Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Property of CaDOG1 Genes
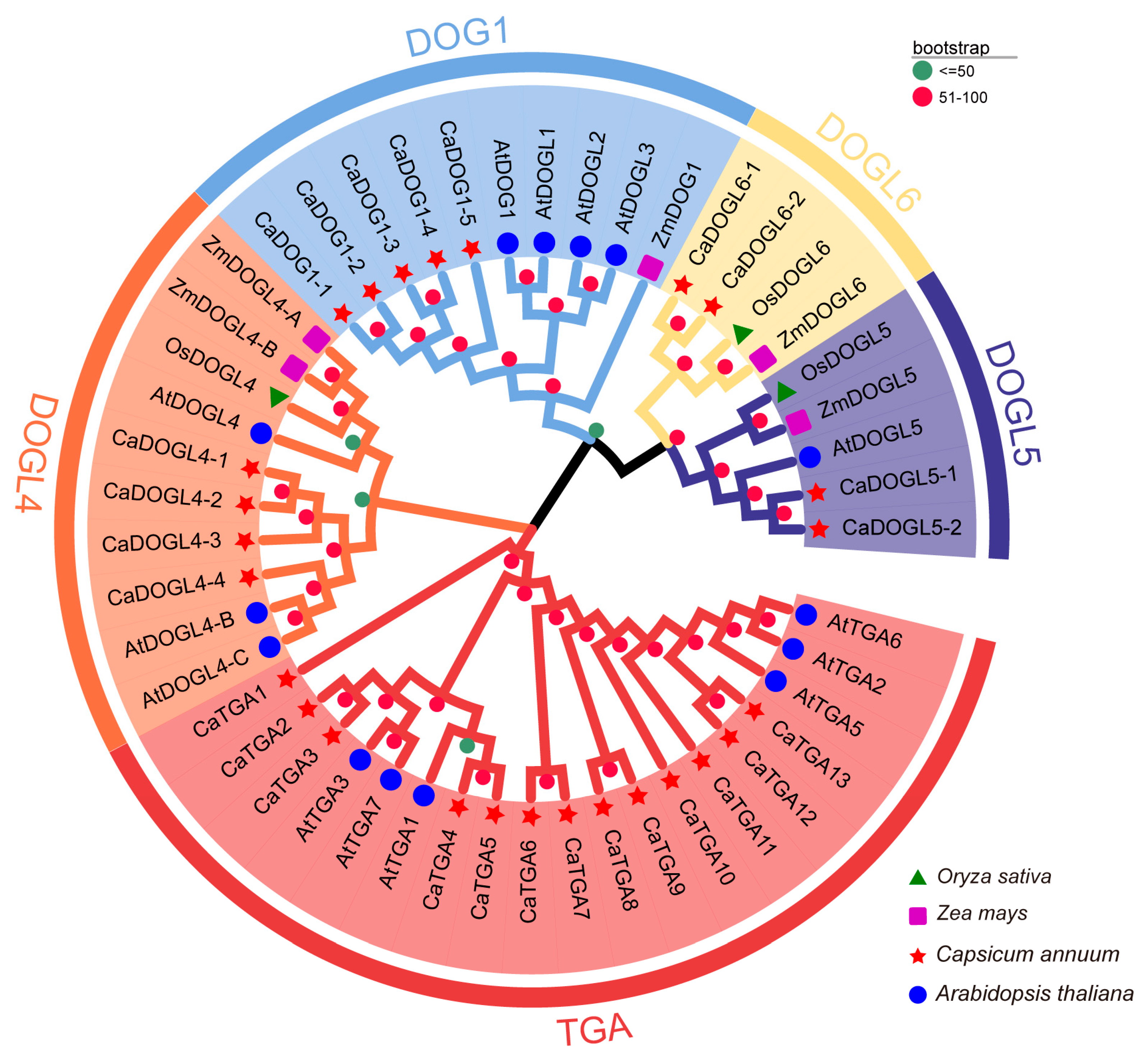
2.2. Phylogenetic Analysis of CaDOG1 Proteins
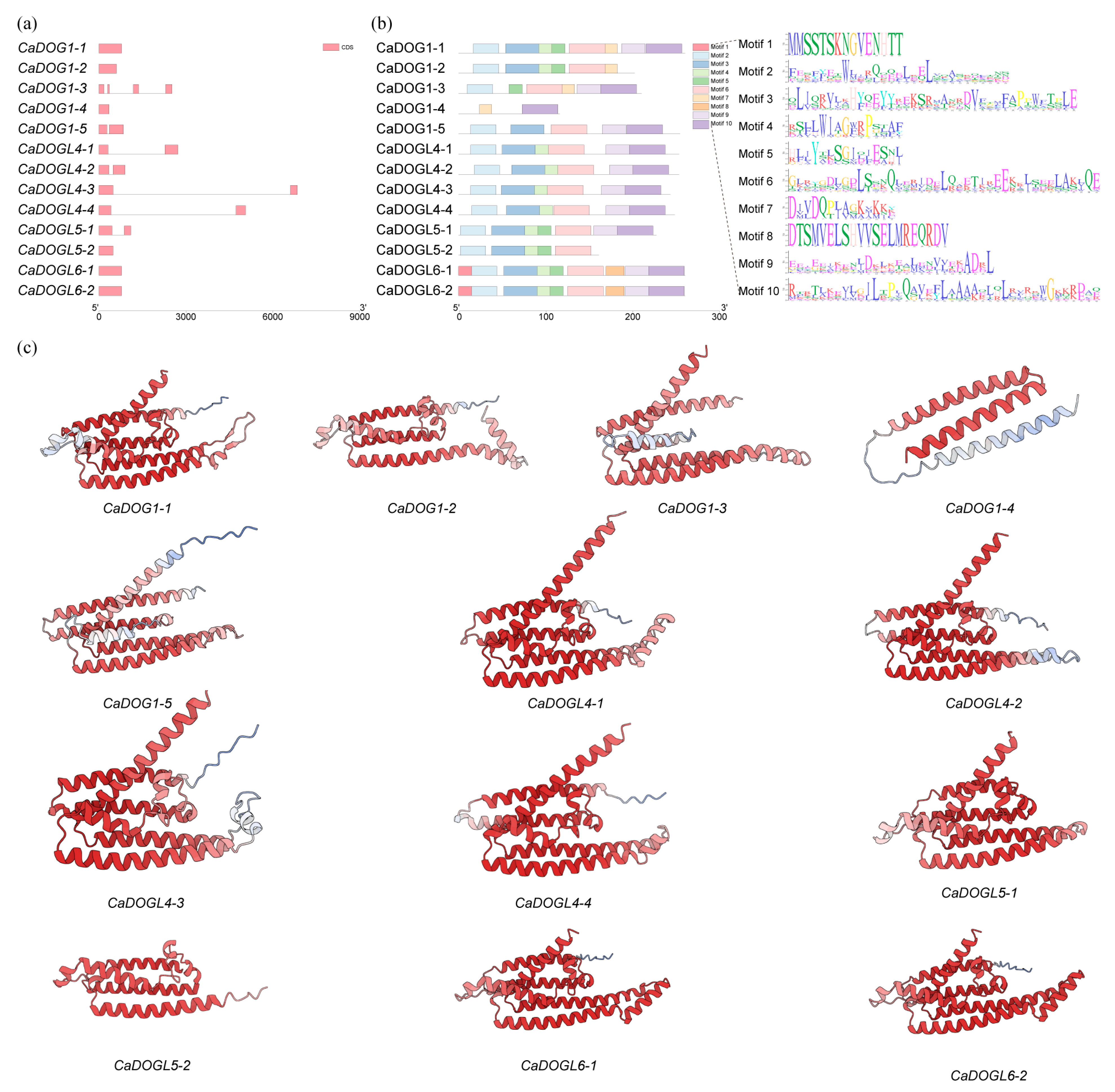
2.3. Intron/Exon Organization and Conserved Motif Analysis of CaDOG1 Genes
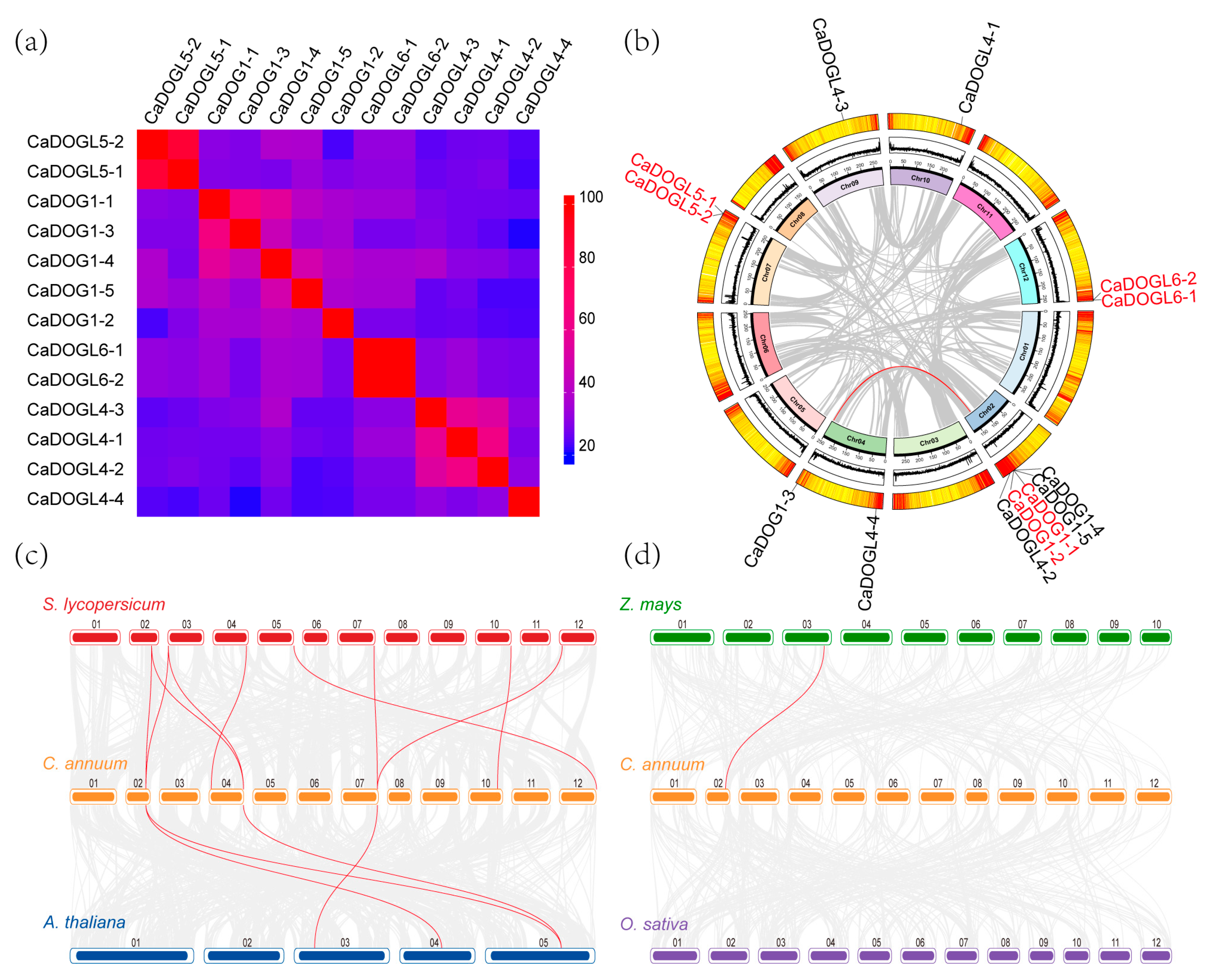
2.4. Identity and Collinearity Analysis of CaDOG1 Family Genes
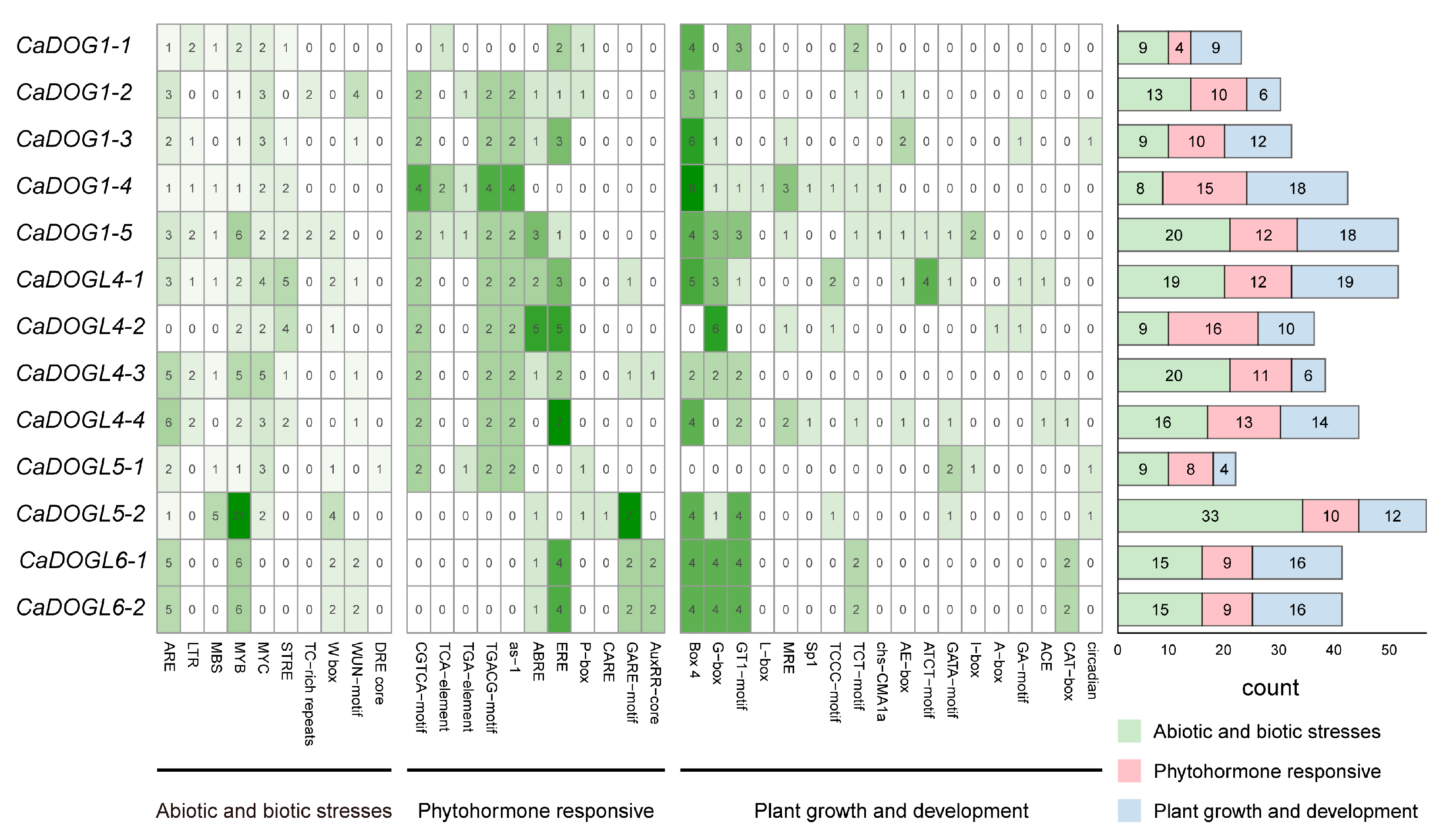
2.5. cis-Element Analyses of CaDOG1 Gene Family
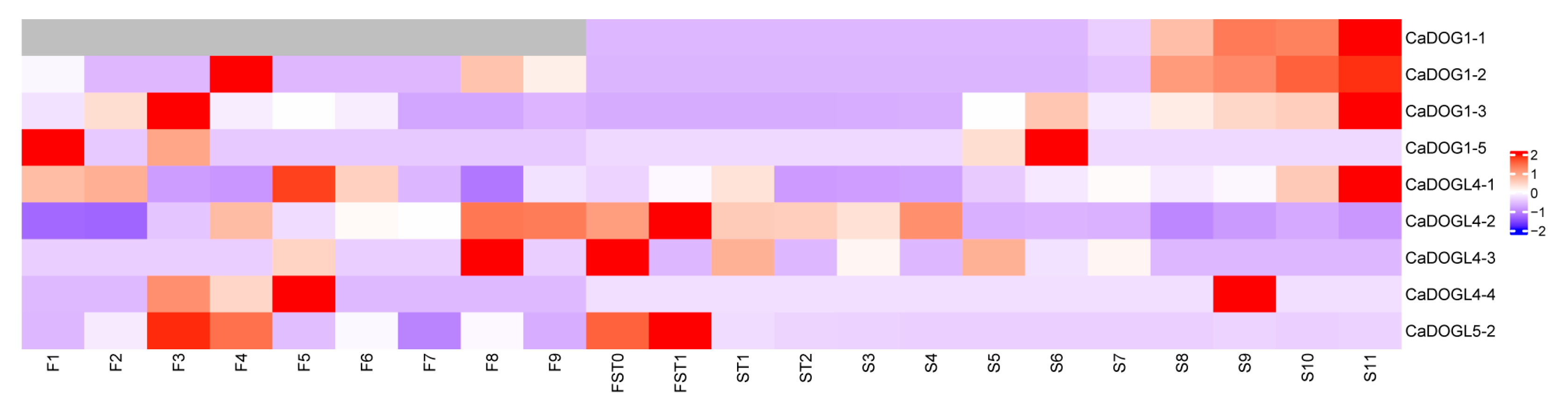
2.6. Tissue-Specific Expression Patterns of CaDOG1 Genes in C. annuum
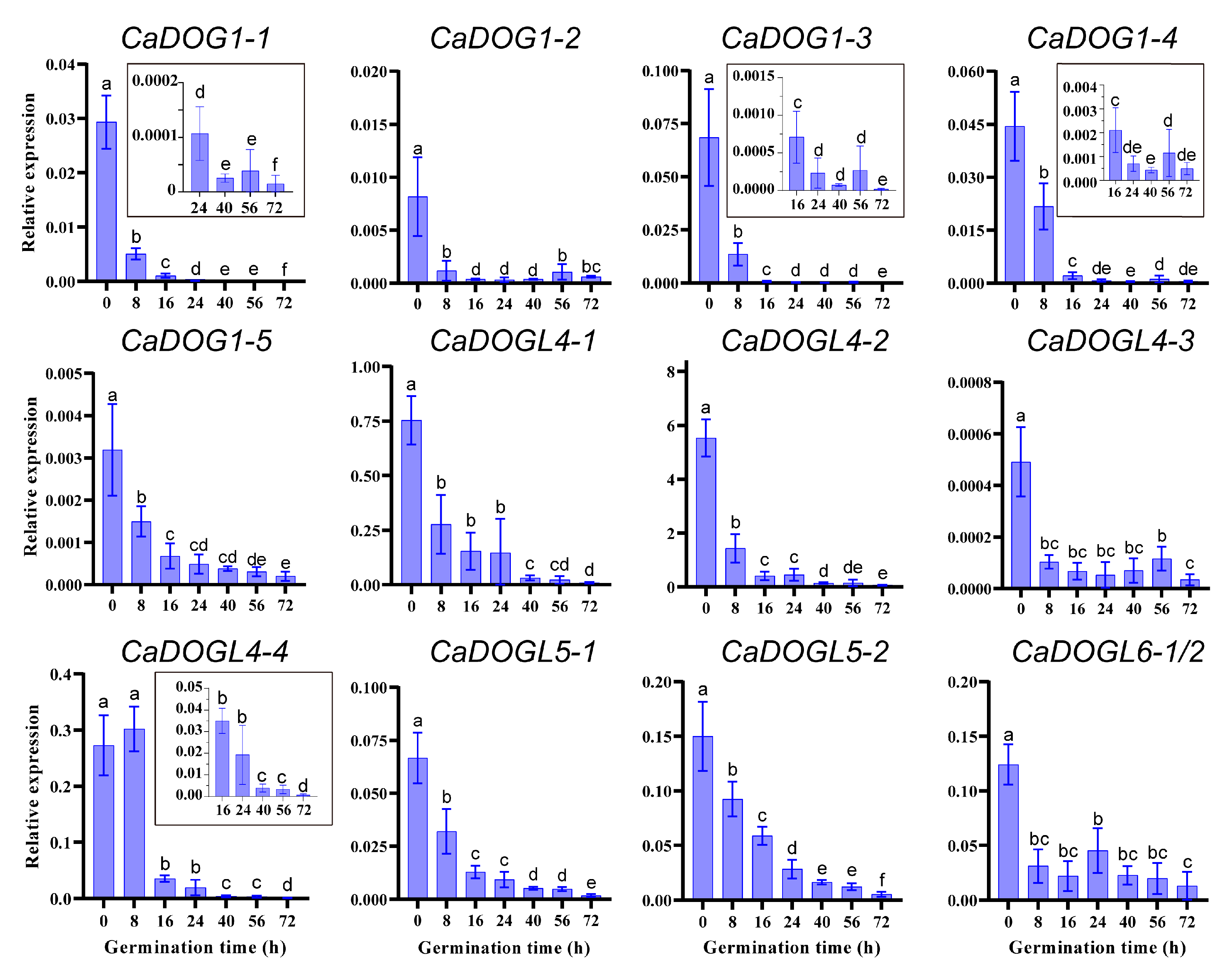
2.7. Expression Patterns of CaDOG1 Genes During Seed Germination
2.8. Hormone-Responsive Expression Patterns of CaDOG1 Genes During Seed Germination
3. Discussion
4. Materials and Methods
4.1. Identification of CaDOG1 Genes
4.2. Gene Structure and Sequence Analysis of CaDOG1 Genes
4.3. cis-Element Analysis of CaDOG1 Promoters
4.4. Phylogenetic Analysis and Collinearity Analysis
4.5. In Silico Expression Analysis of CaDOG1 Genes
4.6. Plant Materials and qRT-PCR Analysis of Gene Expression
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Biol. 2019, 61, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Seed Germination and Dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Hauvermale, A.L.; Tuttle, K.M.; Takebayashi, Y.; Seo, M.; Steber, C.M. Loss of Arabidopsis thaliana Seed Dormancy is Associated with Increased Accumulation of the GID1 GA Hormone Receptors. Plant Cell Physiol. 2015, 56, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Lamont, B.B.; Pausas, J.G. Seed dormancy revisited: Dormancy-release pathways and environmental interactions. Funct. Ecol. 2023, 37, 1106–1125. [Google Scholar] [CrossRef]
