Genome-Wide Identification of the HD-ZIP Transcription Factor Family in Maize and Functional Analysis of the Role of ZmHD-ZIP23 in Seed Size
Abstract
1. Introduction
2. Results
2.1. Identification of ZmHD-ZIP Family Members and Physicochemical Property Analysis
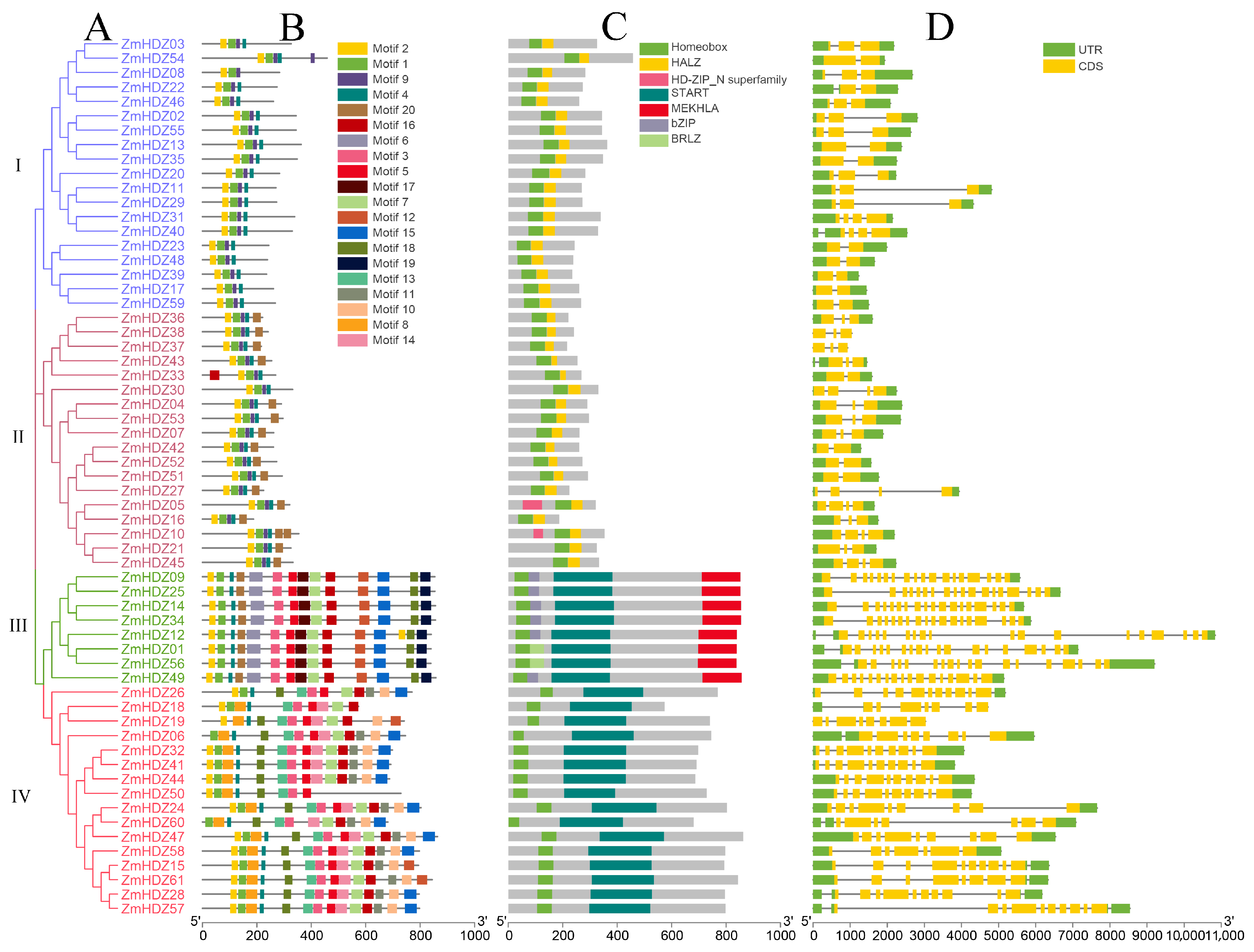
2.2. Analysis of Conserved Motifs, Domains, and Gene Structure of ZmHD-ZIP Family Members
2.3. Phylogenetic Analysis of ZmHD-ZIP Family Members
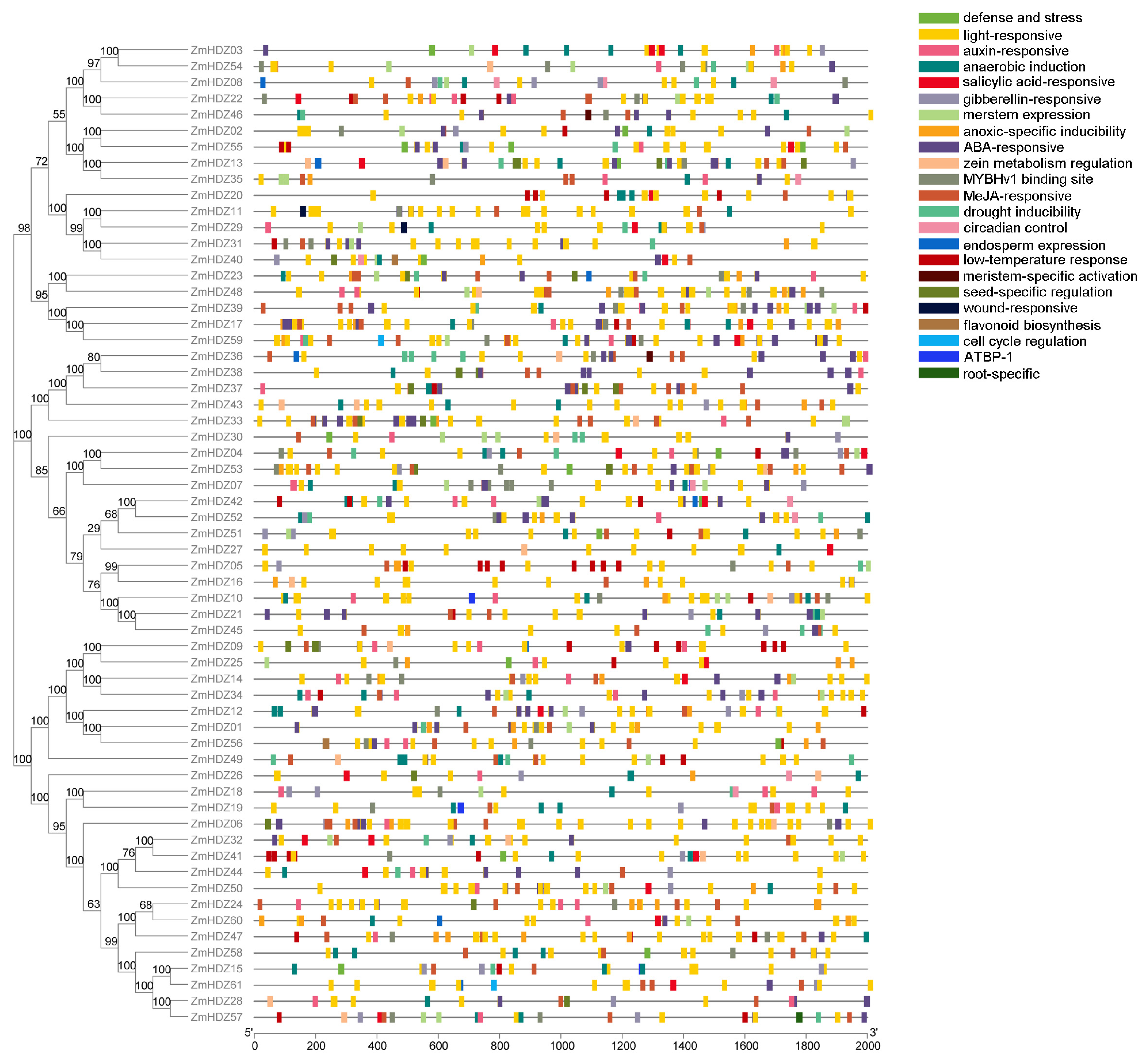
2.4. Cis-Acting Element Analysis of ZmHD-ZIP Genes
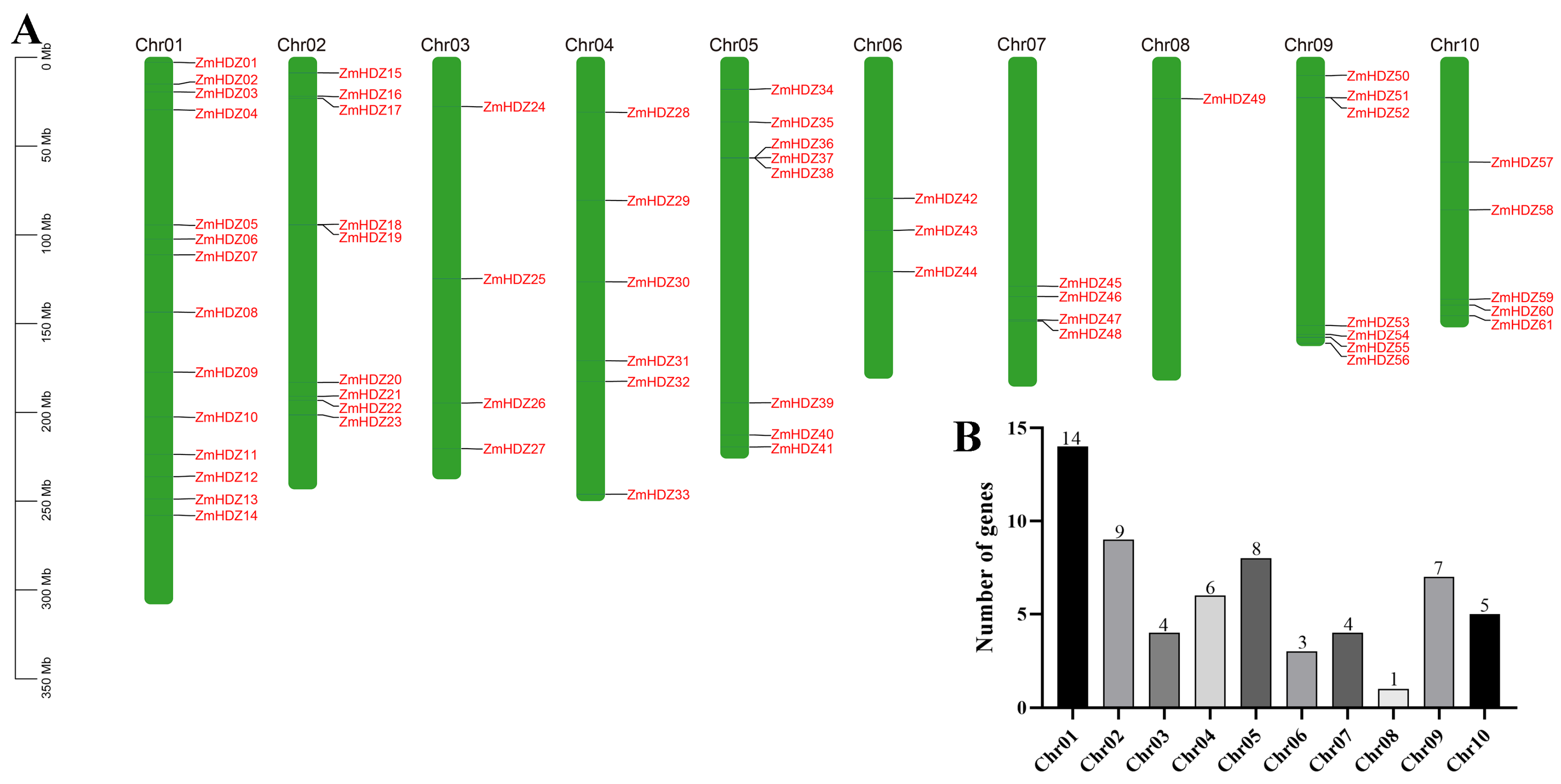
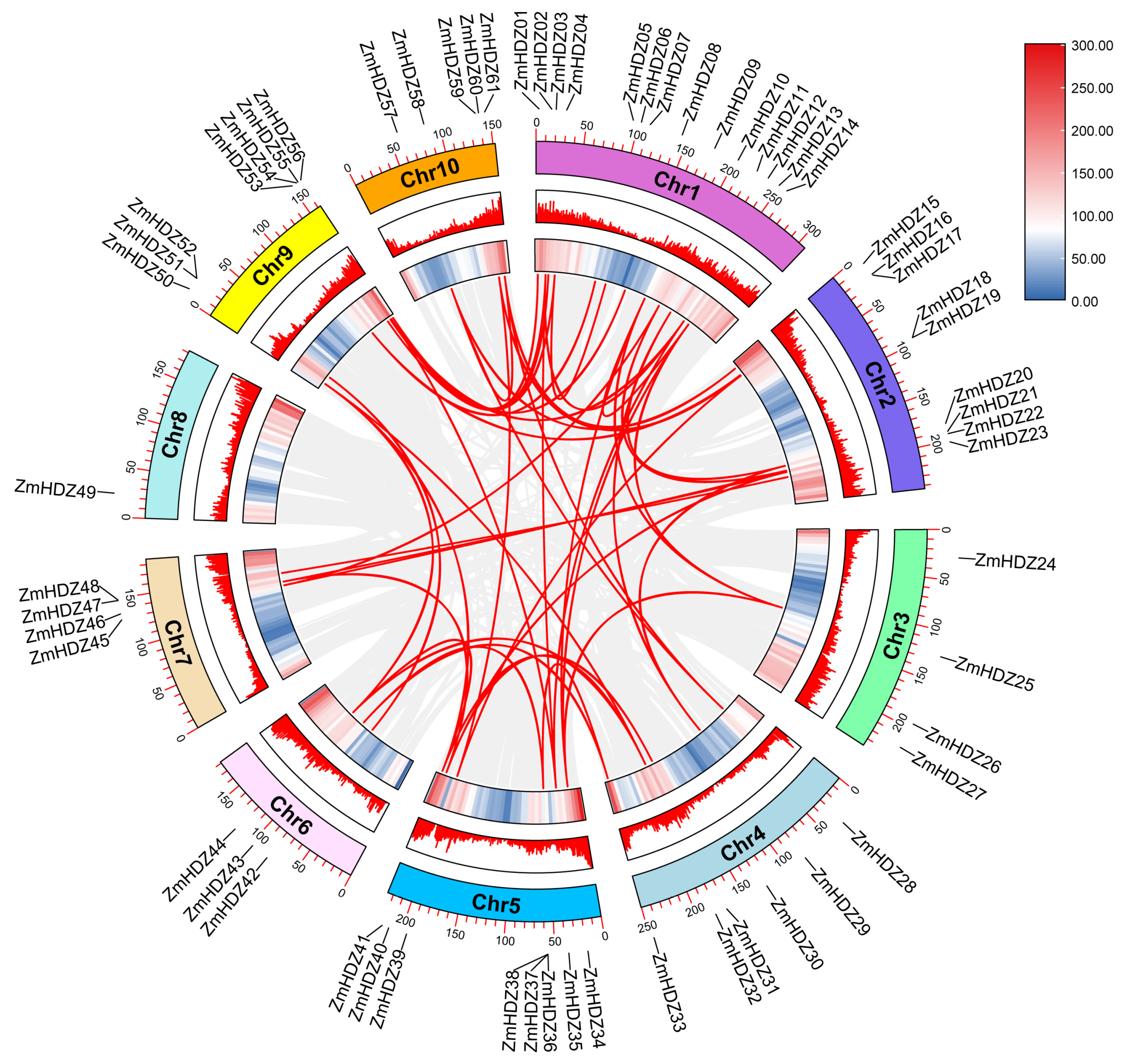
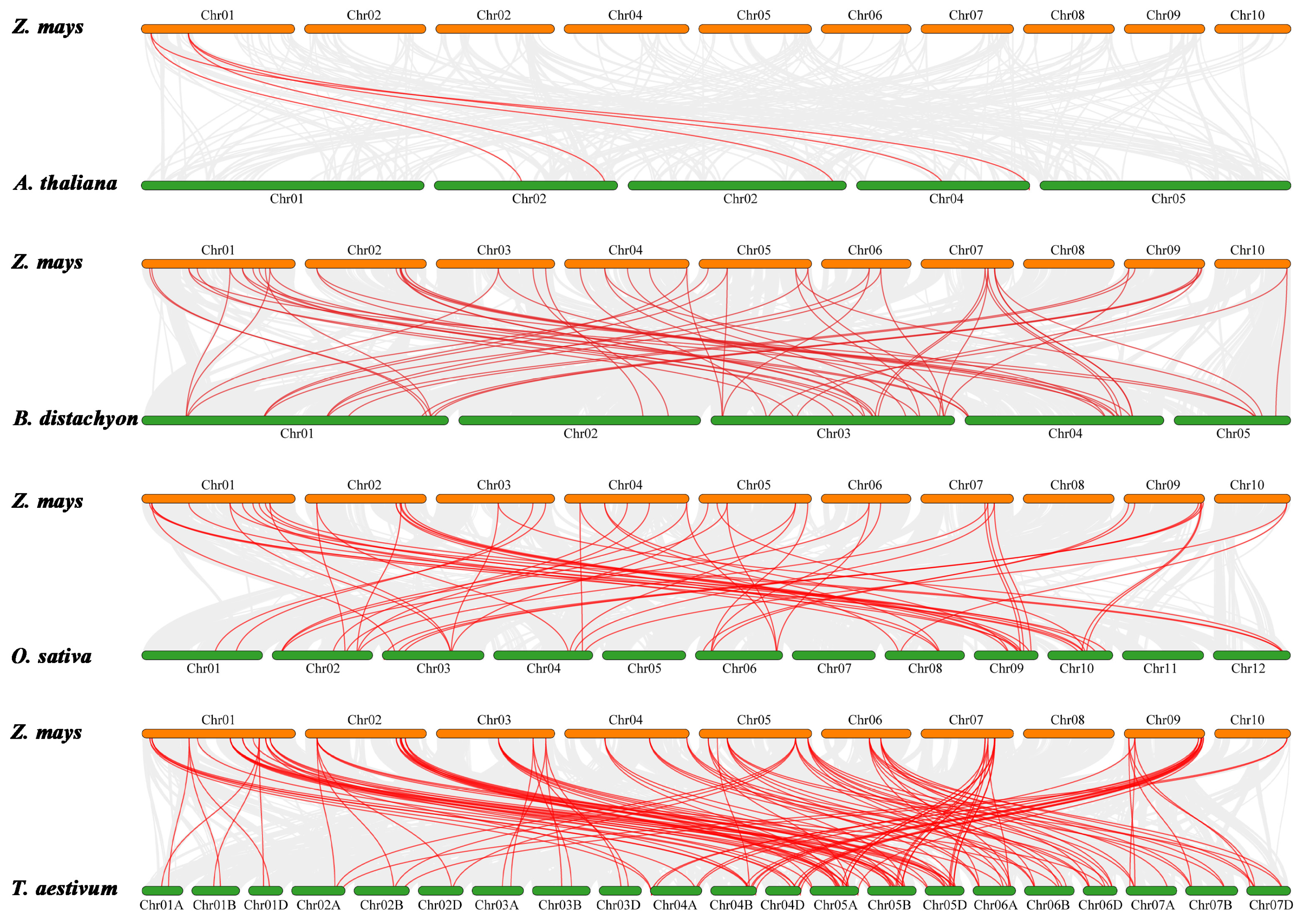
2.5. Chromosome Localization and Collinearity Analysis of ZmHD-ZIP Genes
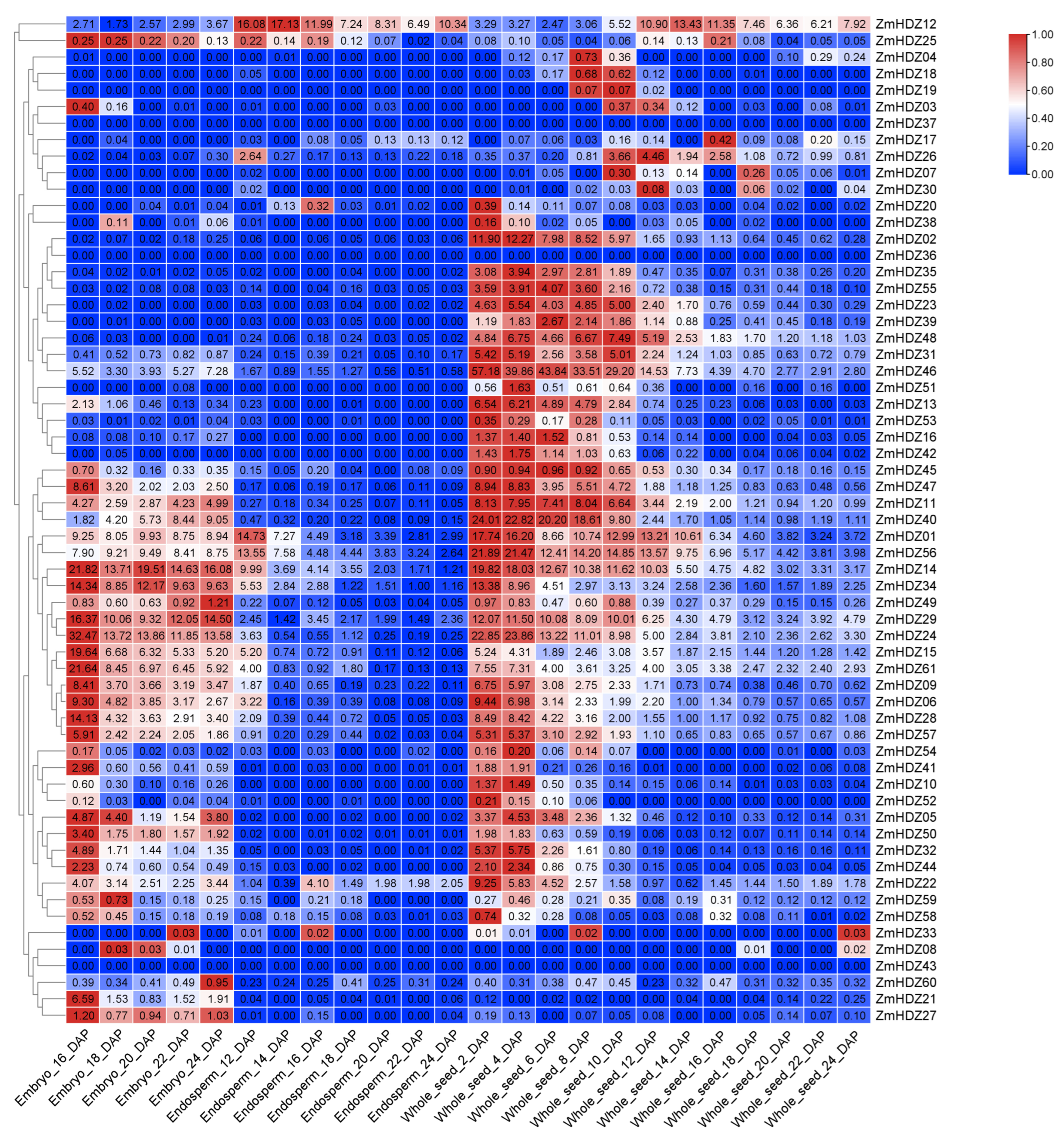
2.6. Expression Pattern Analysis of ZmHD-ZIP Family Members
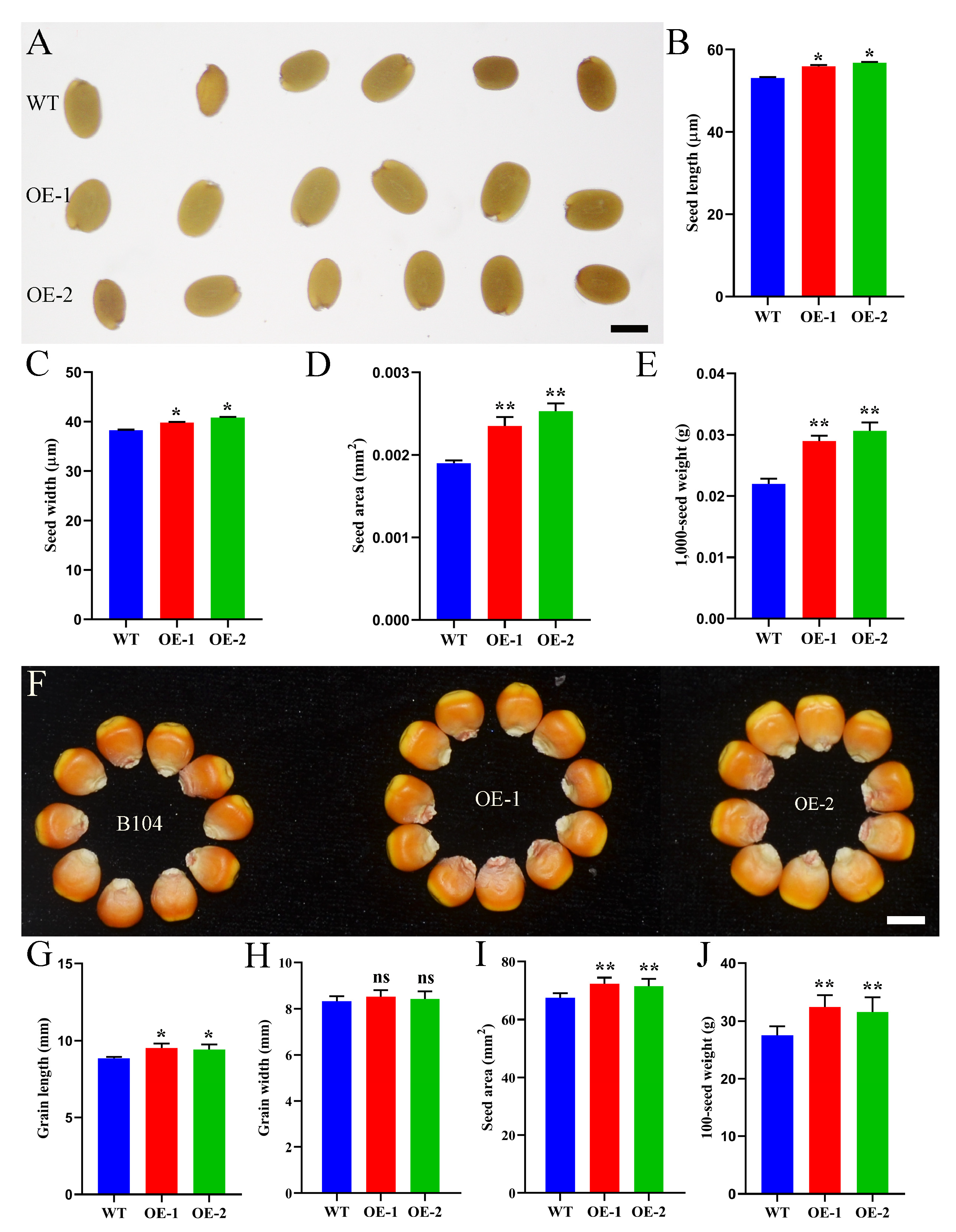
2.7. ZmHD-ZIP23 Positively Regulates Seed Size
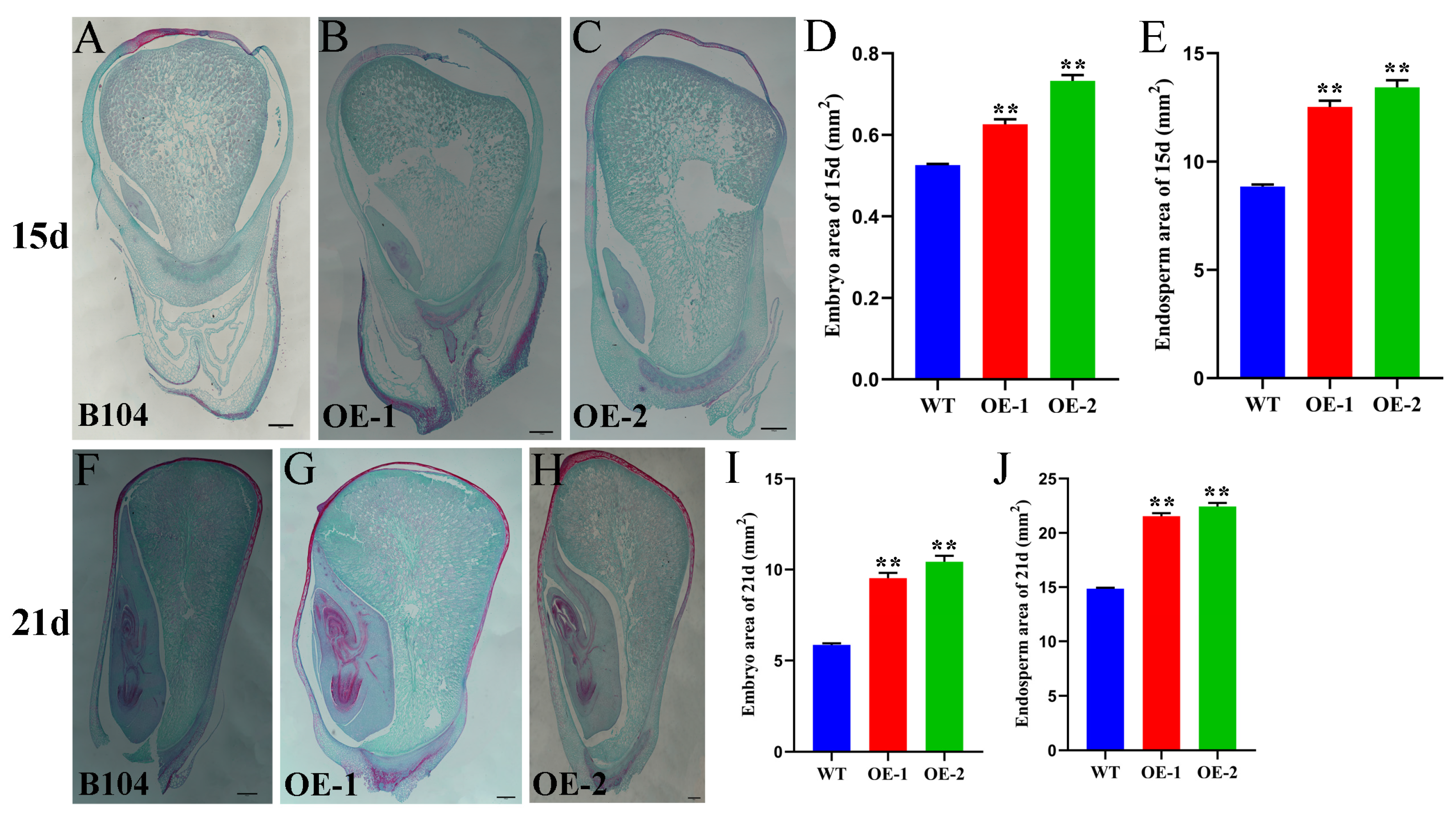
2.8. ZmHD-ZIP23-Overexpressing Maize Has Bigger Embryo and Endosperm
3. Discussion
3.1. Evolutionary and Structural Conservation of the ZmHD-ZIP Family
3.2. Subfamily-Specific Motifs and Functional Implications
3.3. Cis-Elements and Expression Patterns Suggest Multifunctionality
3.4. ZmHD-ZIP23 as a Key Regulator of Seed Size and Yield
3.5. Limitations and Prospects
4. Materials and Methods
4.1. Identification and Analysis of the Physiochemical Properties of the ZmHD-ZIP Family Members
4.2. Analysis of Conserved Motifs, Domains, and Gene Structure
4.3. Phylogenetic Analysis of ZmHD-ZIP Genes
4.4. Cis-Acting Element Analysis of ZmHD-ZIP Genes
4.5. Chromosomal Localization and Synteny Analysis of ZmHD-ZIP Genes
4.6. Expression Pattern Analysis of ZmHD-ZIP Genes
4.7. Vector Construction
4.8. Generation of Transgenic Arabidopsis
4.9. Generation of Transgenic Maize
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef]
