly promoted the study of monolignol glycosylation in ramie and other fiber crops.
Flavonoids are secondary plant metabolites that are widely distributed in nature. They contain at least 10,000 different derivatives with excellent biological functions. The biotransformation of flavonoid aglycones into
O-rutinosides or
O-neohesperidosides in
Citrus plants usually involves two glycosylation reactions involving a series of uridine diphosphate-sugar-dependent glycosyltransferases [
16]. Complementary DNAs (cDNAs) encoding flavonoid diglycosyltransferases from
Citrus have been identified and functionally characterized. These cDNAs include those encoding flavanone/flavone-7-O-glucoside-1,2-rhamnosyltransferase from pummel [
17] and
CsUGT76F1 from
Citrus sinensis (L.) Osbeck [
16]. Similarly, Chen et al. [
18] identified a flavonoid (1–2 rhamnosyltransferase) involved in flavanone neohesperidoside biosynthesis. UGTs are a group of enzymes responsible for the glycosylation of flavonoid glycosides in
Epimedium pubescens Maxim. [
19]. In
Camellia sinensis (L.) Kuntze, two
CsUGT genes (
CsUGT75L12 and
CsUGT79B28) participate in the biosynthesis of the bitter flavonoid 7-O-neohesperidoside via sequential glycosylation and rhamnosylation of flavonoids [
20].
OsUGT88C3 is responsible for the biosynthesis of malvidin 3-
O-galactoside in rice [
21].
RchUGT169 can catalyze the conversion of kaempferol and quercetin to the corresponding flavonoid glycosides in
Rubus chingii Hu through transient expression in tobacco [
22]. In ramie, hydroxycinnamoyl CoA ester is a precursor for
UGT to form chalcone glucose and flavanone glucose (
Figure 1). However, the role of
UGT genes in plant flavanone synthesis has not been thoroughly investigated. Therefore, studying
UGT genes in ramie is necessary.
Despite the widespread identification of UGT family members in various species, limited information is available on this family in ramies. Therefore, a genome-wide analysis was conducted in this study to identify UGT family members in the B. nivea genome. This resulted in the identification of 84 UGT genes. These UGT genes were systematically characterized, including their phylogenetic relationships, gene structures, conserved motifs, cis-acting elements, chromosome distribution, and gene duplication events. Importantly, UGT genes involved in monolignol and flavonoid biosynthesis were identified and characterized. Moreover, expression profiles and accumulation of lignin and flavonoids in ramie during growth were evaluated. In addition, the abiotic stress responses of UGT genes involved in monolignol and flavonoid biosynthesis in ramie were validated. Therefore, comprehensive genome-wide identification and rigorous characterization of these UGT genes in ramie are important for understanding the glycosylation mechanisms and regulation of monolignol and flavonoid accumulation in the plant. Thus, this study provides valuable insights into the further functional characterization of UGT genes in ramie.
2. Results
2.1. Identification and Characterization of BnUGT Genes
Based on 90
Arabidopsis UGT amino acid sequences, 84
UGT genes were identified in
B. nivea (
BnUGT) based on the presence of complete plant secondary product glycosyltransferase (PSPG) motifs. Further characterizations, including protein lengths, MW, pI, GRAVY, index values, and subcellular localization, are presented in
Supplementary Table S1. Protein lengths varied from 119 to 768 amino acids, corresponding to their MWs ranging between 13,162.84 and 84,954.12 Da, and the pI changed from 4.95 to 8.90. The instability index changed from 32.01 to 74.07, the aliphatic index changed from 64.87 to 106.15, and GRAVY changed from −0.533 to 0.132. The above data indicate that the physical and chemical properties of the 84
BnUGT proteins showed substantial differences. Subcellular localization predictions indicated that the cell membrane was the primary cell localization structure for these UGT proteins, followed by the chloroplasts. Notably, two
BnUGT proteins (
Bnt08T012480.1 and
Bnt13T018476.1) were observed over five localization structures.
