Integrated Transcriptomic and Metabolomic Analysis Reveals Nitrogen-Mediated Delay of Premature Leaf Senescence in Red Raspberry Leaves
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials
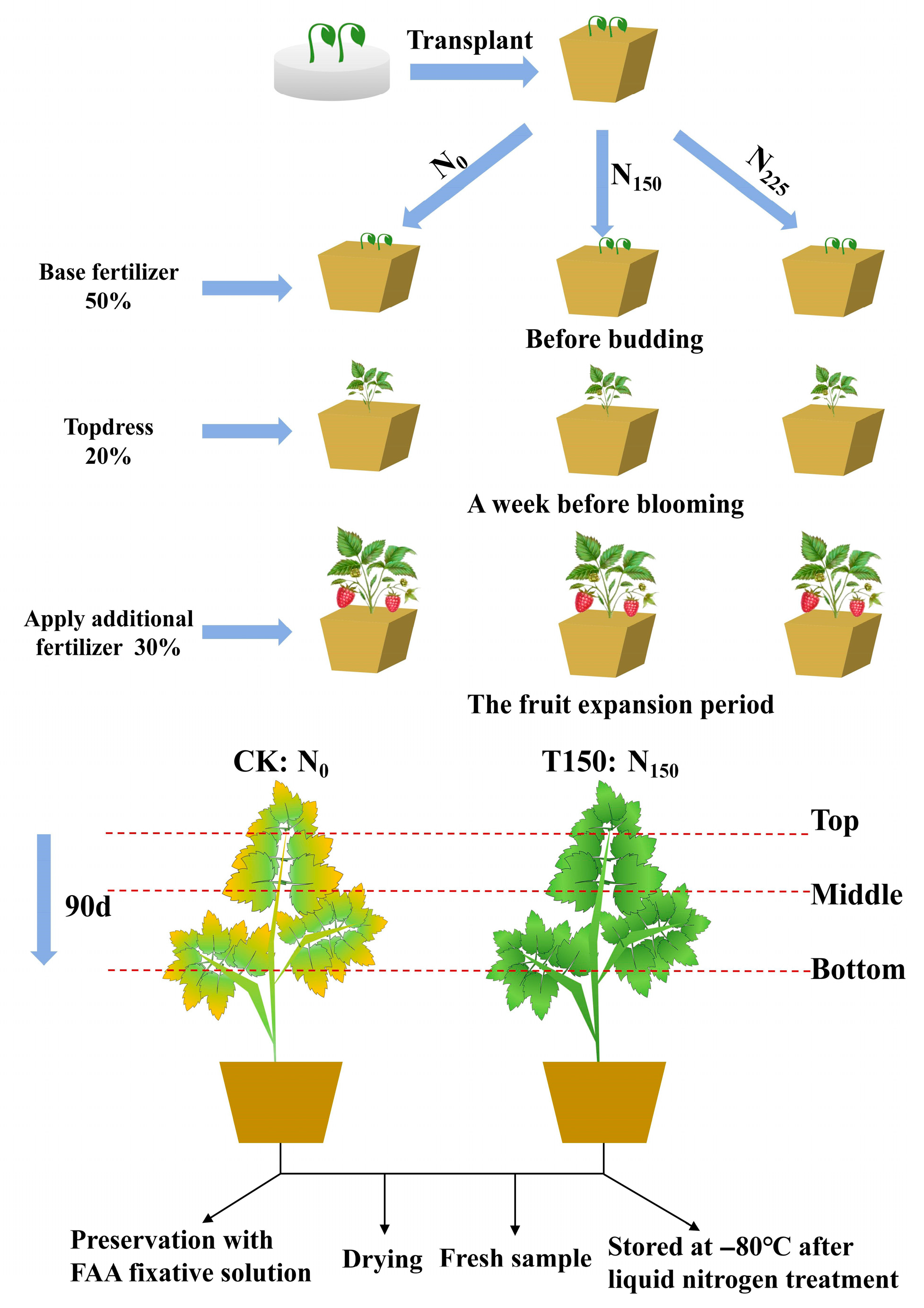
2.2. Experimental Design
2.3. Determination Indicators and Methods
2.3.1. Determination of Differences in Tree Growth and Physiological and Biochemical Indicators
Measurement of Tree Morphological Indicators
Determination of Physiological and Biochemical Indicators
2.3.2. Slice Production Method
2.3.3. Transcriptome Determination
RNA Extraction and Sequencing
Real-Time Quantitative PCR Validation
2.3.4. Metabolome Determination
Sample Extraction Method
Data Analysis Process