- Roodbarkelari, F.; Groot, E.P. Regulatory function of homeodomain-leucine zipper (HD-ZIP) family proteins during embryogenesis. New Phytol. 2017, 213, 95–104. [Google Scholar] [CrossRef]
- Wu, M.; Bian, X.; Huang, B.; Du, Y.; Hu, S.; Wang, Y.; Shen, J.; Wu, S. HD-Zip proteins modify floral structures for self-pollination in tomato. Science 2024, 384, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J. Homeo boxes in the study of development. Science 1987, 236, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Burglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [PubMed]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A Core Regulatory Pathway Controlling Rice Tiller Angle Mediated by the LAZY1-Dependent Asymmetric Distribution of Auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef]
- Hu, Y.; Li, S.; Fan, X.; Song, S.; Zhou, X.; Weng, X.; Xiao, J.; Li, X.; Xiong, L.; You, A.; et al. OsHOX1 and OsHOX28 Redundantly Shape Rice Tiller Angle by Reducing HSFA2D Expression and Auxin Content. Plant Physiol. 2020, 184, 1424–1437. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef]
- Whipple, C.J.; Kebrom, T.H.; Weber, A.L.; Yang, F.; Hall, D.; Meeley, R.; Schmidt, R.; Doebley, J.; Brutnell, T.P.; Jackson, D.P. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 2011, 108, E506–E512. [Google Scholar] [CrossRef]
- Wills, D.M.; Whipple, C.J.; Takuno, S.; Kursel, L.E.; Shannon, L.M.; Ross-Ibarra, J.; Doebley, J.F. From many, one: Genetic control of prolificacy during maize domestication. PLoS Genet. 2013, 9, e1003604. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Hu, Y.; Ma, Q.; Zhao, Y.; Zhou, D.X. Functional analysis of rice HOMEOBOX4 (Oshox4) gene reveals a negative function in gibberellin responses. Plant Mol. Biol. 2008, 66, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cheng, Z.; Liu, L.; Wang, M.; You, X.; Wang, J.; Zhang, F.; Zhou, C.; Zhang, Z.; Zhang, H.; et al. Small Grain and Dwarf 2, encoding an HD-Zip II family transcription factor, regulates plant development by modulating gibberellin biosynthesis in rice. Plant Sci. 2019, 288, 110208. [Google Scholar] [CrossRef]