- Sajeev, N.; Koornneef, M.; Bentsink, L. A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination. Plant Cell 2024, 36, 1358–1376. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Peng, J.; Li, F.; Ali, F.; Wang, Z. Regulation of seed germination: ROS, epigenetic, and hormonal aspects. J. Adv. Res. 2024, 71, 107–125. [Google Scholar] [CrossRef]
- Yang, D.; Zhao, F.; Zhu, D.; Chen, X.; Kong, X.; Wu, Y.; Chen, M.; Du, J.; Qu, L.J.; Wu, Z. Progressive chromatin silencing of ABA biosynthesis genes permits seed germination in Arabidopsis. Plant Cell 2022, 34, 2871–2891. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Ye, N.; Liu, R.; Jia, W.; Zhang, J. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 2009, 183, 1030–1042. [Google Scholar] [CrossRef]
- Iwasaki, M.; Penfield, S.; Lopez-Molina, L. Parental and Environmental Control of Seed Dormancy in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2022, 73, 355–378. [Google Scholar] [CrossRef]
- Koornneef, M.; Bentsink, L.; Hilhorst, H. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002, 5, 33–36. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, Y.; Fan, D.; Zhou, X.; Jiao, Y.; Deng, X.W.; Zhu, D. The noncoding RNA HIDDEN TREASURE 1 promotes phytochrome B-dependent seed germination by repressing abscisic acid biosynthesis. Plant Cell 2023, 35, 700–716. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, L.M.; Volaire, F.A. Are winter and summer dormancy symmetrical seasonal adaptive strategies? The case of temperate herbaceous perennials. Ann. Bot. 2017, 119, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Barral, N.; Rodríguez-Gacio, M.D.C.; Matilla, A.J. Delay of Germination-1 (DOG1): A Key to Understanding Seed Dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, E.; Nonogaki, M.; Yamazaki, S.; Nonogaki, H.; Ohshima, K. Ancient and recent gene duplications as evolutionary drivers of the seed maturation regulators DELAY OF GERMINATION1 family genes. New Phytol. 2021, 230, 889–901. [Google Scholar] [CrossRef]
