Two B-Box Proteins, GhBBX21 and GhBBX24, Antagonistically Modulate Anthocyanin Biosynthesis in R1 Cotton
Abstract
1. Introduction
2. Results
2.1. Comparative Analysis of the Upstream Promoter Regions of GhPAP1D Between GL and R1 Cotton
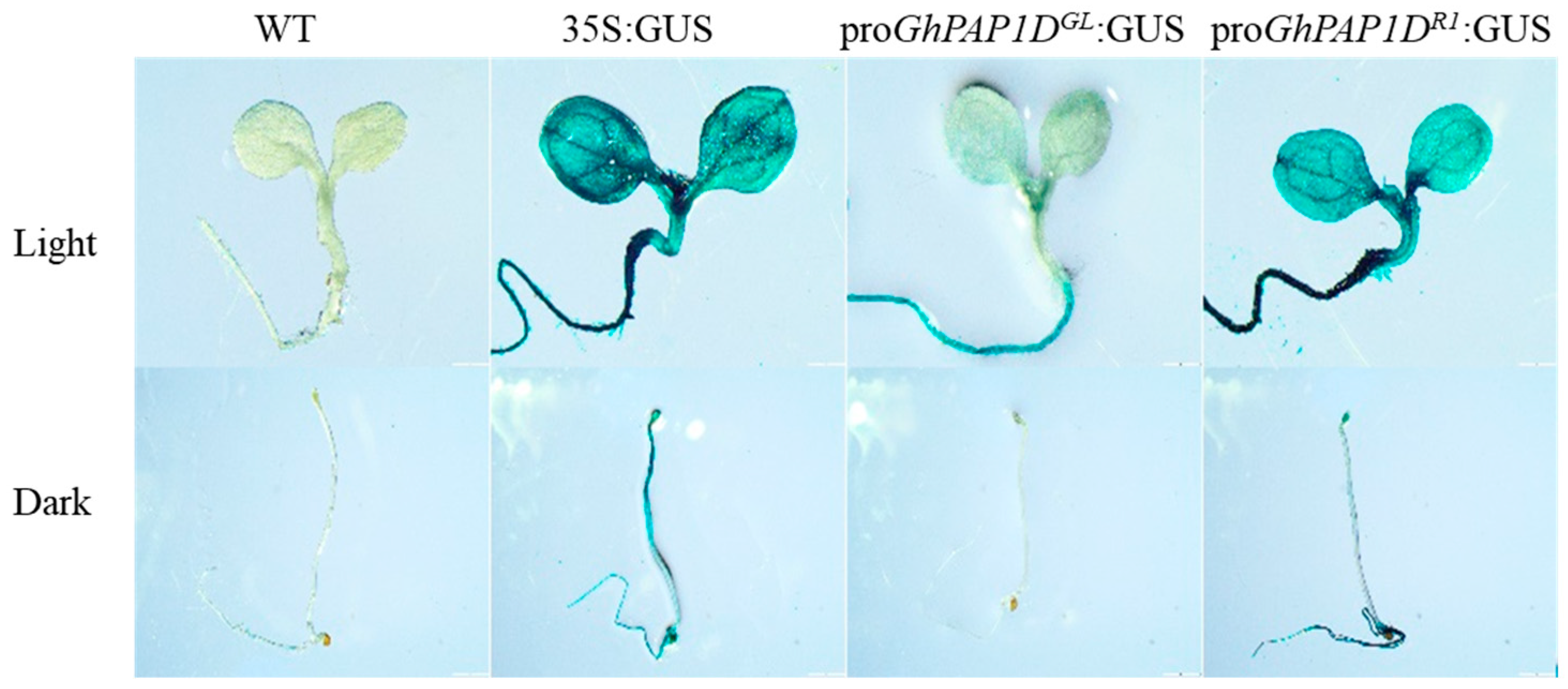
2.2. Activity Analysis of proGhPAP1DGL and proGhPAP1DR1 in Response to Light
2.3. Autoregulation of GhPAP1D in the Dual-Luciferase Transient Tobacco Assays
2.4. Identification of GhHY5 in R1 Cotton
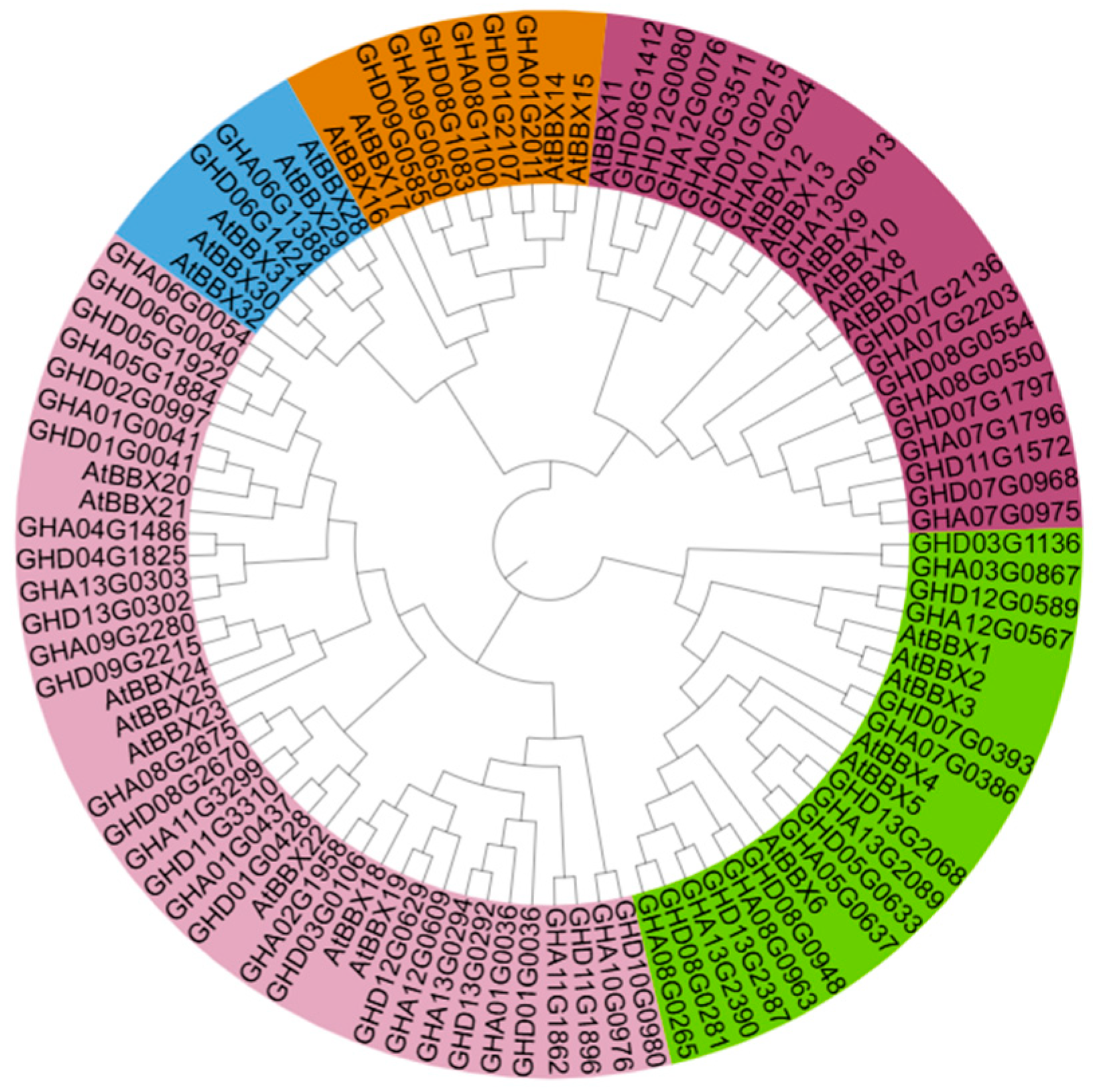
2.5. Identification of BBX Subfamily IV Gene Family from Gossypium hirsutum L.
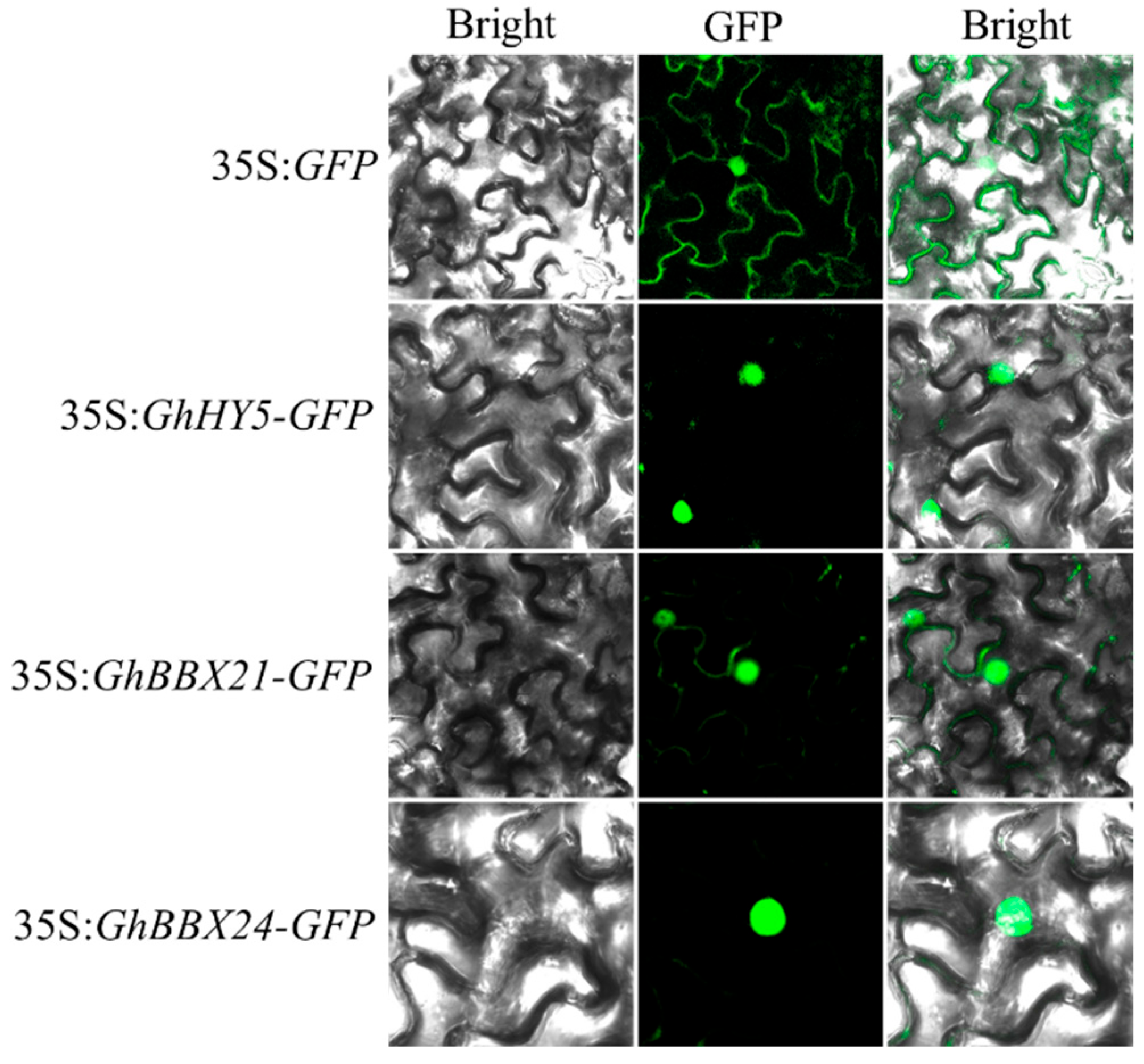
2.6. GhHY5, GhBBX21, and GhBBX24 Are Transcriptional Activators Localized in the Nuclei
2.7. GhBBX21 and GhBBX24 Antagonistically Modulate Anthocyanin Accumulation in Transgenic A. thaliana Seedlings
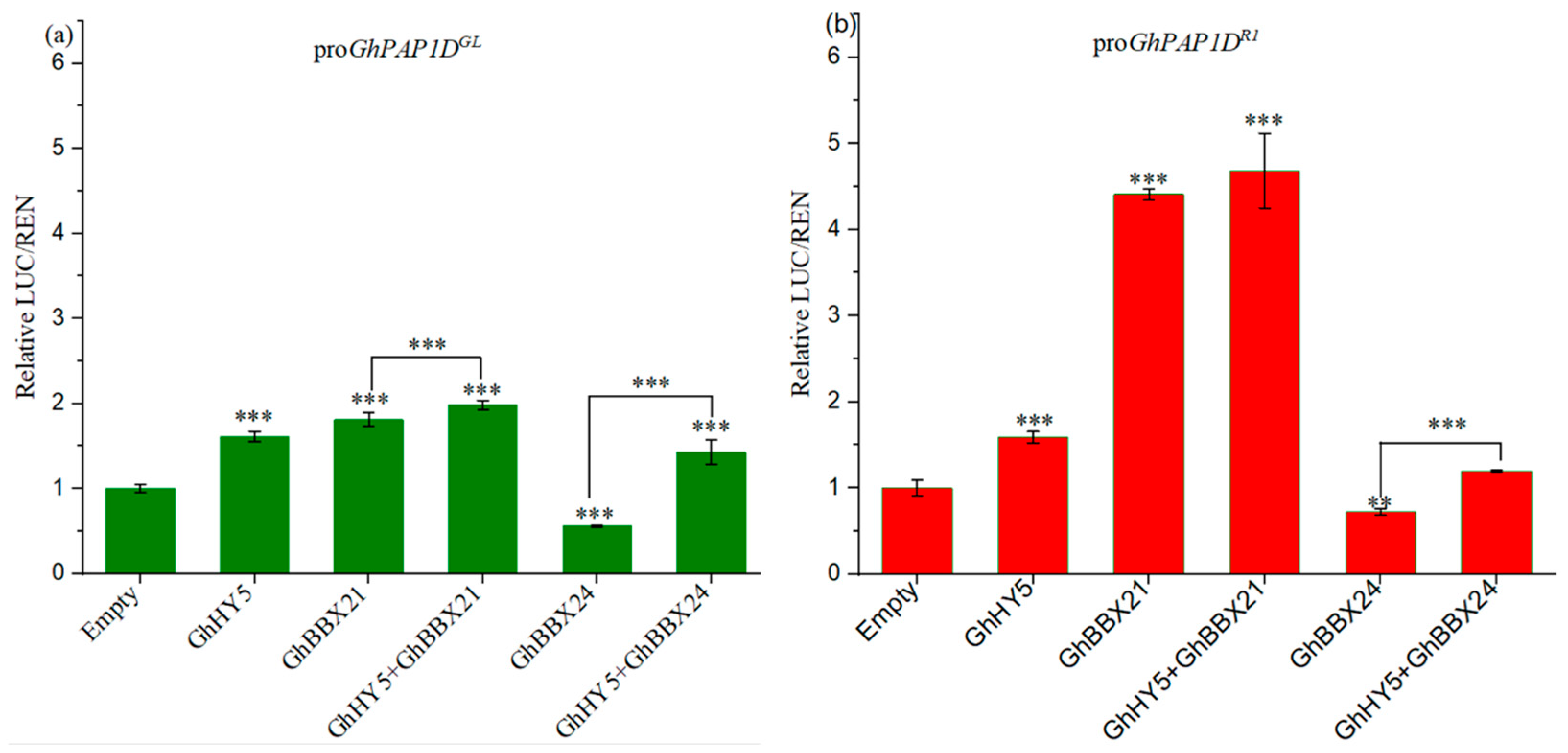
2.8. The Regulation of GhPAP1D by GhHY5, GhBBX21, and GhBBX24
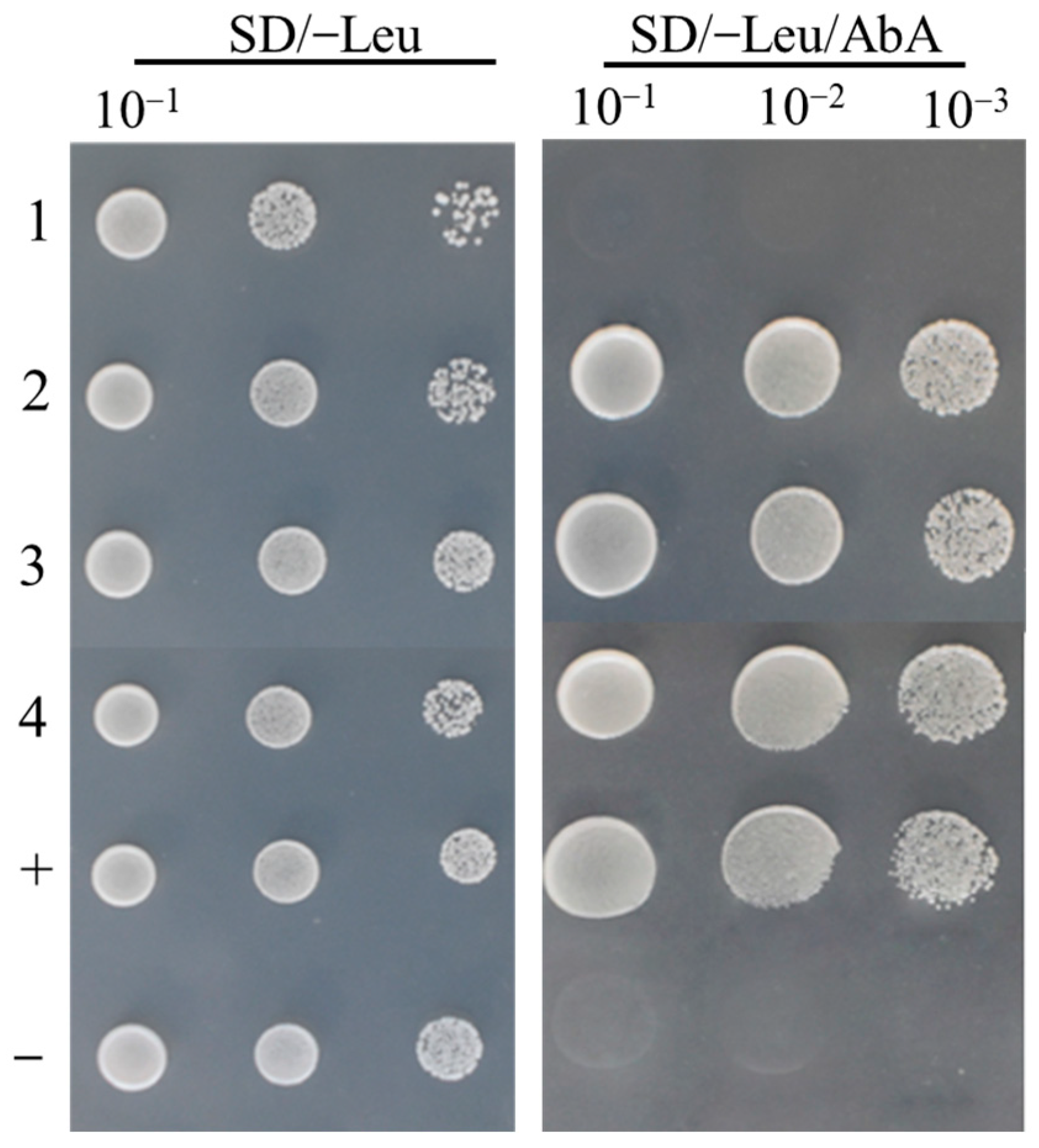
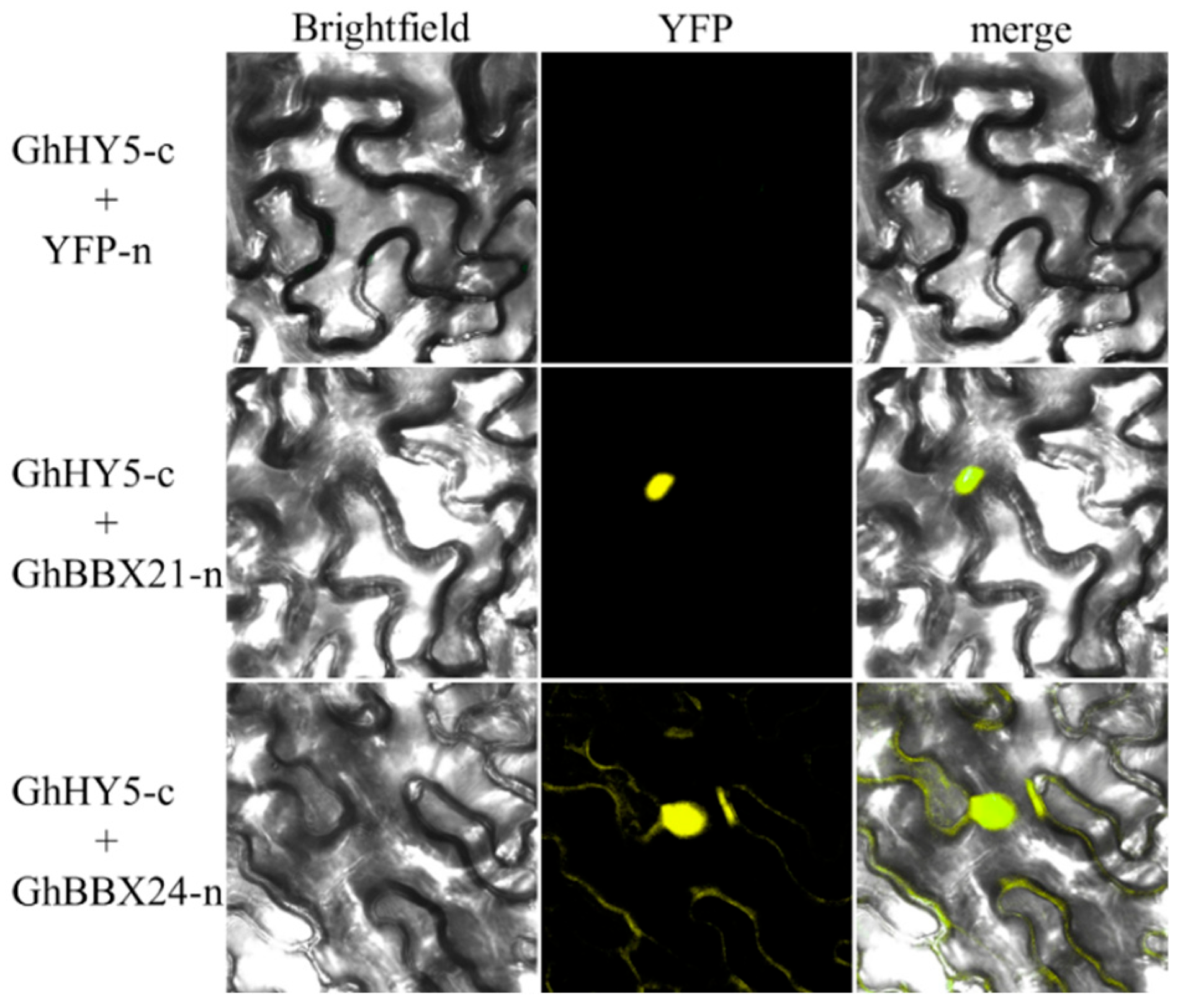
2.9. GhBBX21 and GhBBX24 Proteins Interact with GhHY5
3. Discussion
3.1. Tandem Repeats in the Promoter Region Enhance Light-Responsive Transcription of Ghpap1d