2.2. Phylogenetic and Classification Analysis of BnUGT Genes
To classify and better understand the evolutionary relationships among
BnUGT proteins (
Supplementary Table S2), a phylogenetic tree of
BnUGT genes and other known
UGT proteins from various plants was constructed (
Supplementary Table S3). In total, 84
BnUGT genes in groups A–I, L–P, and R were found (
Figure 2 and
Figure 3a). The study focus was to screen the members of phylogenetic groups involved in monolignol and flavonoid biosynthesis.
The
UGT genes of Group A generally use anthocyanin as the substrate [
23], and
ZmUGT genes from
Z. mays (
GRMZM2G135722_T01 and
GRMZM2G061321_T01) in Group A all encode anthocyanidin 3-O-glucosyltransferase [
24]. Thus, the 14
BnUGTs (such as
Bnt04T006105.1) in this phylogenetic group are likely associated with the glycosylation of anthocyanins.
AtUGT89B1 in Group B is mainly responsible for glycosylation of dihydroxybenzoic acids and flavonoids [
25].
UGT genes in Group F can glycosylate anthocyanidins and flavonols [
23]. In the phylogenetic tree constructed in this study,
AtUGT78D1 (flavonol 3-O-glycosyltransferases) in Group F catalyzes the formation of anthocyanidin or flavonol glycosides [
26]. Therefore, the fourteen
BnUGT genes in Group A,
Bnt09T014140.1 in Group B, and the three
BnUGT genes (
Bnt06T010452.1,
Bnt13T018475.1, and
Bnt13T018476.1) in Group F may catalyze flavonoids to form glycosides.
The
UGT genes responsible for the glycosylation of monolignols and lignin precursors are mainly distributed in Groups D, E, and L [
27].
AtUGT73C5 in Group D catalyzes the formation of cinnamyl alcohol glucoside [
28], and
Pbr032554.1 and
Pbr032553.1 are associated with glycosylation of monolignols or lignin precursors [
12]. The phylogenetic relationships of
BnUGT genes in the same clade, such as
Bnt12T018165.1, which is closest to
AtUGT73C5,
Pbr032554.1, and
Pbr032553.1, suggest that this gene may be associated with monolignol glycosylation.
CsUGT73A20 (
C. sinensis) in Group D catalyzes the formation of flavonol glycosides [
29], and
CtUGT49 (
Carthamus tinctorius L.), which belongs to the
UGT73 family, catalyzes the conversion of naringenin chalcone [
30]. Thus,
BnUGT genes in the same clade, such as
Bnt07T010992.1, which is closest to
CsUGT73A20, may have similar functions.
The highest number of UGTs that catalyze the formation of monolignol/lignin precursor glucose esters or glucosides is in Group E.
AtUGT71C1 catalyzes the glycosylation of caffeic acid [
31], and
Potri.016G014500 was maximally down-regulated under drought conditions. Moreover, five
BnUGT genes (
Bnt02T003078.4,
Bnt04T006292.1,
Bnt05T007747.1,
Bnt05T007753.1, and
Bnt05T007754.1), which are closest to
AtUGT71C1 in the phylogenetic tree, may also be associated with monolignol glycosylation and adverse stress. Among them,
AtUGT72E2 simultaneously catalyzes the glycosylation of lignin monomers (coniferyl alcohol and sinapyl alcohol) [
32] and precursors (coniferyl aldehyde, sinapyl aldehyde, and sinapic acid) [
10], and
AtUGT72D1 catalyzes the glycosylation of sinapic acid [
33]. The phylogenetic relationships of
Bnt14T019888.1 and
Bnt01T001679.1 were the closest to
AtUGT72E2 and
AtUGT72D1. This relationship confirmed that these genes may catalyze similar substrates.
AtUGT72B1 catalyzes the glycosylation of coniferyl aldehyde and alcohol and regulates cell wall development and lignification. Notably,
AtUGT72B1 alters the total amount of lignin in plants [
11].
Potri.014G096100,
Potri.002G168600 [
7],
Pbr014154.1, and
Pbr014155.1 [
12] were considered to be monolignol-glycosylation-related
UGT genes, and the phylogenetic relationships of
Bnt08T012480.1,
Bnt07T010776.1, and
Bnt01T001680.1 were closest to
AtUGT72B1,
Potri.014G096100,
Potri.002G168600,
Pbr014154.1, and
Pbr014155.1. Therefore, it was hypothesized that these
BnUGT genes catalyze glucose conjugation of monolignols.