2.4. Data Processing and Analysis
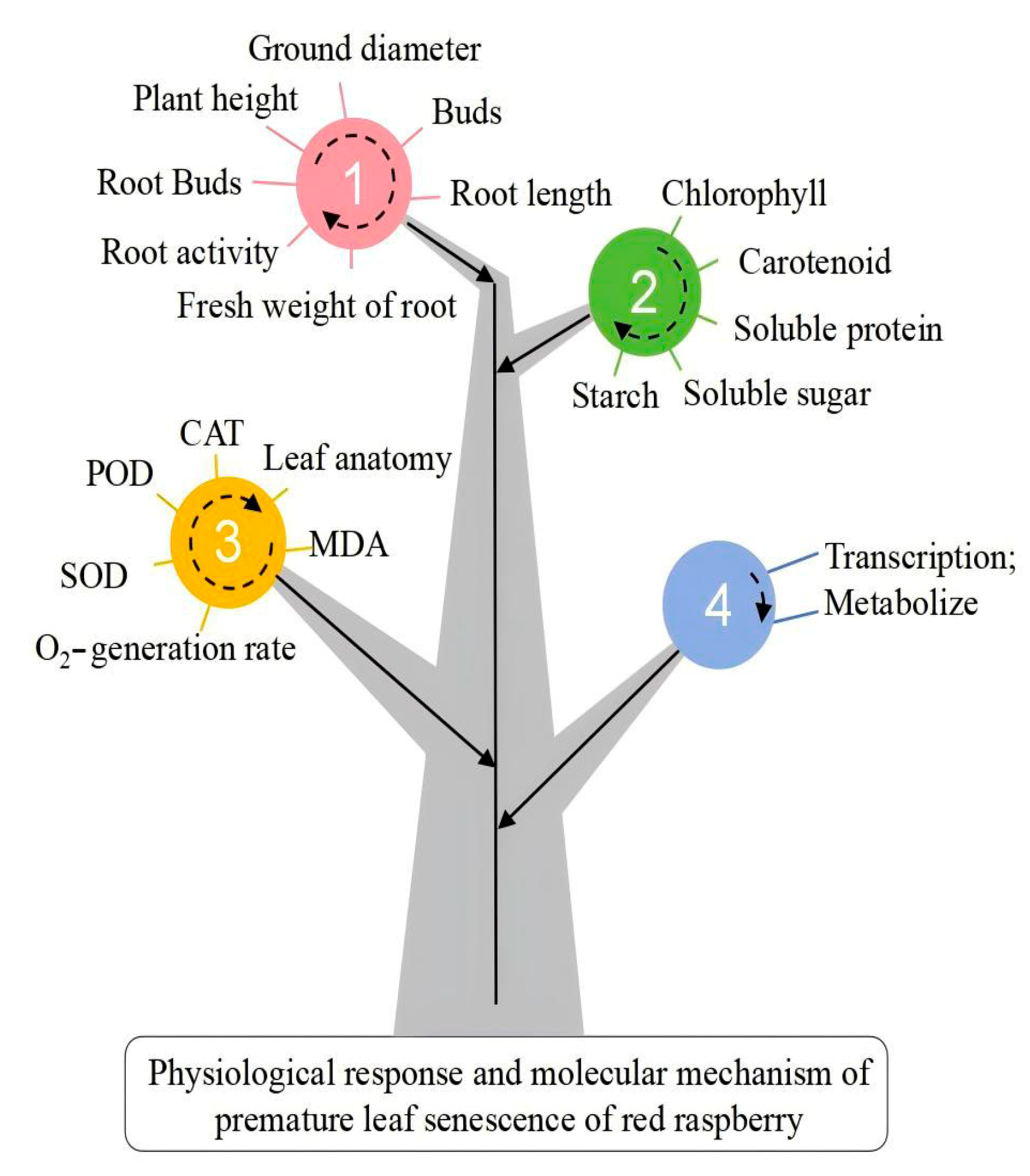
3. Results and Analysis
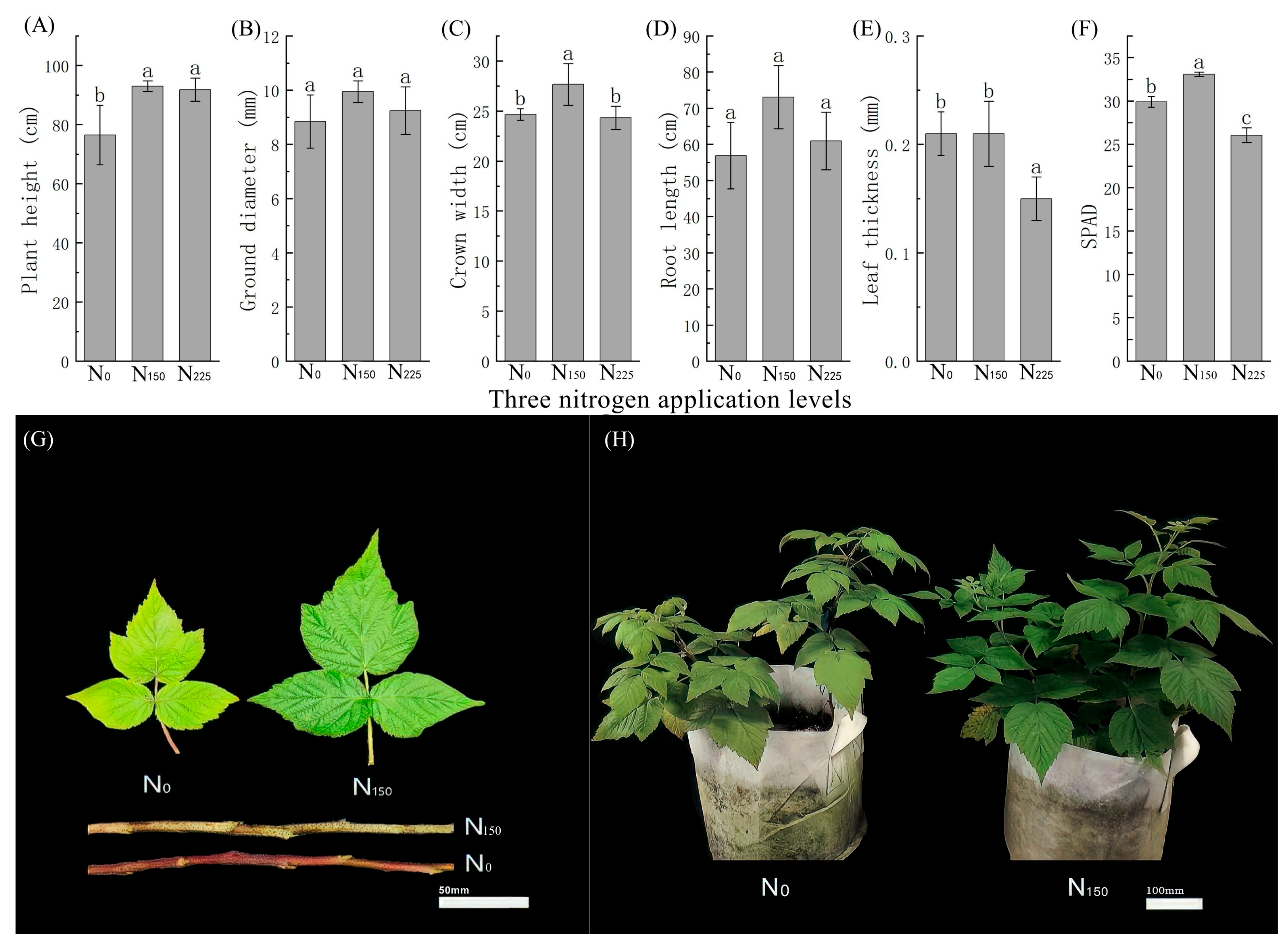
3.1. Growth of Red Raspberries at Three Nitrogen Application Levels
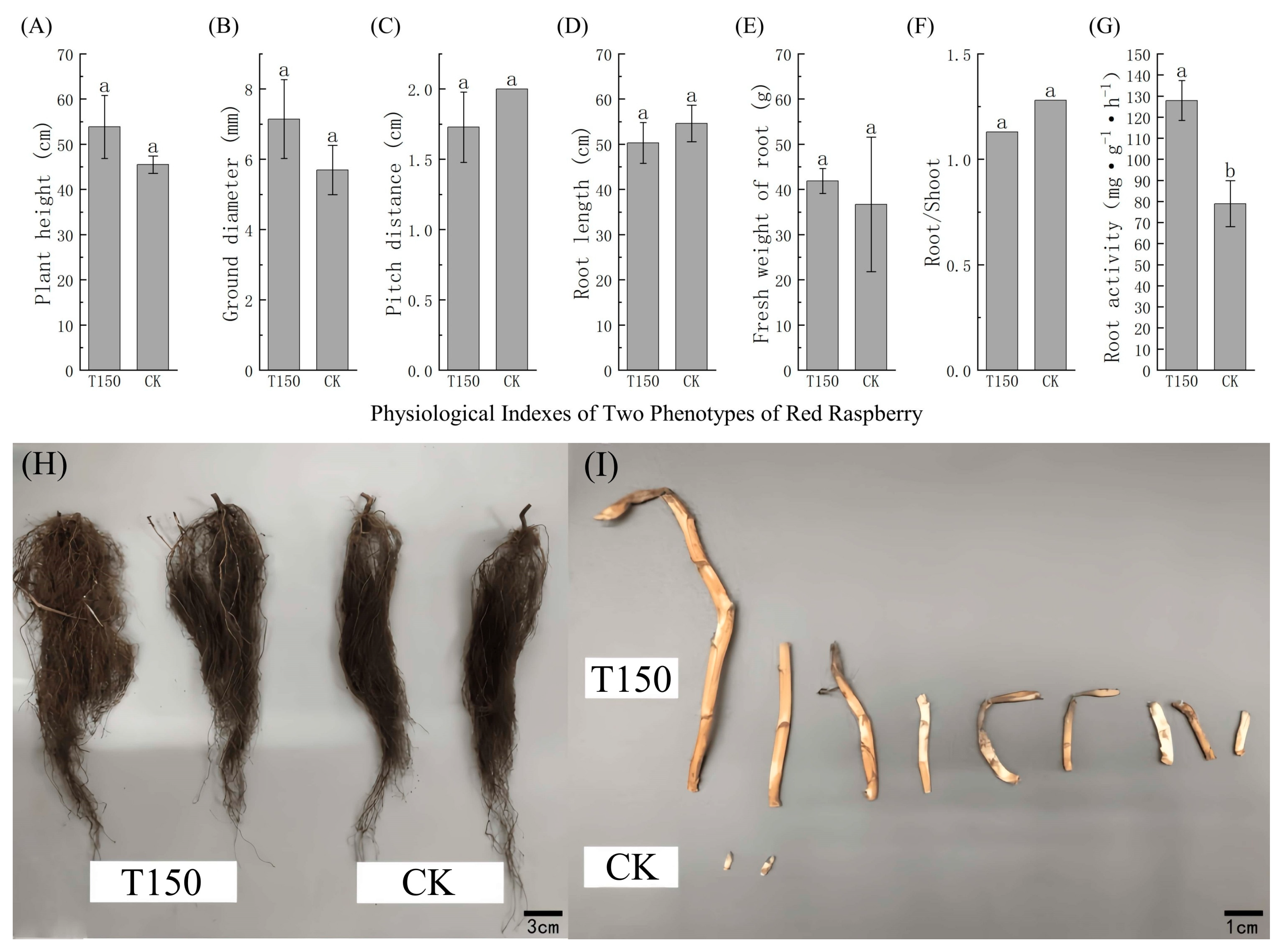
3.2. Differences in Phenotypes and Physiological and Biochemical Parameters Between T150 and CK Plants
3.2.1. Phenotypic Growth Differences Between T150 and CK
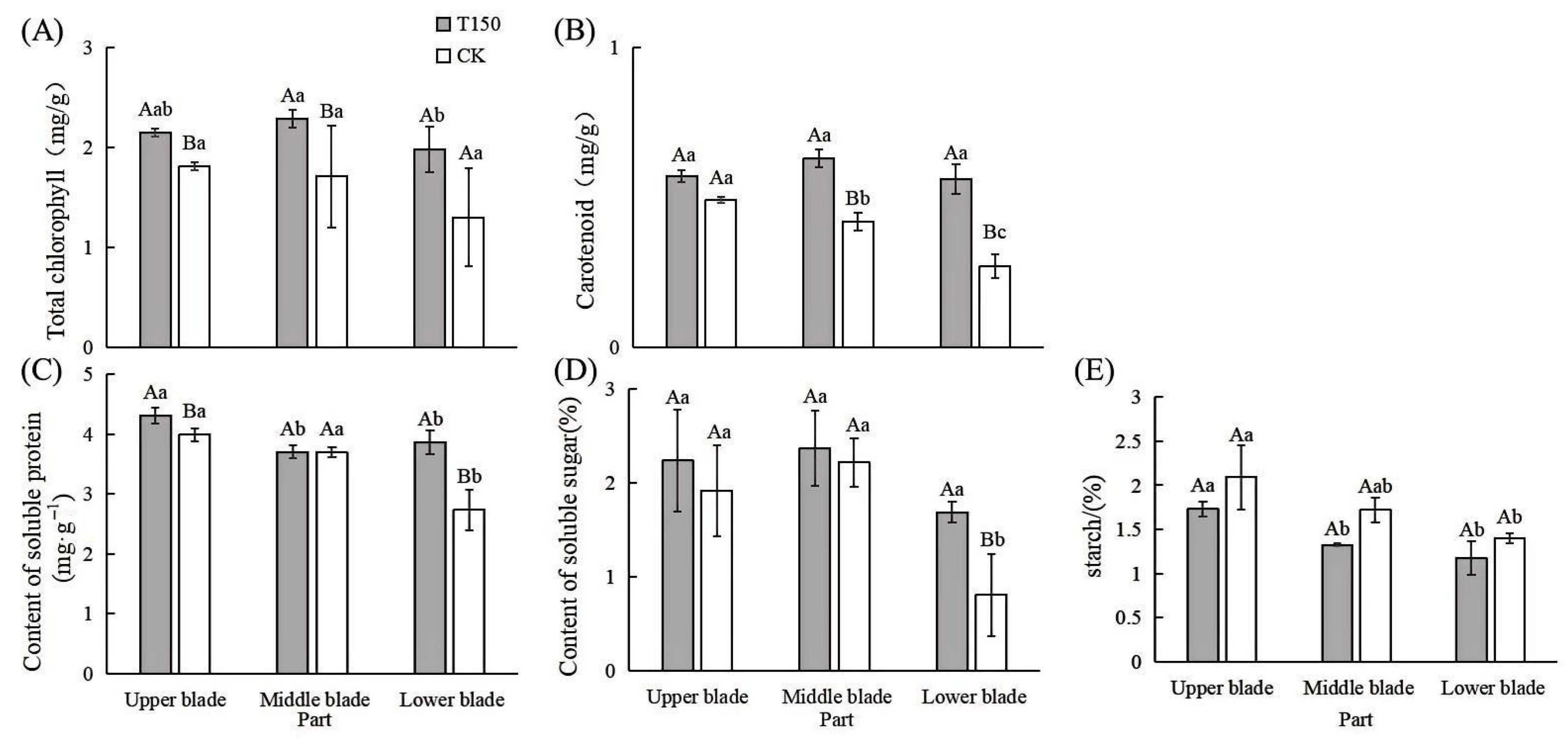
3.2.2. Differences in Photosynthesis and Osmotic Regulation Substances in Leaves of T150 and CK Plants
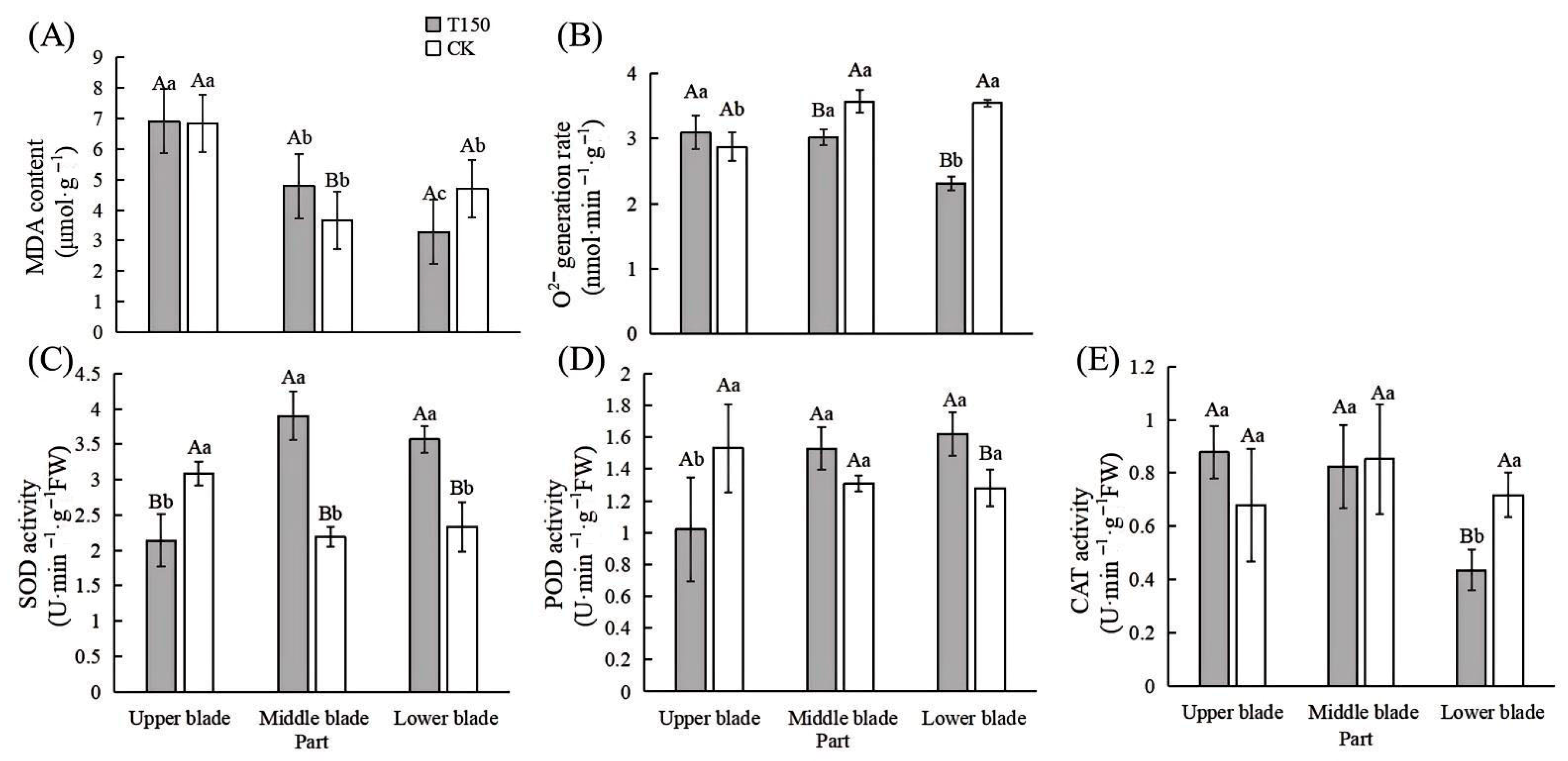
3.2.3. Changes in Membrane Lipid Peroxidation and Antioxidant Enzyme Activities in Leaves of T150 and CK Plants
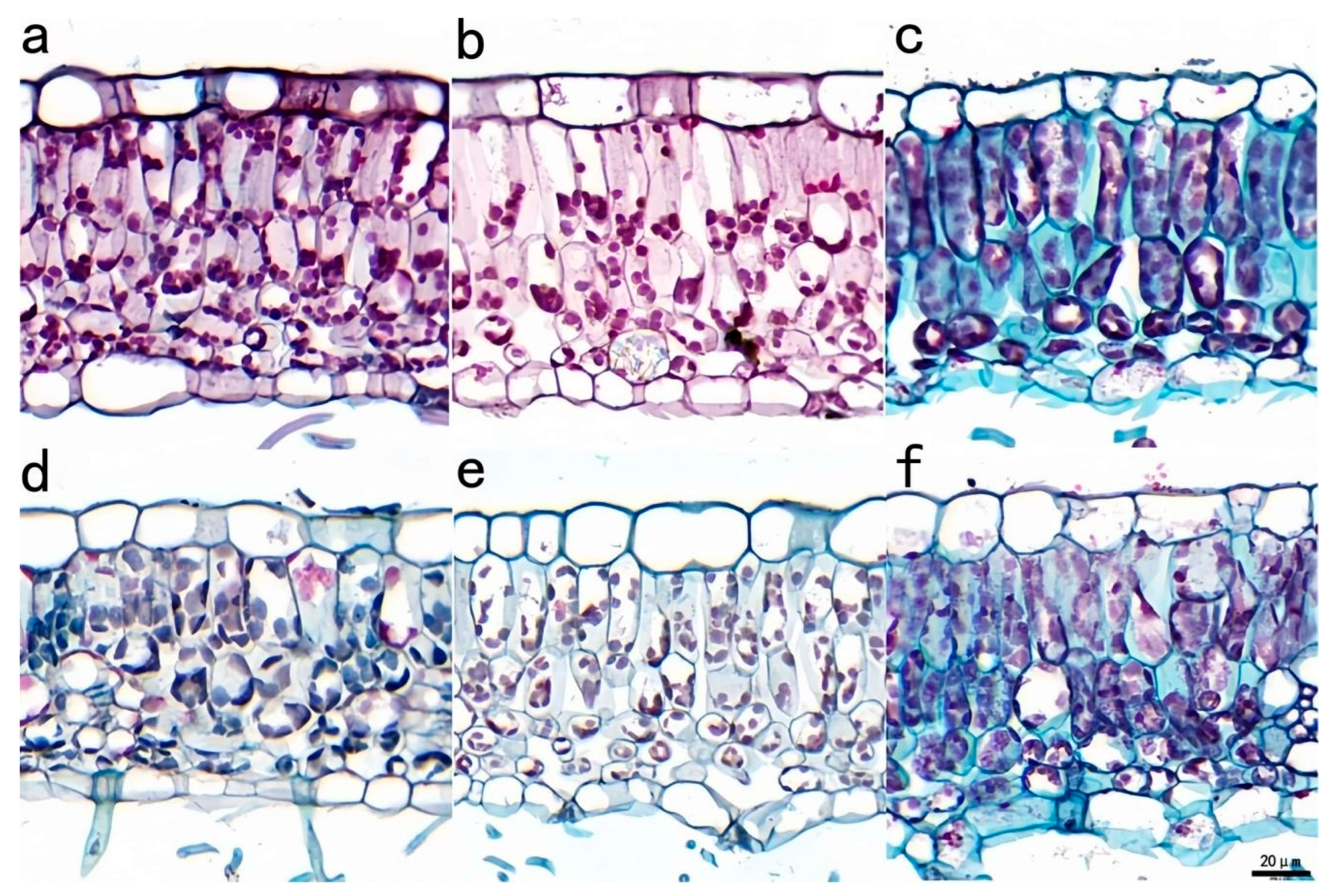