- Cao, Y.; Dou, D.; Zhang, D.; Zheng, Y.; Ren, Z.; Su, H.; Sun, C.; Hu, X.; Bao, M.; Zhu, B.; et al. ZmDWF1 regulates leaf angle in maize. Plant Sci. 2022, 325, 111459. [Google Scholar] [CrossRef]
- Itoh, J.; Hibara, K.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Shen, A.; Xiong, W.; Sun, Q.L.; Luo, Q.; Song, T.; Li, Z.L.; Luan, W.J. Overexpression of OsHox32 Results in Pleiotropic Effects on Plant Type Architecture and Leaf Development in Rice. Rice 2016, 9, 46. [Google Scholar] [CrossRef]
- Fang, J.; Guo, T.; Xie, Z.; Chun, Y.; Zhao, J.; Peng, L.; Zafar, S.A.; Yuan, S.; Xiao, L.; Li, X. The URL1-ROC5-TPL2 transcriptional repressor complex represses the ACL1 gene to modulate leaf rolling in rice. Plant Physiol. 2021, 185, 1722–1744. [Google Scholar] [CrossRef]
- Zou, L.P.; Sun, X.H.; Zhang, Z.G.; Liu, P.; Wu, J.X.; Tian, C.J.; Qiu, J.L.; Lu, T.G. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 2011, 156, 1589–1602. [Google Scholar] [CrossRef]
- Sun, J.; Cui, X.; Teng, S.; Kunnong, Z.; Wang, Y.; Chen, Z.; Sun, X.; Wu, J.; Ai, P.; Quick, W.P.; et al. HD-ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice. Plant Biotechnol. J. 2020, 18, 2559–2572. [Google Scholar] [CrossRef] [PubMed]
- Koppolu, R.; Chen, S.; Schnurbusch, T. Evolution of inflorescence branch modifications in cereal crops. Curr. Opin. Plant Biol. 2022, 65, 102168. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Ma, L.; Sang, X.; Ling, Y.; Wang, Y.; Yu, P.; Zhuang, H.; Huang, J.; Wang, N.; et al. LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc. Natl. Acad. Sci. USA 2017, 114, 9984–9989. [Google Scholar] [CrossRef] [PubMed]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, H.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 2007, 104, 1424–1429. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, S.; Lundqvist, U.; Kakei, Y.; Thirulogachandar, V.; Suzuki, T.; Hori, K.; Wu, J.; Tagiri, A.; Rutten, T.; Koppolu, R.; et al. Extreme Suppression of Lateral Floret Development by a Single Amino Acid Change in the VRS1 Transcription Factor. Plant Physiol. 2017, 175, 1720–1731. [Google Scholar] [CrossRef]