3.2. Autoregulation Is Not the Main Driver of the Increased Expression of GhPAP1D in R1 Cotton
3.3. GhHY5 Binds Constitutively to GhPAP1D Promoter
3.4. GhBBX21 and GhBBX24 Antagonistically Regulate GhPAP1D Expression, with GhBBX21 Significantly Activating the GhPAP1D Transcription in R1 Cotton
3.5. GhHY5 Interacts with Either GhBBX21 or GhBBX24 to Coregulate GhPAP1D Expression in R1 Cotton
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation and Functional Characterization of the GhPAP1D Promoter
4.3. Stable Transformation in A. thaliana and GUS Staining Assays
4.4. GUS Staining Assays
4.5. Phylogenetic Analysis of BBX Family Proteins
4.6. Subcellular Localization and Bimolecular Fluorescence Complementation (BiFC) Assays
4.7. Dual-Luciferase Assays
4.8. Yeast One-Hybrid (Y1H) Assay
4.9. Anthocyanin Content Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ouyang, X.; Zhang, Z.; He, L.; Wang, Y.; Li, Y.; Zhao, J.; Chen, Z.; Wang, C.; Ding, L. Over-expression of the red plant gene R1 enhances anthocyanin production and resistance to bollworm and spider mite in cotton. Mol. Genet. Genom. 2019, 294, 469–478. [Google Scholar] [CrossRef]