3.2.4. Differences in Leaf Anatomical Structure Between T150 and CK Plants
3.3. Transcriptome Data Analysis of Lower Leaves of T150 and CK Plants
3.3.1. Transcriptome Sequencing Data Results and Unigene Functional Annotation and Analysis
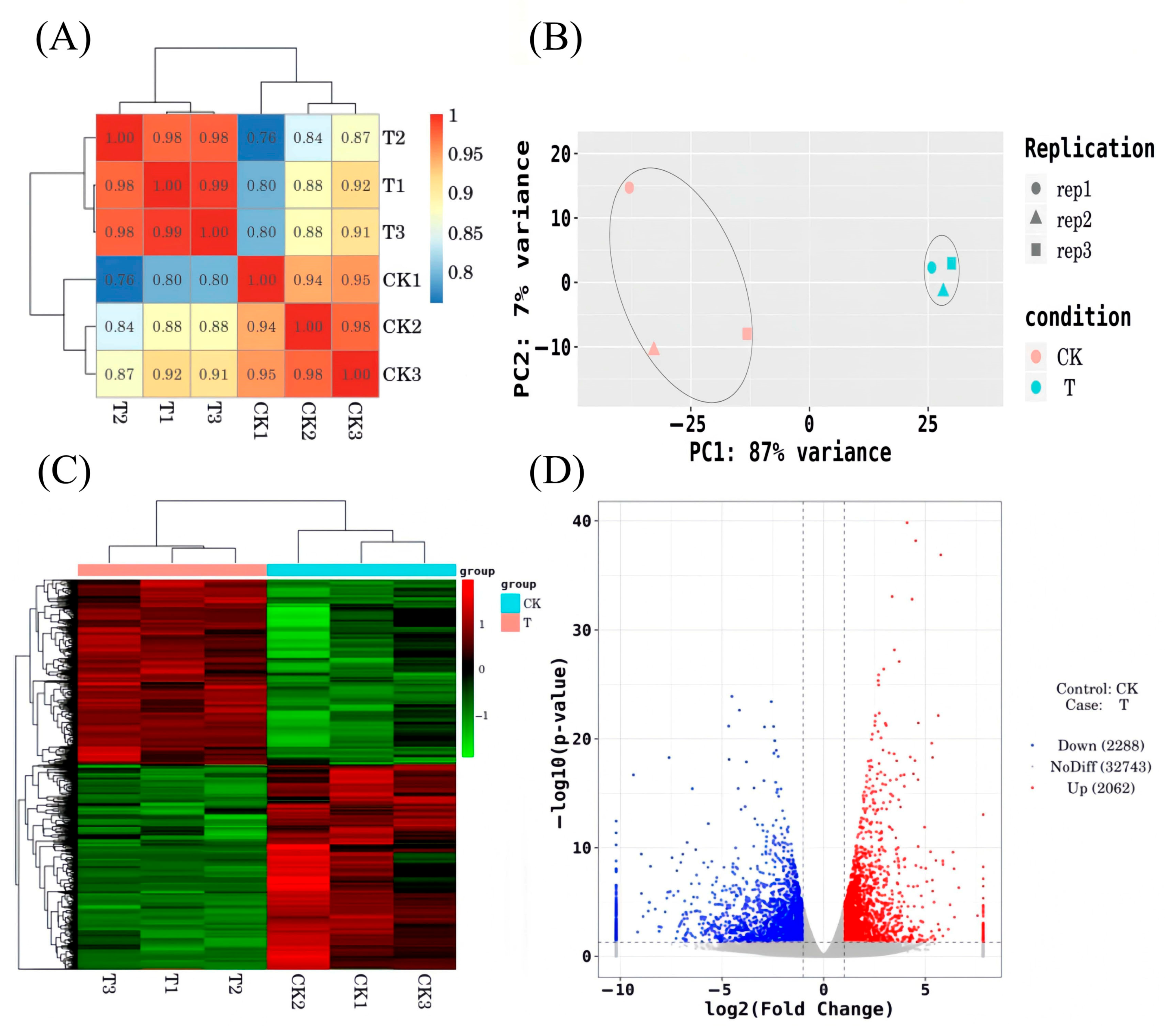
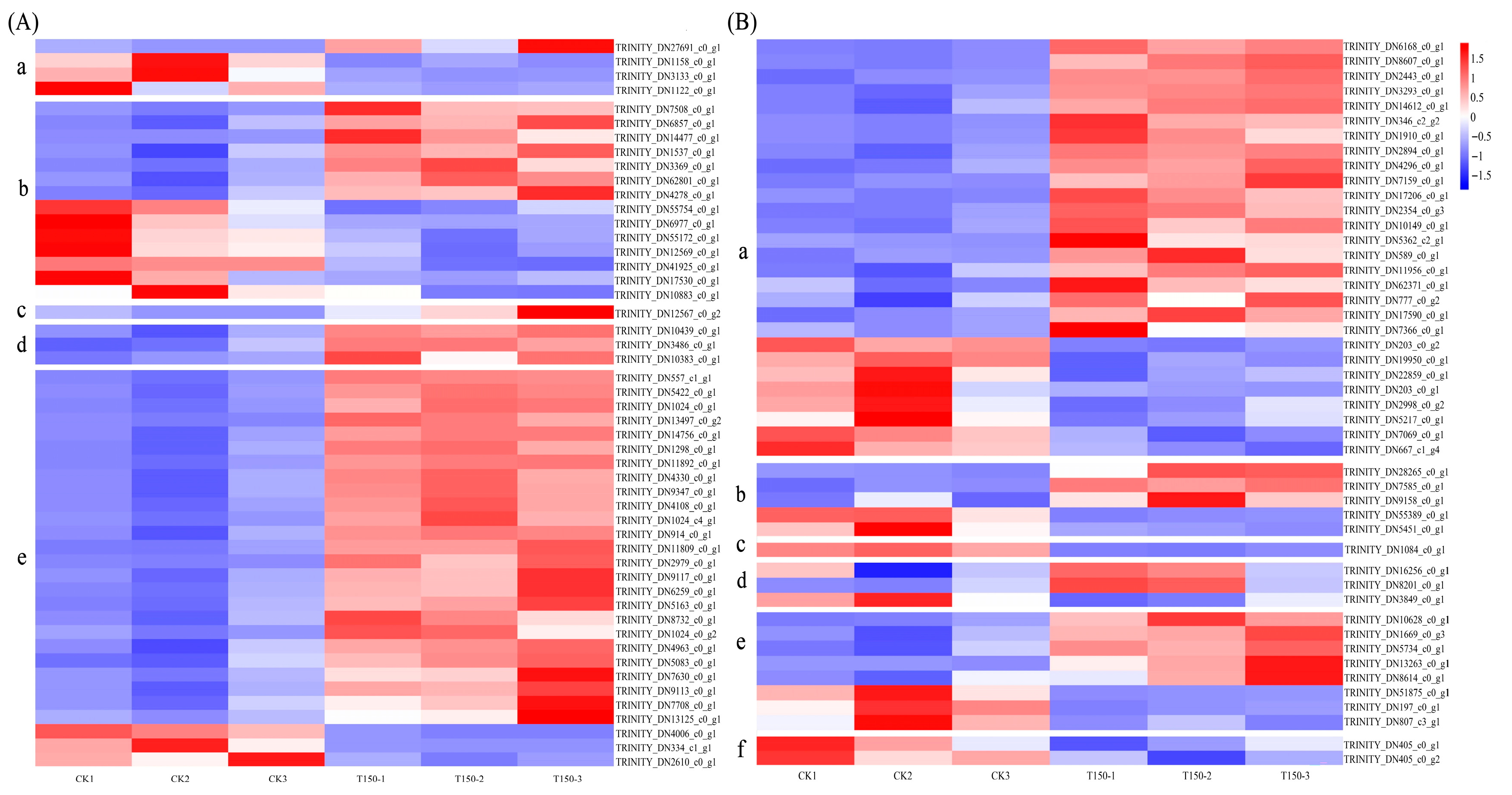
3.3.2. Analysis of Differentially Expressed Genes
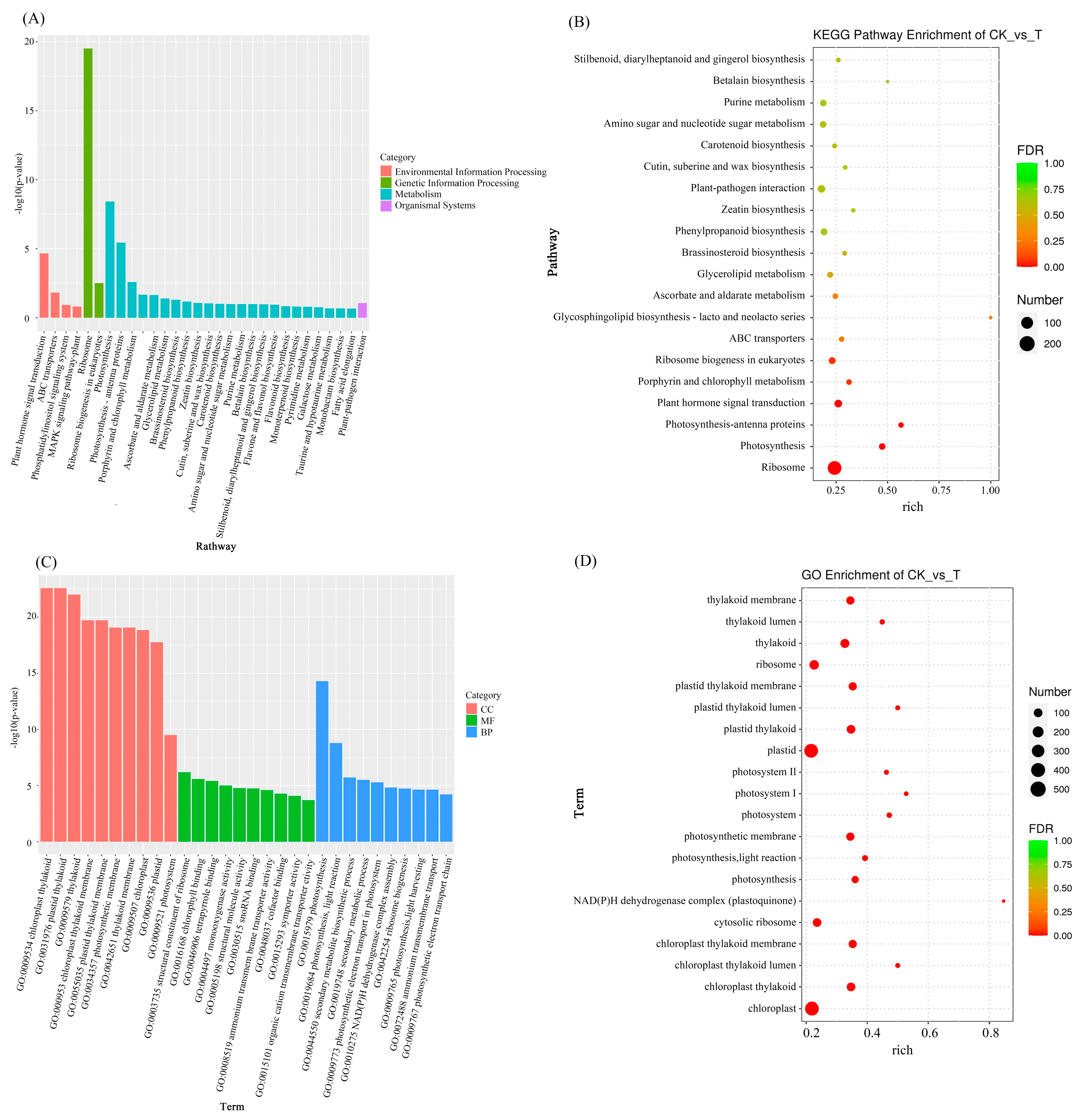
3.3.3. KEGG and GO Enrichment Analysis of Differentially Expressed Genes
3.3.4. Physiological and Biochemical Indicators and Hormone-Related Differentially Expressed Genes
3.4. Metabolome Data Analysis of Lower Leaves of T150 and CK Plants
3.4.1. Metabolite PCA and OPLS-DA Analysis