- Gao, S.; Fang, J.; Xu, F.; Wang, W.; Chu, C. Rice HOX12 Regulates Panicle Exsertion by Directly Modulating the Expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 2016, 28, 680–695. [Google Scholar] [CrossRef]
- Shao, J.; Haider, I.; Xiong, L.; Zhu, X.; Hussain, R.M.F.; Overnas, E.; Meijer, A.H.; Zhang, G.; Wang, M.; Bouwmeester, H.J.; et al. Functional analysis of the HD-Zip transcription factor genes Oshox12 and Oshox14 in rice. PLoS ONE 2018, 13, e0199248. [Google Scholar] [CrossRef]
- Sakuma, S.; Golan, G.; Guo, Z.; Ogawa, T.; Tagiri, A.; Sugimoto, K.; Bernhardt, N.; Brassac, J.; Mascher, M.; Hensel, G.; et al. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proc. Natl. Acad. Sci. USA 2019, 116, 5182–5187. [Google Scholar] [CrossRef]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009, 59, 883–894. [Google Scholar] [CrossRef]
- Ingram, G.C.; Magnard, J.L.; Vergne, P.; Dumas, C.; Rogowsky, P.M. ZmOCL1, an HDGL2 family homeobox gene, is expressed in the outer cell layer throughout maize development. Plant Mol. Biol. 1999, 40, 343–354. [Google Scholar] [CrossRef]
- Khaled, A.S.; Vernoud, V.; Ingram, G.C.; Perez, P.; Sarda, X.; Rogowsky, P.M. Engrailed-ZmOCL1 fusions cause a transient reduction of kernel size in maize. Plant Mol. Biol. 2005, 58, 123–139. [Google Scholar] [CrossRef]
- Javelle, M.; Vernoud, V.; Depege-Fargeix, N.; Arnould, C.; Oursel, D.; Domergue, F.; Sarda, X.; Rogowsky, P.M. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 2010, 154, 273–286. [Google Scholar] [CrossRef]