Members of Group L identify the carboxyl groups of different metabolites, such as phenylpropanoids and auxins [
25].
AtUGT75C1 in Group L was identified as anthocyanin 5-O-glucosyltransferases from various plant species.
UGT75C1 is functionally non-redundant in
A. thaliana because its mutant (ugt75c1) completely lacks anthocyanin 5-O-glucosides associated with flavonoid metabolism [
34].
AtUGT84A1,
AtUGT84A2,
AtUGT84A3, and
AtUGT84A4 in Group L catalyze the formation of cinnamic acid and hydroxycinnamic acids (
p-coumaric acid, caffeic acid, ferulic acid, and sinapic acid) from lignin precursor glucose esters [
35,
36]. This study hypothesized that the ten
BnUGT genes in the same clade can also participate in flavonoid and monolignol metabolism. The study focused on
Bnt06T010117.1 because it was clustered in the phylogenetic tree of
AtUGT75C1 and
AtUGT84A1-4.
In addition, Groups O and P are newly discovered taxa in higher plants; seven members were observed in Group O, and two were observed in Group P.
GRMZM2G110511_T01 and
GRMZM2G168474_T02 in Group O are cis-zeatin
O-glucosyltransferases, and
GRMZM2G082249_T01 (Group M) and
GRMZM5G834303_T01 (Group P) are cytokinin-
O-glucosyltransferases [
3,
24]. Members of these three groups may be closely associated with plant hormone glycosylation. Therefore, this study hypothesized that the
BnUGT genes in groups M, O, and P may be associated with hormone glycosylation. In addition,
Potri.016G105400 from the newly formed phylogenetic Group P was found to be upregulated under drought stress, thereby suggesting that
Bnt03T004065.5 and
Bnt07T011055.1 in Group P may be associated with adverse stress. Moreover, ramie contained one Group R member (
Bnt07T011603.3) but with the absence of Group Q (
Supplementary Table S4).
Based on the phylogenetic tree, seventeen BnUGTs were screened, including ten putative lignin-glycosylation-related BnUGTs (Bnt07T010992.1, Bnt12T018165.1, Bnt02T003078.4, Bnt04T006292.1, Bnt05T007747.1, Bnt05T007753.1, Bnt05T007754.1, Bnt14T019888.1, Bnt08T012480.1, and Bnt06T010117.1); five putative flavonoid-glycosylation-related BnUGTs (Bnt04T006105.1, Bnt09T014140.1, Bnt06T010452.1, Bnt13T018475.1, and Bnt13T018476.1); one putative adverse-stress-related BnUGT in newly formed phylogenetic Group P (Bnt03T004065.5); and one putative plant-hormone-related BnUGT (Bnt02T003773.4). Further analysis was conducted on the expression of these putative BnUGTs in different tissues during the growth and development of ramie and their response to cadmium stress.
2.3. Gene Structure, Conserved Motif, and Conserved Domain Analysis of BnUGT Genes
The motif composition of
BnUGT proteins is shown in
Figure 3b and
Supplementary Figure S1, yielding 12 conserved motifs specific to ramie
UGT proteins. Motif 1 (78/84), motif 2 (81/84), and motif 4 (78/84) were universally present across all
BnUGT proteins, thus representing the conserved
UGT domain. Only
Bnt01T001679.1 (314 aa),
Bnt01T001680.1 (119 aa), and
Bnt06T010519.1 (289 aa) did not contain motif 2, which might be responsible for the short length of the protein sequence. Using SMART to annotate each motif, only motifs 1 and 2 were annotated as uridine diphosphate-glucuronosyltransferase domains (PF00201), and motif 1 contained the complete PSPG-box. Notably, the distribution of some motifs displayed subgroup specificity. For example, the motif composition was divided into two parts in Group A, which is consistent with the evolutionary tree relationship. Members of the same group have similar motifs, which is consistent with the evolutionary relationships between members. Cluster analysis based on similar motif compositions revealed potential functional similarities among
BnUGT proteins. The conserved domains of
BnUGT proteins are shown in
Figure 3c and
Supplementary Table S5. All
BnUGT proteins contain a glycosyltransferase GTB-type superfamily domain. The presence of a conserved PGPS domain within the obtained protein sequences was confirmed.