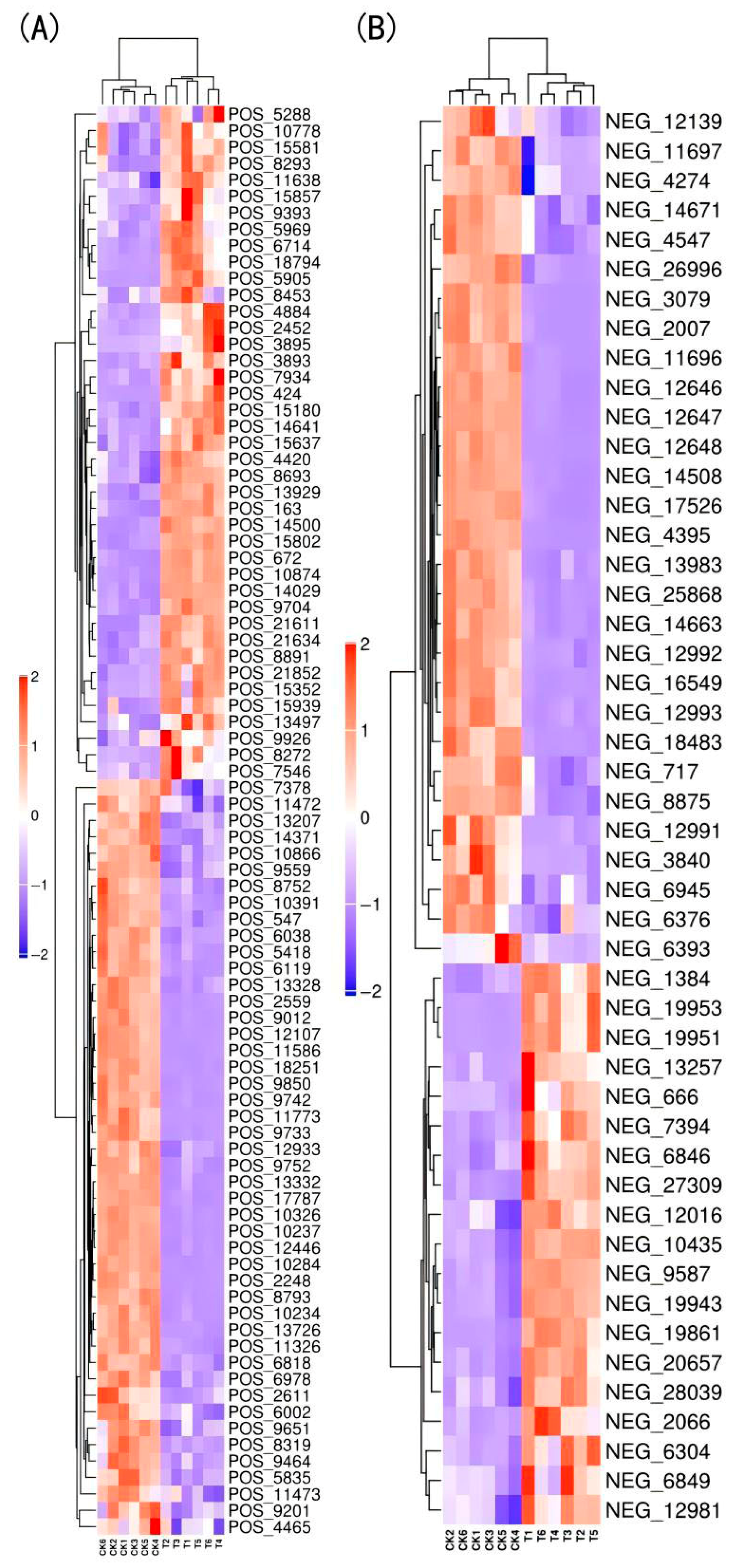
3.4.2. Screening of Differentially Expressed Metabolites (DEMs)
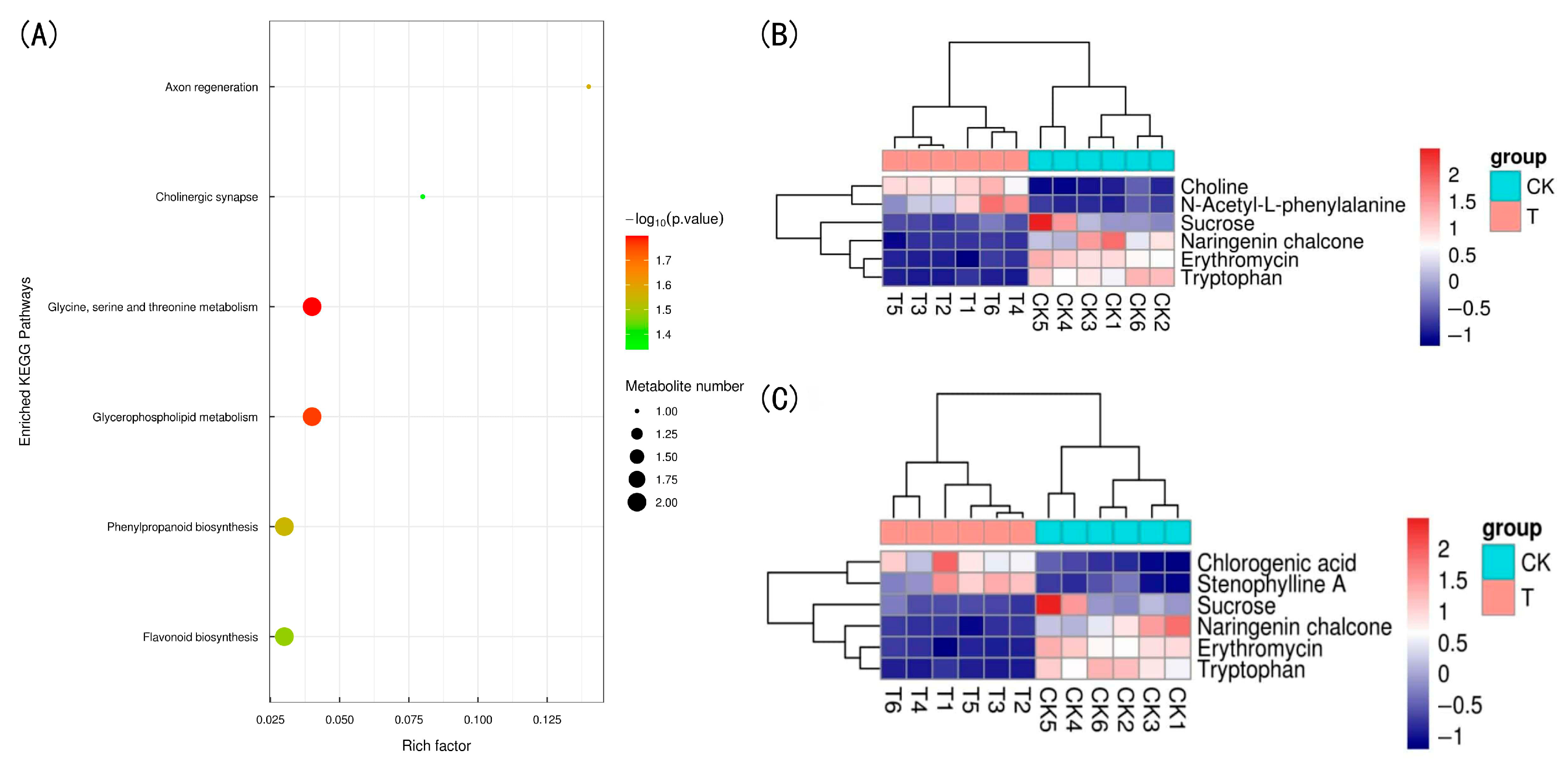
3.4.3. KEGG Pathway Analysis
3.5. Joint Analysis of Metabolome and Transcriptome of Lower Leaves of T150 and CK Plants
4. Discussion
4.1. Apparent Differences and Physiological and Biochemical Changes Between T150 and CK
4.2. Analysis of Differentially Expressed Genes in Premature Leaf Senescence of Red Raspberry
4.3. Analysis of Differential Metabolites in Premature Senescence of Red Raspberry Leaves
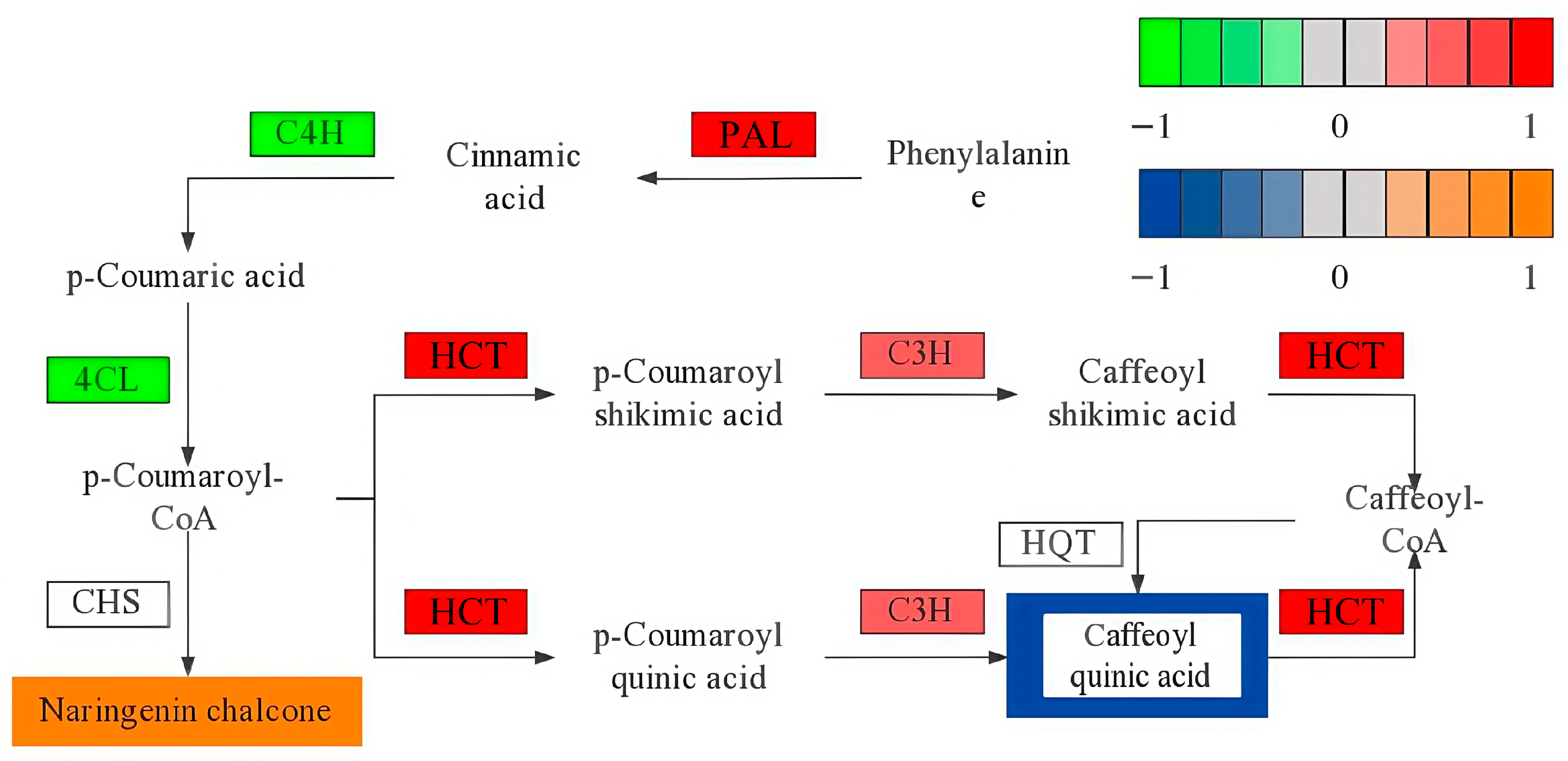
4.4. Analysis of Key Pathways in Premature Senescence of Red Raspberry Leaves
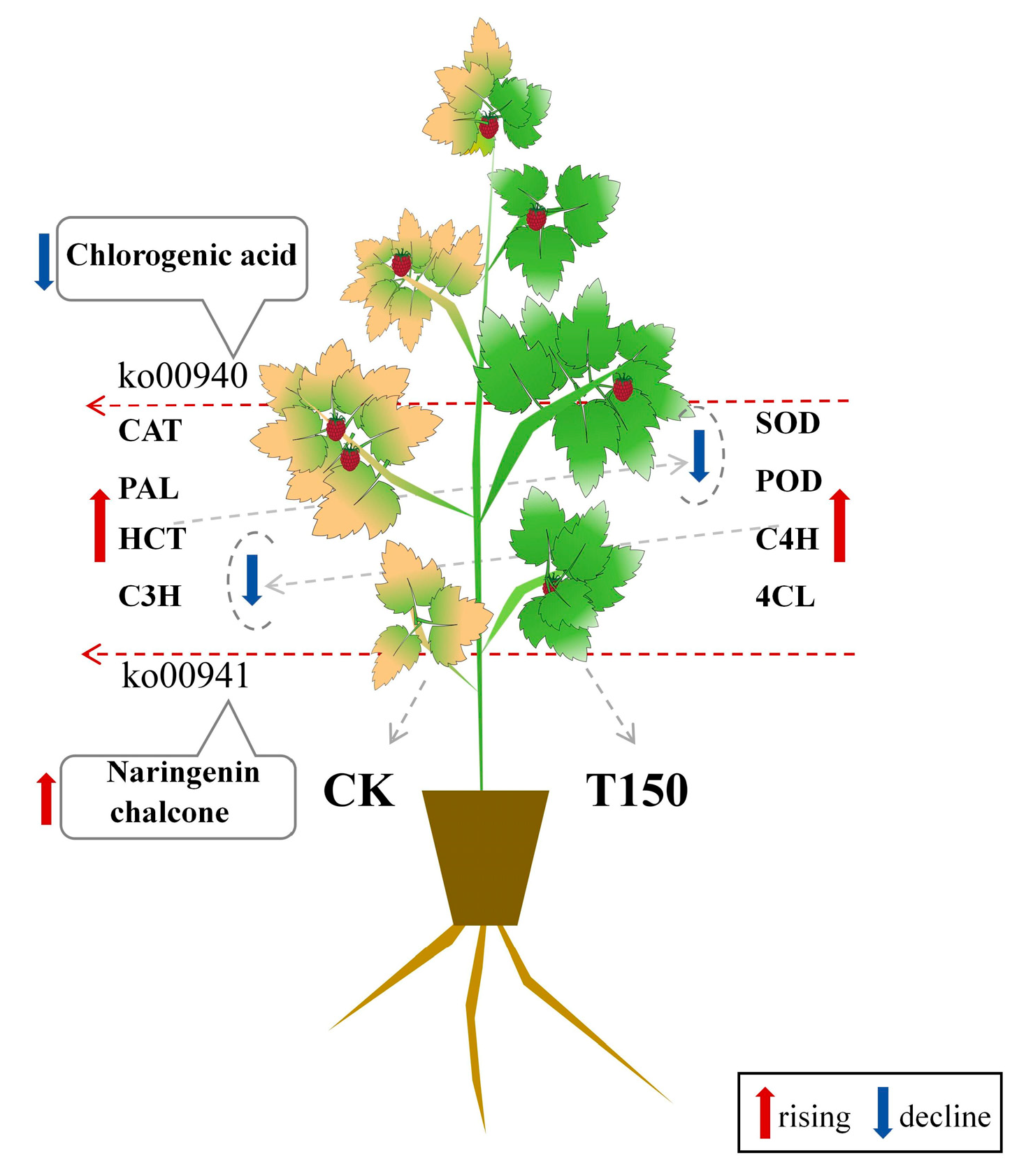