- Depege-Fargeix, N.; Javelle, M.; Chambrier, P.; Frangne, N.; Gerentes, D.; Perez, P.; Rogowsky, P.M.; Vernoud, V. Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J. Exp. Bot. 2011, 62, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Hu, J.; Naqvi, S.; Zhang, Y.; Linze, L.; Iderawumi, A.M. Molecular communication network and its applications in crop sciences. Planta 2022, 255, 128. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide Association Studies in Maize: Praise and Stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef]
- Qiu, X.; Wang, G.; Abou-Elwafa, S.F.; Fu, J.; Liu, Z.; Zhang, P.; Xie, X.; Ku, L.; Ma, Y.; Guan, X.; et al. Genome-wide identification of HD-ZIP transcription factors in maize and their regulatory roles in promoting drought tolerance. Physiol. Mol. Biol. Plants 2022, 28, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nie, L.; Ma, J.; Zhou, B.; Han, X.; Cheng, J.; Lu, X.; Fan, Z.; Li, Y.; Cao, Y. Transcriptomic Variations and Network Hubs Controlling Seed Size and Weight During Maize Seed Development. Front. Plant Sci. 2022, 13, 828923. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, C.; Zhang, X.; Yang, Q.; Shao, R.; Lai, J.; Du, C. Highly interwoven communities of a gene regulatory network unveil topologically important genes for maize seed development. Plant J. 2017, 92, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, Z.; Unver, T.; Zhang, B. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2021, 29, 207–221. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pai, Q.; Yue, L.; Wu, X.; Liu, H.; Wang, W. Cytokinin regulates female gametophyte development by cell cycle modulation in Arabidopsis thaliana. Plant Sci. 2022, 324, 111419. [Google Scholar] [CrossRef] [PubMed]

| Genes | Gene ID | Amino Acids | MW (Da) | pI | Instability Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| ZmHD-ZIP01 | Zm00001eb000690 | 840 | 92,098.18 | 5.84 | 53.84 | −0.125 | Nucleus |
| ZmHD-ZIP02 | Zm00001eb005440 | 344 | 37,604.1 | 6.28 | 58.58 | −0.696 | Nucleus |
| ZmHD-ZIP03 | Zm00001eb006700 | 326 | 35,531.37 | 5.11 | 62.69 | −0.739 | Nucleus |
| ZmHD-ZIP04 | Zm00001eb009550 | 290 | 30,707.68 | 8.69 | 54.27 | −0.492 | Nucleus |