Additionally, the gene structures of
BnUGT family members were obtained. The genes exhibited a diverse range of intron–exon arrangements, and most
BnUGT genes consisted of introns and exons (
Figure 3d). The number of exons in
BnUGT genes varies from one to six. A total of 28 members were intronless genes, and Groups H, I, N, and P did not contain intronless genes. In combination with gene structure and phylogenetic tree analyses, the patterns of exon–intron distribution between
BnUGT genes of the same phylogenetic group were very similar. This phenomenon further revealed the closer evolutionary relationship between these members and simultaneously showed the reliability of the phylogenetic tree construction.
2.4. Chromosomal Distribution and Gene Collinearity Analysis of BnUGT Genes
In total, 84
BnUGT genes were assembled and located on chromosomes 1–14 of ramie (except for chromosome 10) (
Figure 4). Among the chromosomes, chr4 and chr6 harbored the highest number of
BnUGT genes (13/12). Chr1, chr2, and chr12 harbored the same number of
BnUGT genes, all containing nine genes. Two members were observed on chromosomes 9 and 13. The number of
BnUGTs on chromosomes 9 and 13 was minimal, and there were three
BnUGTs on chromosomes 3 and 11. The distribution of different phylogenetic group members on the chromosomes was irregular and mostly existed as gene clusters. The Group O members were all on chromosome 12, while the
BnUGTs on chromosomes 1, 2, 4, and 6 were from five phylogenetic groups. A total of six members of Group A and Group L formed gene clusters on chromosomes 4 and 6, respectively. The members of Group E were mainly distributed on chromosomes 1, 2, 3, 4, 5, 7, and 8; the members of Group A were mainly distributed on chromosomes 1, 2, 4, 8, and 11; and the members of Group D were mainly distributed on chromosomes 2, 6, 7, 12, and 14. In addition, the results suggest that there were three collinear gene pairs in the study:
Bnt01T001679.1 demonstrated a collinear relationship with
Bnt14T019888.1;
Bnt03T004065.5 demonstrated a collinear relationship with
Bnt07T011055.1; and
Bnt14T019754.1 demonstrated a collinear relationship with
Bnt06T010143.2. Comprehensive analysis of collinearity and gene duplication events demonstrated the expansion and diversification of
BnUGT gene members in ramie.
2.5. Analysis of Cis-Acting Elements of Ramie UGT Genes
A total of thirty cis-acting elements were recognized from the promoter region of the
BnUGT genes, which were grouped into seven classes, including nine light-responsive elements (44.88%), seven defense and stress-responsiveness elements (12.66%), four phytohormone-responsive elements (25.49%), four MYB binding sites, four protein binding sites, one auxin-responsive element (8.21%), and one flavonoid biosynthetic binding site (
Supplementary Table S6).
Among all the elements, the G-box had the highest frequency (11.56%), followed by Box4 (10.84%). Additionally, methyl jasmonate response elements (134 CGTCA-motif and 133 TGACG-motif), abscisic acid response elements (218 ABRE), gibberellin-responsive elements (28 P-box, 17 GRAR-motif, and 17 TCTC-box), and salicylic acid response elements (48 TCA-element, 3 SARE, and 1 MRE) were detected in BnUGT genes. Six stress-responsive elements, including ARE (176), LTR (49), TC-rich repeats (40), MBS (29), GC-motif (20), and WUN-motif (4), were retrieved from ramie UGTs and responded to anaerobic induction, low-temperature induction, defense and stress prompts, drought, anoxic induction, and wounding stresses. Several plant-growth-related elements, such as the CAT-box (45), GCN4-motif (17), circadian (13), RY-element (6), MSA-like (3), HD-Zip1 (2), and AACA_motif (1), were obtained from ramie UGT genes. These elements are involved in meristem and endosperm expression, circadian control, seed-specific regulation, cell cycle regulation, palisade mesophyll cells, and negative endosperm-specific expression. Moreover, cis-acting elements associated with flavonoid biosynthetic gene regulation and four MYB-binding sites were identified within the BnUGT promoter sequences. The composition of these cis-acting elements showed that most BnUGT genes were involved in light, stress, hormones, and plant development.