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirakosyan, A.; Seymour, E.M.; Kondoleon, N.; Gutierrez, E.; Wolforth, J.; Bolling, S. The Intake of Red Raspberry Fruit is Inversely Related to Cardiac Risk Factors Associated with Metabolic Syndrome. J. Funct. Foods 2018, 41, 83–89. [Google Scholar] [CrossRef]
- Rao, A.V.; Snyder, D.M. Raspberries and Human Health: A Review. J. Agric. Food Chem. 2010, 58, 3871–3883. [Google Scholar] [CrossRef]
- Galli, R.L.; Carey, A.N.; Luskin, K.A.; Bielinski, D.F.; Shukitt-Hale, B. Red Raspberries Can Improve Motor Function in Aged Rats. J. Berry Res. 2016, 6, 97–103. [Google Scholar] [CrossRef]
- Burton-Freeman, B.M.; Sandhu, A.K.; Edirisinghe, I. Red Raspberries and Their Bioactive Polyphenols: Cardiometabolic and Neuronal Health Links. Adv. Nutr. 2016, 7, 44–65. [Google Scholar] [CrossRef]
- Quintanilla, A.; Mencia, A.; Powers, J.; Rasco, B.; Tang, J.M.; Sablani, S.S. Developing Vacuum-impregnated Dehydrofrozen Red Raspberries with Improved Mechanical Properties. Dry. Technol. 2022, 40, 299–309. [Google Scholar] [CrossRef]
- Simona, O.; Felicia, C. Changes in Total Phenolics and Anthocyanins during Blackberry, Raspberry and Cherry Jam Processing and Storage. Rom. Biotechnol. Lett. 2016, 21, 11232–11237. Available online: https://www.academia.edu/67527231/Changes_in_Total_Phenolics_and_Anthocyanins_during_Blackberry_Raspberry_and_Cherry_Jam_Processing_and_Storage (accessed on 11 August 2023).