| ZmHD-ZIP05 | Zm00001eb023620 | 321 | 34,828.19 | 8.86 | 63.2 | −0.69 | Nucleus |
| ZmHD-ZIP06 | Zm00001eb024680 | 746 | 80,347.87 | 5.95 | 48.17 | −0.188 | Nucleus |
| ZmHD-ZIP07 | Zm00001eb025650 | 262 | 28,241.88 | 8.43 | 48.86 | −0.508 | Nucleus |
| ZmHD-ZIP08 | Zm00001eb027860 | 283 | 30,941.65 | 4.75 | 59.27 | −0.71 | Nucleus |
| ZmHD-ZIP09 | Zm00001eb031670 | 854 | 92,572.62 | 6.54 | 49.04 | −0.125 | Nucleus |
| ZmHD-ZIP10 | Zm00001eb037670 | 354 | 36,671.05 | 8.16 | 56.32 | −0.369 | Nucleus |
| ZmHD-ZIP11 | Zm00001eb042400 | 270 | 29,994.23 | 5 | 56.95 | −0.665 | Nucleus |
| ZmHD-ZIP12 | Zm00001eb045630 | 841 | 92,210.07 | 5.83 | 48.9 | −0.17 | Nucleus |
| ZmHD-ZIP13 | Zm00001eb048440 | 363 | 38,897.15 | 5.74 | 70.68 | −0.666 | Nucleus |
| ZmHD-ZIP14 | Zm00001eb050660 | 856 | 93,167.97 | 6.16 | 47.74 | −0.188 | Nucleus |
| ZmHD-ZIP15 | Zm00001eb070050 | 794 | 85,194.04 | 5.52 | 42.77 | −0.25 | Nucleus |
| ZmHD-ZIP16 | Zm00001eb074830 | 187 | 20,556.37 | 9.06 | 61.44 | −0.639 | Nucleus |
| ZmHD-ZIP17 | Zm00001eb075230 | 261 | 29,384.76 | 5 | 71.3 | −0.715 | Nucleus |
| ZmHD-ZIP18 | Zm00001eb087220 | 574 | 64,224.37 | 5.46 | 48 | −0.279 | Nucleus |
| ZmHD-ZIP19 | Zm00001eb087250 | 741 | 80,705.57 | 5.04 | 47.82 | −0.265 | Nucleus |
| ZmHD-ZIP20 | Zm00001eb098960 | 283 | 31,224.78 | 5.34 | 67.03 | −0.644 | Nucleus |
| ZmHD-ZIP21 | Zm00001eb100690 | 325 | 34,517.69 | 7.6 | 58.03 | −0.515 | Nucleus |
| ZmHD-ZIP22 | Zm00001eb101280 | 274 | 29,643.92 | 4.61 | 61.83 | −0.486 | Nucleus |
| ZmHD-ZIP23 | Zm00001eb103330 | 244 | 26,792.74 | 5.36 | 57.74 | −0.786 | Nucleus |
| ZmHD-ZIP24 | Zm00001eb126140 | 803 | 85,946.16 | 5.44 | 51.43 | −0.145 | Nucleus |
| ZmHD-ZIP25 | Zm00001eb136060 | 854 | 92,757.92 | 6.37 | 49.21 | −0.142 | Nucleus |
| ZmHD-ZIP26 | Zm00001eb151130 | 770 | 83,567.83 | 6.14 | 45.76 | −0.313 | Nucleus |
| ZmHD-ZIP27 | Zm00001eb158680 | 225 | 24,891.12 | 9.27 | 66.93 | −0.837 | Nucleus |
| ZmHD-ZIP28 | Zm00001eb171720 | 796 | 85,608.48 | 5.53 | 46 | −0.259 | Nucleus |