2.6. The Analysis of the Lignin Content, Flavonoid Content, and Putative BnUGT Gene Expression
The lignin content was upregulated in the phloem, xylem, and leaf during the six developmental periods (
Figure 5). In the phloem, the lignin content was not significantly different between 15 and 60 days after emergence; after 75 days of emergence, the lignin accumulation was not significant, and the lignin content reached the maximum value, about 547 mg/g. However, the lignin content of the phloem was not significantly different between 30 and 75 days after emergence; the lignin content 90 days after emergence was significantly higher than that of other periods (except 75 days after emergence). In the leaf, the lignin content was not significantly different between 15 and 60 days after emergence. However, after 60 days of emergence, the lignin accumulation was rapid, and the lignin content reached the maximum value after 75 days of emergence, about 629 mg/g. In the xylem, the lignin content 15 days after emergence was significantly lower than that of other periods. The lignin content was not significantly different between 30 and 45 days after emergence. After 60 days of emergence, the lignin accumulation was significant, reaching a maximum value of about 776 mg/g, which was significantly higher than that of the phloem and leaf.
The flavonoid content showed varying trends across different tissues throughout the six developmental periods (
Figure 5). In the phloem and leaf, the flavonoid content was not significantly different between 15 and 60 days after emergence, flavonoid accumulation was rapid after 60 days of emergence, and the flavonoid content reached the maximum value after 75 days of emergence, about 1.4 mg/g and 2.5 mg/g, respectively. Specifically, the flavonoid content was first downregulated and then upregulated in the xylem, and the flavonoid content reached the maximum value of about 8.8 mg/g after 75 days of germination. The content of flavonoids in the leaf was 6.3 times and 3.5 times that of the phloem and xylem after 75 days of germination, respectively.
The expression levels of seventeen putative
BnUGTs were analyzed during the six developmental periods (
Figure 5). In the phloem, eleven
BnUGT expression patterns were observed; the most common expression pattern involved genes being downregulated and then upregulated, followed by further downregulation and then upregulation, including
Bnt07T010992.1,
Bnt05T007754.1,
Bnt08T012480.1,
Bnt13T018475.1, and
Bnt03T004065.5. The expression level of
Bnt05T007753.1 was upregulated, the expression level of
Bnt14T019888.1 was downregulated, and the expression level of Bnt13T018476.1 was not significantly different between 15 and 90 days after emergence. In the leaf, seven BnUGT expression patterns were observed; the most common expression pattern involved genes being upregulated and then downregulated, followed by further upregulation and then downregulation, including
Bnt07T010992.1,
Bnt12T018165.1,
Bnt02T003078.2,
Bnt04T006292.1,
Bnt05T007754.1,
Bnt08T012480.1,
Bnt06T010117.1, and
Bnt13T018475.1. The expression levels of
Bnt05T007753.1 and
Bnt14T019888.1 were upregulated, and the expression levels of
Bnt04T006105.1,
Bnt09T014140.1, and
Bnt13T018476.1 were downregulated. In the xylem, four
BnUGT expression patterns were observed; the most common expression pattern involved genes being upregulated and then downregulated, followed by further upregulation and then downregulation. This pattern has the same number of genes as the expression pattern, showing initial upregulation and then downregulation, with both containing six genes. The expression levels of
Bnt12T018165.1,
Bnt14T019888.1,
Bnt06T010117.1, and
Bnt02T003773.4 were upregulated. The expression of monolignol-glycosylation-related candidate
BnUGTs (except
Bnt07T010992.1 and
Bnt02T003078.2) in the xylem 75 days after germination was much higher than that during the other five periods. The relative expression ranged from 40 to 90, and the relative expression of
Bnt05T007753.1 reached up to 710. The expression of
Bnt13T018475.1 and
Bnt13T018476.1 (flavonoid-glycosylation-related candidate
BnUGTs) in the xylem 75 days after germination was much higher than that during the other five periods, and the relative expression of
Bnt13T018476.1 reached up to 109 at 60 days after germination.