- Liu, M.; Qin, X.Y.; Wu, X.T. Study on the Technology of Brewing Red Raspberry Wine by Using New Immobilized Yeast Technology. Sci. Rep. 2022, 12, 21344. [Google Scholar] [CrossRef]
- Ispiryan, A.; Viskelis, J.; Viskelis, P. Red Raspberry (Rubus idaeus L.) Seed Oil: A Review. Plants 2021, 10, 944. [Google Scholar] [CrossRef]
- Chang, F.Y.; Zhang, L.Y.; Dong, Q.L.; Luan, H.A.; Jia, P.; Qi, G.H.; Guo, S.P.; Zhang, X.M. The Anatomical Structure Character of Raspberry Stems is A Key Factor Affecting Its Cold Resistance. Flora 2023, 298, 152196. [Google Scholar] [CrossRef]
- Shanks, J.C.H.; Moore, P.P. Resistance of Red Raspberry and Other Rubus Species to Twospotted Spider Mite (Acari: Tetranychidae). J. Econ. Entomol. 1996, 89, 771–774. [Google Scholar] [CrossRef]
- Qiao, Y.L.; Jiang, W.Z.; Lee, J.H.; Park, B.S.; Choi, M.S.; Piao, R.; Woo, M.O.; Roh, J.H.; Han, L.Z.; Paek, N.C.; et al. SPL28 Encodes A Clathrin-associated Adaptor Protein Complex 1, Medium Subunit μ1 (AP1M1) and Is Responsible for Spotted Leaf and Early Senescence in Rice (Oryza sativa). New Phytol. 2010, 185, 258–274. [Google Scholar] [CrossRef]
- Rao, Y.C.; Jiao, R.; Ye, H.F.; Hu, J.; Lu, T.; Wu, X.M.; Fang, Y.X.; Li, S.F.; Lin, H.; Wang, S.; et al. Fine Mapping and Candidate Gene Analysis of Leaf Tip Premature Senescence and Dwarf Mutant dls-1 in Rice. Plant Growth Regul. 2021, 94, 275–285. [Google Scholar] [CrossRef]
- Saglam, N.G. Effect of Epibrassinolide on Pigment Content, Total Protein Amount and Peroxidase Activity in Excised Cucumber Cotyledons. Biotechnol. Biotechnol. Equip. 2013, 27, 3502–3505. [Google Scholar] [CrossRef]
- Farsodia, M.; Mavadiya, P.; Trivedi, M.; Tandel, K.; Vyas, V.; Singh, S.K. Overexpression of Tomato ACL5 Gene in Tobacco Leads to Increased Plant Growth and Delayed the Onset of Leaf Senescence. J. Plant Growth Regul. 2023, 42, 4764–4783. [Google Scholar] [CrossRef]
- Zhao, L.; Duan, X.R.; Zhang, X.P.; Zhang, X.; Song, L.Z.; Chen, P.; Liang, M.; Zhang, C.; Wang, C.Y. Planting Strategy Optimization Can Increase Maize Yield by Delaying Leaf Senescence and Improving Photosynthetic Capacity. Agronomy 2025, 15, 1099. [Google Scholar] [CrossRef]
- Chen, S.; Wang, L.; Feng, W.; Yuan, M.; Li, J.; Xu, H.; Zheng, X.; Zhang, W. Sulfonamides-induced oxidative stress in freshwater microalga Chlorella vulgaris: Evaluation of growth, photosynthesis, antioxidants, ultrastructure, and nucleic acids. Sci. Rep. 2020, 10, 8243. [Google Scholar] [CrossRef]
- Choi, D.S.; Hwang, B.K. Proteomics and Functional Analyses of Pepper Abscisic Acid-Responsive 1 (ABR1), which is Involved in Cell Death and Defense Signaling. Plant Cell 2011, 23, 823–842. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as Sensors and Regulators of Leaf Senescence: Assigning Macro Roles to Small Molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.P.; Yang, Z.; Chen, J.Y.; Wu, S.X.; Zhu, X.Y.; Gao, S.; Gao, J.; Ren, G.D.; Kuai, B.K.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Qiao, H.L.; Liu, Y.W.; Cheng, L.L.; Gu, X.L.; Yin, P.C.; Li, K.; Zhou, S.; Wang, G.; Zhou, C.J. TaWRKY13-A Serves as a Mediator of Jasmonic Acid-Related Leaf Senescence by Modulating Jasmonic Acid Biosynthesis. Front. Plant Sci. 2021, 12, 717233. [Google Scholar] [CrossRef]
- Wang, C.Q.; Dai, S.Y.; Zhang, Z.L.; Lao, W.Q.; Wang, R.Y.; Meng, X.Q.; Zhou, X. Ethylene and Salicylic Acid Synergistically Accelerate Leaf Senescence in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 828–833. [Google Scholar] [CrossRef]
- Ji, Y.B.; Liu, J.; Xing, D. Low Concentrations of Salicylic Acid Delay Methyl Jasmonate-induced Leaf Senescence by Up-regulating Nitric Oxide Synthase Activity. J. Exp. Bot. 2016, 67, 5233–5245. [Google Scholar] [CrossRef]
- Geng, F.; Nie, R.M.; Yang, N.; Cai, L.; Hu, Y.C.; Chen, S.T.; Cheng, X.M.; Wang, Z.L.; Chen, L.Q. Integrated Transcriptome and Metabolome Profiling of Camellia reticulata Reveal Mechanisms of Flower Color Differentiation. Front. Genet. 2022, 13, 1059717. [Google Scholar] [CrossRef]
- Wang, S.S.; Liu, L.; Mi, X.Z.; Zhao, S.Q.; An, Y.L.; Xia, X.B.; Guo, R.; Wei, C.L. Multi-omics Analysis to Visualize the Dynamic Roles of Defense Genes in the Response of Tea Plants to Gray Blight. Plant J. 2021, 106, 862–875. [Google Scholar] [CrossRef]
- Wang, J.G.; Gao, X.M.; Ma, Z.L.; Chen, J.; Liu, Y.N.; Shi, W.Q. Metabolomic and Transcriptomic Profiling of Three Types of Litchi Pericarps Reveals That Changes in the Hormone Balance Constitute the Molecular Basis of the Fruit Cracking Susceptibility of Litchi chinensis cv. Baitangying. Mol. Biol. Rep. 2019, 46, 5295–5308. [Google Scholar] [CrossRef]
- Li, Y.K.; Fang, J.B.; Qi, X.J.; Lin, M.M.; Zhong, Y.P.; Sun, L.M.; Cui, W. Combined Analysis of the Fruit Metabolome and Transcriptome Reveals Candidate Genes Involved in Flavonoid Biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef]
- Mei, X.; Wan, S.H.; Lin, C.Y.; Zhou, C.B.; Hu, L.H.; Deng, C.; Zhang, L.Y. Integration of Metabolome and Transcriptome Reveals the Relationship of Benzenoid-phenylpropanoid Pigment and Aroma in Purple Tea Flowers. Front. Plant Sci. 2021, 12, 762330. [Google Scholar] [CrossRef]
- Wang, F.; Ge, S.F.; Xu, X.X.; Xing, Y.; Du, X.; Zhang, X.; Lv, M.X.; Liu, J.Q.; Zhu, Z.L.; Jiang, Y.M. Multiomics Analysis Reveals New Insights into the Apple Fruit Quality Decline under High Nitrogen Conditions. J. Agric. Food Chem. 2021, 69, 5559–5572. [Google Scholar] [CrossRef]
- Mehta, N.; Meng, Y.F.; Zare, R.; Kamenetsky-Goldstein, R.; Sattely, E. A Developmental gradient Reveals Biosynthetic Pathways to Eukaryotic Toxins in Monocot Geophytes. Cell 2024, 187, 5620–5637.e10. [Google Scholar] [CrossRef]
- Han, Z.G.; Xu, Z.W.; Xu, Y.; Lin, J.H.; Chen, X.L.; Wang, Y.; Yu, Q.X.; Li, C.; Chen, D.H.; Hu, H.L.; et al. Phylogenomics Reveal DcTPS-mediated Terpenoid Accumulation and Environmental Response in Dendrobium catenatum. Ind. Crops Prod. 2024, 208, 117799. [Google Scholar] [CrossRef]
- Zhang, P.Y.; Zhang, R.Y.; Sun, M.N.; Li, H.; Guo, S.P.; Qi, G.H.; Zhang, X.M. Effects of Leaf Presenility on Nutrition and Main Enzyme Activity in Red Raspberry (Rubus idaeus L.). Non-Wood For. Res. 2020, 38, 164–171. [Google Scholar] [CrossRef]
- Vendruscolo, R.G.; Fernandes, A.S.; Fagundes, M.B.; Zepka, L.Q.; de Menezes, C.R.; Jacob–Lopes, E.; Wagner, R. Development of a new method for simultaneous extraction of chlorophylls and carotenoids from microalgal biomass. J. Appl. Phycol. 2021, 33, 1987–1997. [Google Scholar] [CrossRef]
- Gao, J.F. Guidance of Plant Physiology Experiment; Higher Education Press: Beijing, China, 2006; pp. 59–61, 142–147, 210–214, 217–218. Available online: http://hn.sslibrary.com/showbook.do?dxNumber=11711260&d=14549C52DD36C7F26691A929688F26A4&fFenleiID=0Q9450 (accessed on 15 October 2023).
- Li, H.S. Principles and Techniques of Plant Physiological Biochemical Experiment; Higher Education Press: Beijing, China, 2000; pp. 164–169, 184–185, 195–197, 260–263. Available online: https://m.book118.com/try_down/128136143074006072.pdf (accessed on 18 October 2023).