- Kendall, S.L.; Hellwege, A.; Marriot, P.; Whalley, C.; Graham, I.A.; Penfield, S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 2011, 23, 2568–2580. [Google Scholar] [CrossRef]
- Chiang, G.C.K.; Bartsch, M.; Barua, D.; Nakabayashi, K.; Debieu, M.; Kronholm, I.; Koornneef, M.; Soppe, W.J.J.; Donohue, K.; De Meaux, J. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol. Ecol. 2011, 20, 3336–3349. [Google Scholar] [CrossRef]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Teng, S.; Rognoni, S.; Bentsink, L.; Smeekens, S. The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J. 2008, 55, 372–381. [Google Scholar] [CrossRef]
- Li, X.; Chen, T.; Li, Y.; Wang, Z.; Cao, H.; Chen, F.; Li, Y.; Soppe, W.J.J.; Li, W.; Liu, Y. ETR1/RDO3 Regulates Seed Dormancy by Relieving the Inhibitory Effect of the ERF12-TPL Complex on DELAY OF GERMINATION1 Expression. Plant Cell 2019, 31, 832–847. [Google Scholar] [CrossRef]
- Martínez-Berdeja, A.; Stitzer, M.C.; Taylor, M.A.; Okada, M.; Ezcurra, E.; Runcie, D.E.; Schmitt, J. Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 2526–2534. [Google Scholar] [CrossRef]
- Jing, H.; Liu, W.; Qu, G.-P.; Niu, D.; Jin, J.B. SUMOylation of AL6 regulates seed dormancy and thermoinhibition in Arabidopsis. New Phytol. 2025, 245, 1040–1055. [Google Scholar] [CrossRef] [PubMed]
- Yatusevich, R.; Fedak, H.; Ciesielski, A.; Krzyczmonik, K.; Kulik, A.; Dobrowolska, G.; Swiezewski, S. Antisense transcription represses Arabidopsis seed dormancy QTL DOG1 to regulate drought tolerance. EMBO Rep. 2017, 18, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zheng, C.; Chandrasekaran, U.; Yu, L.; Liu, C.; Pu, T.; Wang, X.; Du, J.; Liu, J.; Yang, F.; et al. Identification and Bioinformatic Analysis of the GmDOG1-Like Family in Soybean and Investigation of Their Expression in Response to Gibberellic Acid and Abscisic Acid. Plants 2020, 9, 937. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.S.; Kim, J.-B.; Kim, D.Y.; Seo, Y.W.; Hong, M.J. Unveiling differential expression profiles of the wheat DOG1 gene family and functional analysis of the association between TaDOG1-1 and heat stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2024, 207, 108325. [Google Scholar] [CrossRef]
- Zhu, H.; Xie, W.; Xu, D.; Miki, D.; Tang, K.; Huang, C.F.; Zhu, J.K. DNA demethylase ROS1 negatively regulates the imprinting of DOGL4 and seed dormancy in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E9962–E9970. [Google Scholar] [CrossRef]
- Sall, K.; Dekkers, B.J.W.; Nonogaki, M.; Katsuragawa, Y.; Koyari, R.; Hendrix, D.; Willems, L.A.J.; Bentsink, L.; Nonogaki, H. DELAY OF GERMINATION 1-LIKE 4 acts as an inducer of seed reserve accumulation. Plant J. 2019, 100, 7–19. [Google Scholar] [CrossRef]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef]
- Qiao, H.; Zhou, X.; Yi, Y.; Wei, L.; Xu, X.; Jin, P.; Su, W.; Weng, Y.; Yu, D.; He, S.; et al. Molecular mechanism of vivipary as revealed by the genomes of viviparous mangroves and non-viviparous relatives. Curr. Biol. 2024, 34, 3707–3721.e7. [Google Scholar] [CrossRef] [PubMed]
- Vollmeister, E.; Phokas, A.; Meyberg, R.; Böhm, C.V.; Peter, M.; Kohnert, E.; Yuan, J.; Grosche, C.; Göttig, M.; Ullrich, K.K.; et al. A DELAY OF GERMINATION 1 (DOG1)-like protein regulates spore germination in the moss Physcomitrium patens. Plant J. 2024, 117, 909–923. [Google Scholar] [CrossRef]
- Huo, H.; Wei, S.; Bradford, K.J. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations (FAO). World Food and Agriculture—Statistical Yearbook 2024; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2024. [Google Scholar]