2.7. Correlation Between Lignin Content and Putative BnUGT Expression
The correlations between the expression levels of seventeen putative
BnUGTs and lignin content in the phloem, xylem, and leaf during the six developmental stages were analyzed (
Figure 6a). The lignin content was upregulated in the phloem, xylem, and leaf as the plant grew and developed; however, the expression levels of
BnUGTs were different (
Figure 5). In the phloem, only
Bnt05T007753.1 expression showed a significant positive correlation with lignin content, whereas
Bnt14T019888.1 expression showed a significant negative correlation with lignin content. In the xylem, the expression of
Bnt12T018165.1,
Bnt14T019888.1,
Bnt06T010117.1, and
Bnt02T003773.4 showed a significant positive correlation with lignin content. In the leaf, the expression of
Bnt05T007753.1 and
Bnt05T007754.1 showed a significant positive correlation with lignin content, and
Bnt14T019888.1 expression showed an extremely significant positive correlation with lignin content.
2.8. Correlation Between Flavonoid Content and Putative BnUGT Gene Expression
The correlations between the expression levels of seventeen putative
BnUGTs and flavonoid content in the phloem, xylem, and leaf during the six developmental stages were analyzed (
Figure 6b). In the phloem, only
Bnt13T018476.1 expression showed a significant positive correlation with flavonoid content. In the xylem,
Bnt06T010117.1 expression showed a significant positive correlation with flavonoid content, and
Bnt14T019888.1 expression showed an extremely significant positive correlation with flavonoid content. In the leaf, the expression of
Bnt04T006105.1 and
Bnt09T014140.1 showed a significant negative correlation with flavonoid content, whereas the expression of
Bnt05T007753.1 and
Bnt14T019888.1 showed an extremely significant positive correlation with flavonoid content. It is worth noting that the expression of
Bnt06T010117.1 showed a strong positive correlation with lignin and flavonoid content in the xylem (
Figure 6).
2.9. The Expression Patterns of Putative BnUGTs Under Cd Stresses
To determine whether putative
BnUGT genes were involved in abiotic stress responses, the expression of ten monolignol-glycosylation-related genes, two flavonoid-glycosylation-related
BnUGT genes (
Bnt04T006105.1 and
Bnt09T014140.1), and one adverse-stress-related
BnUGT (
Bnt03T004065.5) was analyzed using qRT-PCR (
Figure 7). Eight
BnUGT expression patterns were identified in ramie roots in response to increasing Cd stress. The expression of
Bnt04T006105.1 was upregulated, and that of
Bnt08T012480.1 and
Bnt03T004065.5 was downregulated. Four
BnUGT expression patterns were observed in ramie stems in response to increasing Cd stress. Specifically, the expression of nine
BnUGT genes was first upregulated and then downregulated; only the expression of
Bnt05T007747.1 was upregulated. The expression of
Bnt02T003078.2,
Bnt04T006292.1,
Bnt06T010117.1,
Bnt04T006105.1, and
Bnt02T003773.4 under 20 mg/L Cd treatment was much higher than at other Cd concentrations. In particular, the expression of
Bnt06T010117.1 increased 106-fold. Among the six
BnUGT expression patterns in ramie leaves under increasing Cd stress, only the expression of
Bnt04T006105.1 and
Bnt07T010992.1 was upregulated and downregulated, respectively. The expression of
Bnt04T006105.1 under 40 mg/L Cd treatment was 10–50 times higher than at other Cd concentrations. Moreover, the expression of
Bnt02T003078.2,
Bnt04T006292.1,
Bnt05T007747.1,
Bnt05T007753.1, and
Bnt08T012480.1 under 20 mg/L Cd treatment was much higher than at other Cd concentrations. In addition, all genes had the lowest expression in the roots compared to stems and leaves (except
Bnt07T010992.1), and the expression of
Bnt05T007753.1 in leaves was much higher compared to roots and stems.