- Larcher, L.; Boeger, M. Arquitetura foliar de Odontonema strictum (Nees) O. Kuntze (Acanthaceae) em duas condições de luminosidade. Hoehnea 2009, 36, 321–327. [Google Scholar] [CrossRef][Green Version]
- da Silva, B.P.M.; Fanalli, S.L.; Gomes, J.D.; de Almeida, V.V.; Fukumasu, H.; Freitas, F.A.O.; Moreira, G.C.M.; Silva-Vignato, B.; Reecy, J.M.; Koltes, J.E.; et al. Brain Fatty Acid and Transcriptome Profiles of Pig Fed Diets with Different Levels of Soybean Oil. BMC Genom. 2023, 24, 91. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling of Animal and Human Tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Guo, X.L.; Zhang, Z.; Li, J.Y.; Zhang, S.Q.; Sun, W.; Xiao, X.C.; Sun, Z.C.; Xue, X.Z.; Wang, Z.M.; Zhang, Y.H. Phenotypic and Transcriptome Profiling of Spikes Reveals the Regulation of Light Regimens on Spike Growth and Fertile Floret Number in Wheat. Plant Cell Environ. 2024, 47, 1575–1591. [Google Scholar] [CrossRef]
- Guo, S.; Arshad, A.; Yang, L.; Qin, Y.S.; Mu, X.H.; Mi, G.H. Comparative Transcriptome Analysis Reveals Common and Developmental Stage-specific Genes That Respond to Low Nitrogen in Maize Leaves. Plants 2022, 11, 1550. [Google Scholar] [CrossRef]
- Lei, L.; Wu, D.; Cui, C.; Gao, X.; Yao, Y.J.; Dong, J.; Xu, L.S.; Yang, M.M. Transcriptome Analysis of Early Senescence in the Post-anthesis Flag Leaf of Wheat (Triticum aestivum L.). Plants 2022, 11, 2593. [Google Scholar] [CrossRef]
- Semwal, V.K.; Khanna-Chopra, R. Reproductive Sink Enhanced Drought Induced Senescence in Wheat Fertile Line is Associated with Loss of Antioxidant Competence Compared to Its CMS Line. Physiol. Mol. Biol. Plants 2018, 24, 591–604. [Google Scholar] [CrossRef]
- Tian, X.Q.; Zhao, Y.N.; Chen, Z.Y.; Li, Z.; Liu, Y.H. Effects of Nitrogen Fertilization on Yield and Nitrogen Utilization of Film Side Planting Rapeseed (Brassica napus L.) Under Different Soil Fertility Conditions. J. Soil. Sci. Plant Nutr. 2023, 23, 368–379. [Google Scholar] [CrossRef]
- Rasmussen, I.S.; Dresboll, D.B.; Thorup-Kristensen, K. Winter Wheat Cultivars and Nitrogen (N) Fertilization-Effects on Root Growth, N Uptake Efficiency and N Use Efficiency. Eur. J. Agron. 2015, 68, 38–49. [Google Scholar] [CrossRef]
- Chun, L.; Mi, G.H.; Li, J.S.; Chen, F.J.; Zhang, F.S. Genetic Analysis of Maize Root Characteristics in Response to Low Nitrogen Stress. Plant Soil. 2005, 276, 369–382. [Google Scholar] [CrossRef]
- Qin, L.; Walk, T.C.; Han, P.P.; Chen, L.Y.; Zhang, S.; Li, Y.S.; Hu, X.J.; Xie, L.H.; Yang, Y.; Liu, J.P.; et al. Adaption of Roots to Nitrogen Deficiency Revealed by 3D Quantification and Proteomic Analysis. Plant Physiol. 2019, 179, 329–347. [Google Scholar] [CrossRef]
- Shazadi, K.; Christopher, J.T.; Chenu, K. Does late water deficit induce root growth or senescence in wheat? Front. Plant Sci. 2024, 15, 1351436. [Google Scholar] [CrossRef]
- Pommel, B.; Gallais, A.; Coque, M.; Quillere, I.; Hirel, B.; Prioul, J.L.; Andrieu, B.; Floriot, M. Carbon and Nitrogen Allocation and Grain Filling in Three Maize Hybrids Differing in Leaf Senescence. Eur. J. Agron. 2005, 24, 203–211. [Google Scholar] [CrossRef]
- Yang, C.Q.; Fang, X.; Wu, X.M.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Transcriptional Regulation of Plant Secondary MetabolismF. J. Integr. Plant Biol. 2012, 54, 10–13. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Xing, G.; Zhang, Z.; Sun, H.Y.; Zhao, K.R.; Chen, Y.D. Combined Analysis of Transcriptome and Metabolome Reveals the Effects of Novel Aroma-producing Naumovozyma castellii on Feng-flavor Baijiu. LWT-Food Sci. Technol. 2024, 211, 116901. [Google Scholar] [CrossRef]
- Cheng, A.X.; Lou, Y.G.; Mao, Y.B.; Lu, S.; Wang, L.J.; Chen, X.Y. Plant Terpenoids: Biosynthesis and Ecological Functions. J. Integr. Plant Biol. 2007, 49, 179–186. [Google Scholar] [CrossRef]
- Sangiorgio, D.; Cellini, A.; Spinelli, F.; Pastore, C.; Farneti, B.; Savioli, S.; Rodriguez-Estrada, M.T.; Donati, I. Contribution of Fruit Microbiome to Raspberry Volatile Organic Compounds Emission. Postharvest Biol. Technol. 2022, 183, 111742. [Google Scholar] [CrossRef]
- Seleem, D.; Pardi, V.; Murata, R.M. Review of Flavonoids: A Diverse Group of Natural Compounds with Anti-Candida albicans Activity in vitro. Arch. Oral. Biol. 2017, 76, 76–83. [Google Scholar] [CrossRef]
- Duda-Madej, A.; Viscardi, S.; Niezgódka, P.; Szewczyk, W.; Winska, K. The Impact of Plant-derived Polyphenols on Combating Efflux-mediated Antibiotic Resistance. Int. J. Mol. Sci. 2025, 26, 4030. [Google Scholar] [CrossRef]
- Gifford, I.; Battenberg, K.; Vaniya, A.; Wilson, A.; Tian, L.; Fiehn, O.; Berry, A.M. Distinctive Patterns of Flavonoid Biosynthesis in Roots and Nodules of Datisca glomerata and Medicago spp. Revealed by Metabolomic and Gene Expression Profiles. Front. Plant Sci. 2018, 9, 1463. [Google Scholar] [CrossRef]
- Wang, X.Q.; Cao, X.Y.; Shang, Y.; Bu, H.D.; Wang, T.Y.; Lyu, D.G.; Du, G.D. Preharvest Application of Prohydrojasmon Affects Color Development, Phenolic Metabolism, and Pigment-related Gene Expression in Red Pear (Pyrus ussuriensis). J. Sci. Food Agric. 2020, 100, 4766–4775. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Duan, Y.H.; Li, Y.J.; Zhang, P.; Li, Q.; Yu, L.Y.; Han, C.C.; Huo, J.C.; Chen, W.S.; Xiao, Y. CYP98A Monooxygenases: A Key Enzyme Family in Plant Phenolic Compound Biosynthesis. Hortic. Res. 2025, 12, uhaf074. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An Important Role of the Pepper Phenylalanine Ammonia-lyase Gene (PAL1) in Salicylic Acid-dependent Signalling of the Defence Response to Microbial Pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Ohkawara, T.; Takeda, H.; Nishihira, J. Protective Effect of Chlorogenic Acid on the Inflammatory Damage of Pancreas and Lung in Mice with L-arginine-induced Pancreatitis. Life Sci. 2017, 190, 91–96. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Z.; Yan, M.; Zhao, H.T.; He, Z.X.; Zhu, M.K. Responses of Intestinal Antioxidant Capacity, Morphology, Barrier Function, Immunity, and Microbial Diversity to Chlorogenic Acid in Late-Peak Laying Hens. Animals 2024, 14, 2957. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.B.; Zhang, Y.; Liu, Y.; Xu, M.X.; Yao, Z.C.; Zhang, X.D.; Sun, Y.; Zhou, T.L.; Shen, M. Effects of Chlorogenic Acid on Antimicrobial, Antivirulence, and Anti-quorum Sensing of Carbapenem-resistant Klebsiella pneumoniae. Front. Microbiol. 2022, 13, 997310. [Google Scholar] [CrossRef]
- Pietta, P.G. Flavonoids as Antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef]
- Li, P.; Ding, L.; Zhang, L.; He, J.; Huan, Z.W. Weisiensin B Inhibits Primary and Lateral Root Development by Interfering with Polar Auxin Transport in Arabidopsis thaliana. Plant Physiol. Biochem. 2019, 139, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.L.; Fan, Z.Q.; Shan, W.; Yin, X.R.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Association of BrERF72 with Methyl Jasmonate-induced Leaf Senescence of Chinese Flowering Cabbage through Activating JA Biosynthesis-related Genes. Hortic. Res. 2018, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Tang, Y.; Pan, L.; Wang, C.; Guo, Y.A.; Yang, H.N.; Li, Z.R.; Bai, L.Y.; Wang, L.F. Effect of Fulvic Acid on Barnyardgrass (Echinochloa crus-galli) Seedling Growth under Flooding Conditions. Weed Sci. 2021, 69, 192–202. [Google Scholar] [CrossRef]
- Saraste, A. Morphologic Criteria and Detection of Apoptosis. Herz 1999, 24, 189–195. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, Q.; Chang, F.; Jia, P.; Fu, Z.; Zhao, J.; Gao, Y.; Luan, H.; Wang, Y.; Dong, Q.; Qi, G.; et al. Integrated Transcriptomic and Metabolomic Analysis Reveals Nitrogen-Mediated Delay of Premature Leaf Senescence in Red Raspberry Leaves. Plants 2025, 14, 2388. https://doi.org/10.3390/plants14152388
Huo Q, Chang F, Jia P, Fu Z, Zhao J, Gao Y, Luan H, Wang Y, Dong Q, Qi G, et al. Integrated Transcriptomic and Metabolomic Analysis Reveals Nitrogen-Mediated Delay of Premature Leaf Senescence in Red Raspberry Leaves. Plants. 2025; 14(15):2388. https://doi.org/10.3390/plants14152388
Chicago/Turabian StyleHuo, Qiang, Feiyang Chang, Peng Jia, Ziqian Fu, Jiaqi Zhao, Yiwen Gao, Haoan Luan, Ying Wang, Qinglong Dong, Guohui Qi, and et al. 2025. "Integrated Transcriptomic and Metabolomic Analysis Reveals Nitrogen-Mediated Delay of Premature Leaf Senescence in Red Raspberry Leaves" Plants 14, no. 15: 2388. https://doi.org/10.3390/plants14152388
APA StyleHuo, Q., Chang, F., Jia, P., Fu, Z., Zhao, J., Gao, Y., Luan, H., Wang, Y., Dong, Q., Qi, G., & Zhang, X. (2025). Integrated Transcriptomic and Metabolomic Analysis Reveals Nitrogen-Mediated Delay of Premature Leaf Senescence in Red Raspberry Leaves. Plants, 14(15), 2388. https://doi.org/10.3390/plants14152388