- Carrizo García, C.; Barfuss, M.H.J.; Sehr, E.M.; Barboza, G.E.; Samuel, R.; Moscone, E.A.; Ehrendorfer, F. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 2016, 118, 35–51. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Chen, S.; Liu, Y.; Zhang, L.; Yang, X.; Yu, H.; Cao, Y.; Zhang, L.; Cai, C.; et al. The gap-free assembly of pepper genome reveals transposable-element-driven expansion and rapid evolution of pericentromeres. Plant Commun. 2025, 6, 101177. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2024, 53, D444–D456. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Müller, K.; Wunchova, A.; Rott, A.; Leubner-Metzger, G. Cross-species approaches to seed dormancy and germination: Conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol. Biol. 2010, 73, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E. Letting Sleeping DOGs Lie: Regulation of DOG1 during Seed Dormancy. Plant Cell 2019, 31, 1218–1219. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Liu, X.; Wu, K. Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant Sci. 2015, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Tsuchiya, W.; Suzuki, N.; Hirayama, T.; Yamazaki, T. Identification and characterization of functional DOG1 residues regulating the abscisic acid response in Arabidopsis. Plant J. 2025, 122, e70180. [Google Scholar] [CrossRef]
- Dekkers, B.J.; He, H.; Hanson, J.; Willems, L.A.; Jamar, D.C.; Cueff, G.; Rajjou, L.; Hilhorst, H.W.; Bentsink, L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016, 85, 451–465. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Steinbrecher, T.; Mummenhoff, K.; Tarkowská, D.; Turečková, V.; Ignatz, M.; Sperber, K.; Voegele, A.; de Jong, H.; et al. DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 2014, 111, E3571–E3580. [Google Scholar] [CrossRef]