- Buitrago, S.; Yang, X.; Wang, L.; Pan, R.; Zhang, W. Evolutionary analysis of anthocyanin biosynthetic genes: Insights into abiotic stress adaptation. Plant Mol. Biol. 2024, 115, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.-B.; Choi, G.; Park, Y.-I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Zhao, Y.; Yang, J.; He, Y.; Li, G.; Ma, W.; Huang, X.; Su, J. Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear ‘Yunhongli No. 1. Plant Physiol. Biochem. 2020, 154, 665–674. [Google Scholar] [CrossRef]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.-H. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin-Wang, K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple B-Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Liu, C.; Zhang, Y.; Li, Y.; Yi, K.; Zhao, X.; Cui, M.L. The Promoter Structure Differentiation of a MYB Transcription Factor RLC1 Causes Red Leaf Coloration in Empire Red Leaf Cotton under Light. PLoS ONE 2013, 8, e77891. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef]
- Espley, R.V.; Brendolise, C.; Chagné, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P.; et al. Multiple Repeats of a Promoter Segment Causes Transcription Factor Autoregulation in Red Apples. Plant Cell 2009, 21, 168–183. [Google Scholar] [CrossRef]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.a.L.; Desai, M.; Rubio, V.; Holm, M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box Protein Involved in Light-Dependent Development and Gene Expression, Undergoes COP1-Mediated Ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX Protein BBX25 Interacts with HY5, Negatively Regulating BBX22 Expression to Suppress Seedling Photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. Preview 2018, 176, 2963–2976. [Google Scholar] [CrossRef]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-Box Protein in Arabidopsis That Activates Transcription and Positively Regulates Light-Mediated Development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef]
- Araguirang, G.E.; Richter, A.S. Activation of anthocyanin biosynthesis in high light—What is the initial signal? New Phytol. 2022, 236, 2037–2043. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Fu, Y.; Li, C. Environmental Stimuli and Phytohormones in Anthocyanin Biosynthesis: A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 16415. [Google Scholar] [CrossRef]