| ZmHD-ZIP29 | Zm00001eb179000 | 272 | 30,035.08 | 4.79 | 54.13 | −0.679 | Nucleus |
| ZmHD-ZIP30 | Zm00001eb182650 | 331 | 35,536.87 | 5.49 | 70.03 | −0.438 | Nucleus |
| ZmHD-ZIP31 | Zm00001eb189900 | 339 | 36,998.47 | 4.62 | 53.68 | −0.723 | Nucleus |
| ZmHD-ZIP32 | Zm00001eb193330 | 698 | 76,174.03 | 6.19 | 53.03 | −0.164 | Nucleus |
| ZmHD-ZIP33 | Zm00001eb208000 | 269 | 28,268.55 | 9.65 | 61.44 | −0.516 | Nucleus |
| ZmHD-ZIP34 | Zm00001eb218730 | 856 | 93,155.96 | 6.16 | 47.24 | −0.187 | Nucleus |
| ZmHD-ZIP35 | Zm00001eb223300 | 348 | 37,530.79 | 6.01 | 65.65 | −0.689 | Nucleus |
| ZmHD-ZIP36 | Zm00001eb226570 | 221 | 23,801.92 | 9.2 | 54.34 | −0.533 | Nucleus |
| ZmHD-ZIP37 | Zm00001eb226580 | 216 | 23,703.89 | 9.72 | 68.41 | −0.643 | Nucleus |
| ZmHD-ZIP38 | Zm00001eb226590 | 241 | 25,859.12 | 9.48 | 61.57 | −0.606 | Nucleus |
| ZmHD-ZIP39 | Zm00001eb248930 | 235 | 26,413.07 | 5.01 | 56.92 | −0.814 | Nucleus |
| ZmHD-ZIP40 | Zm00001eb253470 | 330 | 35,891.27 | 4.58 | 56.81 | −0.636 | Nucleus |
| ZmHD-ZIP41 | Zm00001eb256420 | 692 | 75,813.55 | 6.15 | 48.09 | −0.205 | Nucleus |
| ZmHD-ZIP42 | Zm00001eb270270 | 261 | 27,367.76 | 9.5 | 64.05 | −0.419 | Nucleus |
| ZmHD-ZIP43 | Zm00001eb273120 | 254 | 27,552.84 | 9.11 | 71.41 | −0.651 | Nucleus |
| ZmHD-ZIP44 | Zm00001eb278870 | 687 | 74,909.39 | 5.77 | 49.66 | −0.166 | Nucleus |
| ZmHD-ZIP45 | Zm00001eb314500 | 333 | 36,021.26 | 6.98 | 74.68 | −0.656 | Nucleus |
| ZmHD-ZIP46 | Zm00001eb315750 | 261 | 28,461.52 | 4.76 | 57.29 | −0.703 | Nucleus |
| ZmHD-ZIP47 | Zm00001eb319090 | 863 | 91,541.38 | 5.88 | 47.53 | −0.246 | Nucleus |
| ZmHD-ZIP48 | Zm00001eb319390 | 239 | 26,235.01 | 5.37 | 46.67 | −0.853 | Nucleus |
| ZmHD-ZIP49 | Zm00001eb337970 | 858 | 92,028.36 | 5.72 | 45.27 | 0.019 | Nucleus |
| ZmHD-ZIP50 | Zm00001eb373520 | 729 | 81,395.84 | 10.54 | 67.18 | −0.725 | Nucleus |
| ZmHD-ZIP51 | Zm00001eb377480 | 293 | 31,098.95 | 9.37 | 48.67 | −0.532 | Nucleus |