3. Discussion
The
UGT multigene family has been identified in several plant species, including eudicots and monocots. In total, 84
BnUGT family members were first identified in the
B. nivea genome, and the number of
BnUGTs in ramie is smaller than that of other plants, such as
A. thaliana (107) [
25],
P. trichocarpa (192) [
3], and
Camellia sinensis (132) [
37]. However, there was little difference in the number of UGTs in
Citrus sinensis genes (
Supplementary Table S4) [
18]. The
UGTs responsible for the glycosylation modification of monolignol are mainly distributed in Groups D, E, and L [
35], in which 33
BnUGTs were identified in ramie. The
UGTs responsible for the glycosylation modification of flavonoids are mainly distributed in Groups A, B, and F, in which 18
BnUGTs were identified in this study (
Figure 2). Groups D and E are relatively large phylogenetic groups of plants whose members can recognize a range of substrates, such as terpenoids, flavonoids, benzoates, and lignin metabolism intermediates [
25,
32]. The number of
BnUGTs in Groups D, E, and L (
Supplementary Table S4) was higher compared to other groups, similar to other plants. Notably, Group O and Group P are newly discovered taxa in higher plants. Unlike
Arabidopsis [
25], these two groups also exist in ramie (
Supplementary Table S4), which is consistent with poplar [
3]. Groups Q and R were absent in
A. thaliana [
25],
P. trichocarpa [
3], and
Linum usitatissimum L. [
38]. Ramie and
C. sinensis [
37] were similar in the absence of Group Q. However, ramie contained one Group R member, which may be due to the distinct S/G/H lignin ratio in ramie (6:3:1) compared to that of model plants. As a fiber crop, members of Groups J and K were present in
L. usitatissimum but not in ramie (
Supplementary Table S4). This highlights lineage-specific gene loss or divergence and further underscores species-specific adaptations in UGT function [
15].
Motifs are the locally conserved regions in the gene sequence and may play an essential role in specific biological functions. Conserved motifs are commonly regarded as key indicators for analyzing the expansion of gene families. Despite significant variation among these groups, motif 1, motif 2, and motif 4 exhibit high conservation in ramie
UGTs, suggesting their potential importance in the glycosylation function of UGT enzymes, which is consistent with their role in substrate recognition [
23]. This finding diverges from a previous report on
Ci. sinensis (motif 1 and motif 3) [
18]. The expansion of
UGT families in ramie, particularly on chromosomes 4 and 6 (
Figure 4), likely reflects gene duplication events that drive functional diversification. Moreover, we found three collinear gene pairs in this study, indicating that the arrangement order of
BnUGTs on chromosomes was maintained. The number of
BnUGT gene pairs in ramie is smaller than that of other plants, such as
Ci. sinensis (27) [
18], and all 191
UGTs in
Populus trichocarpa were duplicated from each other [
3]. This may indicate that
BnUGTs in ramie have different biological functions than in other plants. These genomic features provide a foundation for further comparative studies aimed at elucidating the evolutionary dynamics of
UGT genes in plants.
Monolignol glycosylation is essential for lignin biosynthesis and influences fiber quality and plant cell wall integrity. The lignin content was upregulated in the phloem, xylem, and leaf during the six developmental periods. Lignin accumulation was rapid after 60 days of emergence, and the lignin content reached the maximum value after 75 days of emergence in the phloem and leaf. This may be related to the functional requirements and physiological states of the phloem and leaf at different stages [
39]. The phloem and leaf may focus more on nutrient transport between 15 and 60 days after emergence. The lignin content gradually increases after 60 days of emergence due to the demand for mechanical protection; thus, the best time for fiber harvest is before 75 days after emergence. Lignin content reaches its peak at 60 days after emergence, indicating rapid lignin accumulation in the xylem to establish a robust structure, facilitating mechanical support and water transport, which are necessary for rapid plant development [
1]. Lignin accumulation gradually slows down after 60 days of emergence. The expression patterns of 17 putative
BnUGTs were characterized in three tissues during six developmental stages, and the results indicated that
BnUGTs were grouped into 11, 7, and 4 classes in the phloem, leaf, and xylem based on their expression profiles, respectively.
Bnt05T007753.1 expression showed a significant positive correlation with lignin content in the phloem and leaf. In addition, the expression level of
Bnt05T007753.1 in the xylem reached up to 710 after 75 days of germination.