- Pan, C.; Liao, Y.; Shi, B.; Zhang, M.; Zhou, Y.; Wu, J.; Wu, H.; Qian, M.; Bai, S.; Teng, Y. Blue light-induced MiBBX24 and MiBBX27 simultaneously promote peel anthocyanin and flesh carotenoid biosynthesis in mango. Plant Physiol. Biochem. 2025, 219, 109315. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, L.; Wang, Y.; Zhang, L.; Xu, S.; Wang, X.; He, W.; Zhang, Y.; Lin, Y.; Wang, Y. The blue light signal transduction module FaCRY1-FaCOP1-FaHY5 regulates anthocyanin accumulation in cultivated strawberry. Front. Plant Sci. 2023, 14, 1144273. [Google Scholar] [CrossRef]
- Xu, D. COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 2020, 228, 1748–1753. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, B.; Yao, T.; Shen, C.; Wen, T.; Zhang, R.; Li, Y.; Le, Y.; Li, Z.; Zhang, X. Re enhances anthocyanin and proanthocyanidin accumulation to produce red foliated cotton and brown fiber. Plant Physiol. 2022, 189, 1466–1481. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, E.; Soltani, B.M.; Yadollahi, A.; Hosseini, E. Independence of color intensity variation in red flesh apples from the number of repeat units in promoter region of the MdMYB10 gene as an allele to MdMYB1 and MdMYBA. Iran. J. Biotechnol. 2012, 10, 153–160. [Google Scholar]