| ZmHD-ZIP52 | Zm00001eb377500 | 272 | 28,026.55 | 9.45 | 67.6 | −0.318 | Nucleus |
| ZmHD-ZIP53 | Zm00001eb399360 | 296 | 31,560.54 | 9.21 | 61.43 | −0.651 | Nucleus |
| ZmHD-ZIP54 | Zm00001eb401450 | 458 | 50,056.7 | 8.61 | 58.17 | −0.24 | Nucleus |
| ZmHD-ZIP55 | Zm00001eb402220 | 344 | 37,573.13 | 6.04 | 59.47 | −0.639 | Nucleus |
| ZmHD-ZIP56 | Zm00001eb404260 | 839 | 92,090.13 | 5.94 | 54.2 | −0.138 | Nucleus |
| ZmHD-ZIP57 | Zm00001eb413140 | 798 | 85,633.3 | 5.43 | 47.99 | −0.295 | Nucleus |
| ZmHD-ZIP58 | Zm00001eb416980 | 797 | 85,648.79 | 5.97 | 44.99 | −0.256 | Nucleus |
| ZmHD-ZIP59 | Zm00001eb427650 | 268 | 29,290.62 | 5.32 | 67.04 | −0.63 | Nucleus |
| ZmHD-ZIP60 | Zm00001eb428740 | 681 | 74,199.73 | 6.21 | 42.4 | 0.003 | Nucleus |
| ZmHD-ZIP61 | Zm00001eb431350 | 844 | 91,063.09 | 5.68 | 49.88 | −0.209 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhang, X.; Liu, M.; Jin, Y.; Pai, Q.; Wu, X.; Sun, D. Genome-Wide Identification of the HD-ZIP Transcription Factor Family in Maize and Functional Analysis of the Role of ZmHD-ZIP23 in Seed Size. Plants 2025, 14, 2477. https://doi.org/10.3390/plants14162477
Zhang J, Zhang X, Liu M, Jin Y, Pai Q, Wu X, Sun D. Genome-Wide Identification of the HD-ZIP Transcription Factor Family in Maize and Functional Analysis of the Role of ZmHD-ZIP23 in Seed Size. Plants. 2025; 14(16):2477. https://doi.org/10.3390/plants14162477
Chicago/Turabian StyleZhang, Jinghua, Xuan Zhang, Mengru Liu, Yichen Jin, Qiaofeng Pai, Xiaolin Wu, and Doudou Sun. 2025. "Genome-Wide Identification of the HD-ZIP Transcription Factor Family in Maize and Functional Analysis of the Role of ZmHD-ZIP23 in Seed Size" Plants 14, no. 16: 2477. https://doi.org/10.3390/plants14162477
APA StyleZhang, J., Zhang, X., Liu, M., Jin, Y., Pai, Q., Wu, X., & Sun, D. (2025). Genome-Wide Identification of the HD-ZIP Transcription Factor Family in Maize and Functional Analysis of the Role of ZmHD-ZIP23 in Seed Size. Plants, 14(16), 2477. https://doi.org/10.3390/plants14162477