- Ashikawa, I.; Abe, F.; Nakamura, S. DOG1-like genes in cereals: Investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 2013, 208, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Barral, N.; Matilla, A.J.; García-Ramas, C.; Rodríguez-Gacio Mdel, C. ABA-stimulated SoDOG1 expression is after-ripening inhibited during early imbibition of germinating Sisymbrium officinale seeds. Physiol. Plant 2015, 155, 457–471. [Google Scholar] [CrossRef]
- Mönke, G.; Altschmied, L.; Tewes, A.; Reidt, W.; Mock, H.P.; Bäumlein, H.; Conrad, U. Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 2004, 219, 158–166. [Google Scholar] [CrossRef]
- Ruta, V.; Longo, C.; Lepri, A.; De Angelis, V.; Occhigrossi, S.; Costantino, P.; Vittorioso, P. The DOF Transcription Factors in Seed and Seedling Development. Plants 2020, 9, 218. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, X.; Zhang, S.; Shan, S.; Xiang, Y. DELAY OF GERMINATION 1, the Master Regulator of Seed Dormancy, Integrates the Regulatory Network of Phytohormones at the Transcriptional Level to Control Seed Dormancy. Curr. Issues Mol. Biol. 2022, 44, 6205–6217. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Yoshida, T.; Murayama, M.; Asami, T.; Shinozaki, K.; Hirayama, T. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol. 2004, 45, 1485–1499. [Google Scholar] [CrossRef]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef]
- Montez, M.; Majchrowska, M.; Krzyszton, M.; Bokota, G.; Sacharowski, S.; Wrona, M.; Yatusevich, R.; Massana, F.; Plewczynski, D.; Swiezewski, S. Promoter-pervasive transcription causes RNA polymerase II pausing to boost DOG1 expression in response to salt. EMBO J. 2023, 42, e112443. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef]
- Ashikawa, I.; Abe, F.; Nakamura, S. Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 2010, 179, 536–542. [Google Scholar] [CrossRef]