Bnt14T019888.1 expression showed a significant negative correlation with lignin content in the phloem but a significant positive correlation with lignin content in the leaf and xylem. These results align with those of previous studies on
Arabidopsis and poplar, where
UGT72 family members glycosylated monolignols to modulate lignin composition [
7,
10,
11]. In addition, the gene module PdeWRKY65-UGT75L28 negatively regulates petiole lignification by modulating the glycosylation of coniferyl aldehydes in poplars [
40]. We inferred that
Bnt05T007753.1 may catalyze the glycosylation of caffeic acid [
31] and subsequently participate in lignification, and
Bnt14T019888.1 simultaneously catalyzes the glycosylation of lignin monomers [
32] and precursors (coniferyl aldehyde, sinapyl aldehyde, and sinapic acid) [
10,
33], thus reducing the amount of precursor material needed for lignin synthesis and consequently hampering lignin synthesis. Therefore, based on the phylogenetic tree, we further confirmed that
Bnt05T007753.1 and
Bnt14T019888.1 may play key roles in regulating lignin glycosylation during ramie growth.
Flavonoid glycosylation enhances solubility and stability and contributes to the medicinal properties of ramie [
41]. The flavonoid content in the phloem and leaf during the six developmental periods was upregulated but showed no significant change from 15 to 60 days after germination. The flavonoid content significantly increased after 60 days of germination, which may be due to the rapid growth of seedlings as a result of consuming precursor substances, leading to flavonoid accumulation. However, the flavonoid content in the xylem was first downregulated and then upregulated during the six developmental periods. It is possible that the flavonoid was consumed during the initial growth [
42]. The negative correlation between
Bnt04T006105.1/
Bnt09T014140.1 expression and flavonoid content in leaves suggests complex regulatory mechanisms, possibly involving substrate competition or feedback inhibition, as observed in citrus flavonoid glycosyltransferases [
16,
18]. In addition, the evolutionary relationship suggests that
Bnt14T019888.1 and
Bnt06T010117.1 may participate in flavonoid and monolignol metabolism. We found that the expression of
Bnt06T010117.1 showed a significant positive correlation with both lignin and flavonoid content in the xylem. In the leaf and xylem,
Bnt14T019888.1 expression showed a significant positive correlation with both lignin and flavonoid content, respectively. Therefore,
Bnt14T019888.1 and
Bnt06T010117.1 may have the function of both
UGT75 and
UGT84 in
A. thaliana. Specifically, the UGT glycosylates anthocyanin [
34] and catalyzes the formation of cinnamic acid and hydroxycinnamic acids (
p-coumaric acid, caffeic acid, ferulic acid, and sinapic acid) of lignin precursor glucose esters [
35,
36].
Glycosylation is a common modification in the synthesis of plant secondary metabolites and plays a critical role in normal plant growth and stress responses as an evolutionary mechanism [
43,
44]. This study revealed that
BnUGT genes respond dynamically to Cd stress.
Bnt04T006105.1 (Group P) was upregulated in the stems and leaves under Cd treatment, thus suggesting its role in detoxification or stress signaling, similar to UGTs in
P. trichocarpa that mitigate abiotic stress [
3]. Citrus flavonoids are secondary metabolites that play crucial roles in the response to biotic and abiotic stresses, such as pathogen defense [
38,
45] and stress tolerance, and they have medicinal properties [
18]. In the stem, the expression of
Bnt06T010117.1 increased 106-fold under 20 mg/L Cd treatment, and the expression of
Bnt06T010117.1 showed a significant positive correlation with both lignin and flavonoid content. Therefore, we inferred that
Bnt06T010117.1 may play a role in the response to stress by regulating lignin and flavonoid glycosylation in ramie. The presence of stress-responsive
cis-elements, such as ABRE and TC-rich repeats, in
BnUGT promoters further supported their involvement in stress adaptation. Additionally,
BnUGTs in Group M (
Bnt02T003773.4) and P were linked to hormone glycosylation (e.g., cytokinins) [
24], which may influence growth and stress responses. For example,
Bnt02T003773.4 expression in the stem under 20 mg/L Cd treatment was much higher than that under other Cd concentrations.
Four candidate BnUGTs (Bnt05T007753.1, Bnt14T019888.1, Bnt06T010117.1, and Bnt04T006105.1) were screened from the genome of ramie in this study. However, further enzymatic assays and functional validation of these four candidate BnUGTs are needed to further confirm their roles in lignin/flavonoid glycosylation and stress responses.