- Jiang, W.; Liu, T.; Nan, W.; Jeewani, D.C.; Niu, Y.; Li, C.; Wang, Y.; Shi, X.; Wang, C.; Wang, J. Two transcription factors TaPpm1 and TaPpb1 co-regulate anthocyanin biosynthesis in purple pericarps of wheat. J. Exp. Bot. 2018, 69, 2555–2567. [Google Scholar] [CrossRef]
- Liang, A.; Zhao, J.; Li, X.; Yan, F.; Chen, Z.; Chen, X.; Wang, Y.; Li, Y.; Wang, C.; Xiao, Y. Up-regulation of GhPAP1A results in moderate anthocyanin accumulation and pigmentation in sub-red cotton. Mol. Genet. Genom. 2020, 295, 1393–1400. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Zhang, W.; Zheng, H.; Xu, X.; Li, H.; Sun, C.; Hu, H.; Zhao, W.; Ma, R. Promoter replication of grape MYB transcription factor is associated with a new red flesh phenotype. Plant Cell Rep. 2024, 43, 136. [Google Scholar] [CrossRef]
- Fu, J.; Liao, L.; Jin, J.; Lu, Z.; Sun, J.; Song, L.; Huang, Y.; Liu, S.; Huang, D.; Xu, Y.; et al. A transcriptional cascade involving BBX22 and HY5 finely regulates both plant height and fruit pigmentation in citrus. J. Integr. Plant Biol. 2024, 66, 1752–1768. [Google Scholar] [CrossRef]
- Bursch, K.; Niemann, E.T.; Nelson, D.C.; Johansson, H. Karrikins control seedling photomorphogenesis and anthocyanin biosynthesis through a HY5-BBX transcriptional module. Plant J. 2021, 107, 1346–1362. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Gong, Z.; Yan, Y.; Zhang, J.; Shao, A.; Li, H.; Wang, P.; Zhang, S.; Cheng, C.; Zhang, J. ChBBX6 and ChBBX18 are positive regulators of anthocyanins biosynthesis and carotenoids degradation in Cerasus humilis. Int. J. Biol. Macromol. 2024, 282, 137195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, J.; Tian, S.; Hao, W.; Du, L. Two B-Box Proteins, MaBBX20 and MaBBX51, Coordinate Light-Induced Anthocyanin Biosynthesis in Grape Hyacinth. Int. J. Mol. Sci. 2022, 23, 5678. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, Y.; Sun, Y.; Zhang, X.; Du, B.; Turupu, M.; Yao, Q.; Gai, S.; Tong, S.; Huang, J. Two B-box proteins, PavBBX6/9, positively regulate light-induced anthocyanin accumulation in sweet cherry. Plant Physiol. 2023, 192, 2030–2048. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2008, 16, 735–743. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Hu, C.D.; Chinenov, Y.; Kerppola, T.K. Visualization of Interactions among bZIP and Rel Family Proteins in Living Cells Using Bimolecular Fluorescence Complementation. Mol. Cell 2002, 9, 789–798. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Zhu, H.F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a Single-Repeat R3 MYB, Is a Negative Regulator of Anthocyanin Biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Zhang, K.; Fu, C.; Wu, C.; Zuo, D.; Cheng, H.; Lv, L.; Zhao, H.; Wang, J.; Wu, C.; et al. Two B-Box Proteins, GhBBX21 and GhBBX24, Antagonistically Modulate Anthocyanin Biosynthesis in R1 Cotton. Plants 2025, 14, 2367. https://doi.org/10.3390/plants14152367
Li S, Zhang K, Fu C, Wu C, Zuo D, Cheng H, Lv L, Zhao H, Wang J, Wu C, et al. Two B-Box Proteins, GhBBX21 and GhBBX24, Antagonistically Modulate Anthocyanin Biosynthesis in R1 Cotton. Plants. 2025; 14(15):2367. https://doi.org/10.3390/plants14152367
Chicago/Turabian StyleLi, Shuyan, Kunpeng Zhang, Chenxi Fu, Chaofeng Wu, Dongyun Zuo, Hailiang Cheng, Limin Lv, Haiyan Zhao, Jianshe Wang, Cuicui Wu, and et al. 2025. "Two B-Box Proteins, GhBBX21 and GhBBX24, Antagonistically Modulate Anthocyanin Biosynthesis in R1 Cotton" Plants 14, no. 15: 2367. https://doi.org/10.3390/plants14152367
APA StyleLi, S., Zhang, K., Fu, C., Wu, C., Zuo, D., Cheng, H., Lv, L., Zhao, H., Wang, J., Wu, C., Guo, X., & Song, G. (2025). Two B-Box Proteins, GhBBX21 and GhBBX24, Antagonistically Modulate Anthocyanin Biosynthesis in R1 Cotton. Plants, 14(15), 2367. https://doi.org/10.3390/plants14152367