- Ashikawa, I.; Mori, M.; Nakamura, S.; Abe, F. A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1-like genes. Transgenic Res. 2014, 23, 621–629. [Google Scholar] [CrossRef]
- Atwell, S.; Huang, Y.S.; Vilhjálmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef]
- Cao, Y.; Dai, Y.; Cui, S.; Ma, L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 2008, 20, 2586–2602. [Google Scholar] [CrossRef] [PubMed]
- Peeters, A.J.; Blankestijn-De Vries, H.; Hanhart, C.J.; Léon-Kloosterziel, K.M.; Zeevaart, J.A.; Koornneef, M. Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant 2002, 115, 604–612. [Google Scholar] [CrossRef]
- Liu, Y.; Koornneef, M.; Soppe, W.J.J. The Absence of Histone H2B Monoubiquitination in the Arabidopsis hub1 (rdo4) Mutant Reveals a Role for Chromatin Remodeling in Seed Dormancy. Plant Cell 2007, 19, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.R.; Yang, S.F. Release of heat pretreatment-induced dormancy in lettuce seeds by ethylene or cytokinin in relation to the production of ethylene and the synthesis of 1-aminocyclopropane-1-carboxylic acid during germination. J. Plant Growth Regul. 1983, 2, 185–192. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, L.; Dong, D.; Xu, J.; Li, H.; Deng, Q.; Zhang, Y.; Huang, W.; Zhang, H.; Guo, Y.D. The transcription factor SlLBD40 regulates seed germination by inhibiting cell wall remodeling enzymes during endosperm weakening. Plant Physiol. 2025, 197, kiaf022. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Basse, B.; Olsen, E.; Celis, J.E. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 1994, 15, 529–539. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Raffeiner, M.; Üstün, S.; Guerra, T.; Spinti, D.; Fitzner, M.; Sonnewald, S.; Baldermann, S.; Börnke, F. The Xanthomonas type-III effector XopS stabilizes CaWRKY40a to regulate defense responses and stomatal immunity in pepper (Capsicum annuum). Plant Cell 2022, 34, 1684–1708. [Google Scholar] [CrossRef]
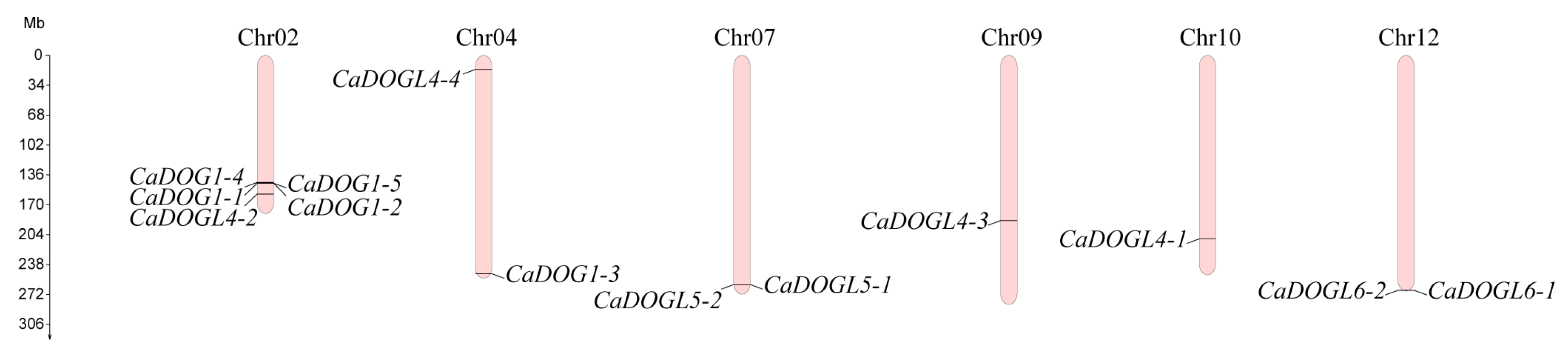

| Name | Gene ID | Chr. | AA | MW (kDa) | pI | Gravy | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| CaDOG1-1 | ZLC02G0015410 | 2 | 260 | 29.80 | 5.48 | −0.560 | cyto |
| CaDOG1-2 | ZLC02G0015420 | 2 | 202 | 22.69 | 5.11 | −0.484 | cyto |
| CaDOG1-3 | ZLC04G0026780 | 4 | 210 | 24.11 | 5.19 | −0.605 | cyto_nucl |
| CaDOG1-4 | ZLC02G0015240 | 2 | 116 | 13.31 | 6.06 | −0.810 | nucl |
| CaDOG1-5 | ZLC02G0015390 | 2 | 254 | 28.90 | 6.00 | −0.647 | nucl |
| CaDOGL4-1 | ZLC10G0015190 | 10 | 253 | 29.08 | 4.55 | −0.516 | cyto |
| CaDOGL4-2 | ZLC02G0022320 | 2 | 253 | 28.94 | 4.83 | −0.352 | chlo |
| CaDOGL4-3 | ZLC09G0015000 | 9 | 243 | 28.28 | 5.83 | −0.456 | mito |
| CaDOGL4-4 | ZLC04G0008420 | 4 | 248 | 28.39 | 5.30 | −0.385 | cyto |
| CaDOGL5-1 | ZLC07G0022880 | 7 | 227 | 26.41 | 5.15 | −0.425 | cyto |
| CaDOGL5-2 | ZLC07G0022870 | 7 | 161 | 18.88 | 6.15 | −0.361 | mito |
| CaDOGL6-1 | ZLC12G0030560 | 12 | 260 | 30.24 | 5.86 | −0.664 | nucl |
| CaDOGL6-2 | ZLC12G0030550 | 12 | 260 | 30.24 | 5.86 | −0.664 | nucl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Sun, J.; Zhang, F.; Dong, C. Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination. Plants 2025, 14, 1913. https://doi.org/10.3390/plants14131913
Zhao Z, Sun J, Zhang F, Dong C. Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination. Plants. 2025; 14(13):1913. https://doi.org/10.3390/plants14131913
Chicago/Turabian StyleZhao, Zhichao, Jingbo Sun, Feng Zhang, and Chunjuan Dong. 2025. "Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination" Plants 14, no. 13: 1913. https://doi.org/10.3390/plants14131913
APA StyleZhao, Z., Sun, J., Zhang, F., & Dong, C. (2025). Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination. Plants, 14(13), 1913. https://doi.org/10.3390/plants14131913






