Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea
Abstract
1. Introduction
2. Result
2.1. Microscopic Observation of Symptoms and Resistant Spectrum Detection of Ganpi2 to Pg Isolates
2.2. RNA-Seq Data, Assessment of BGIseq500 Sequence, and Aligning to Reference Genomes
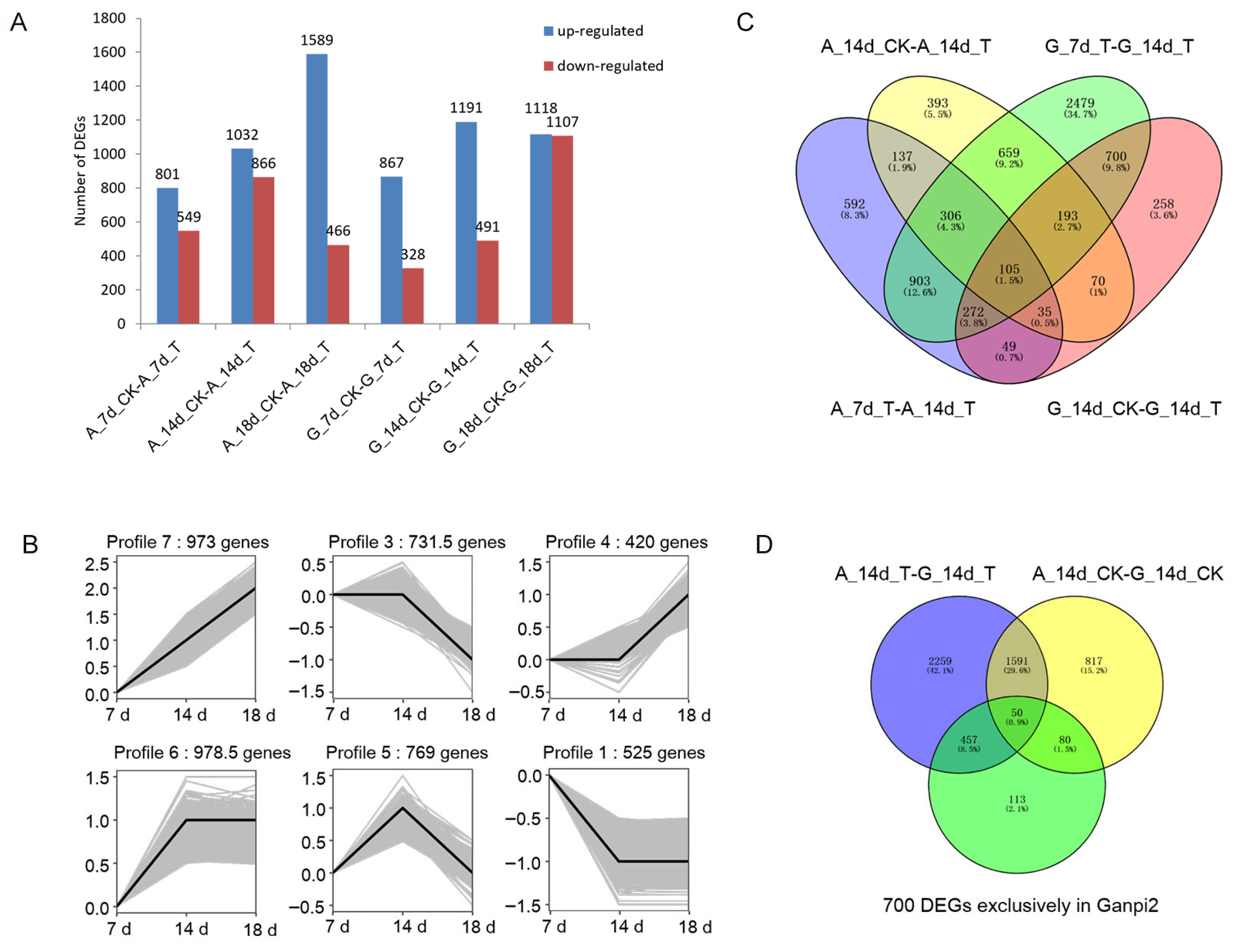
2.3. Expression Pattern Analysis and Functional Enrichment of DEGs
2.4. Detection of Candidate Genes Related to Ganpi2 Immune Response
2.4.1. DEGs Might Be Related to Resistance to Pg
2.4.2. DEGs Involved in Plant-Pathogen Interactions
2.4.3. DEGs Involved in Kinase Activation
2.4.4. DEGs Related to Resistance and TF
2.5. qRT-PCR Verification of Differentially Expressed Genes
2.6. Cloning and Characterization of HvLRR_8-1 Gene
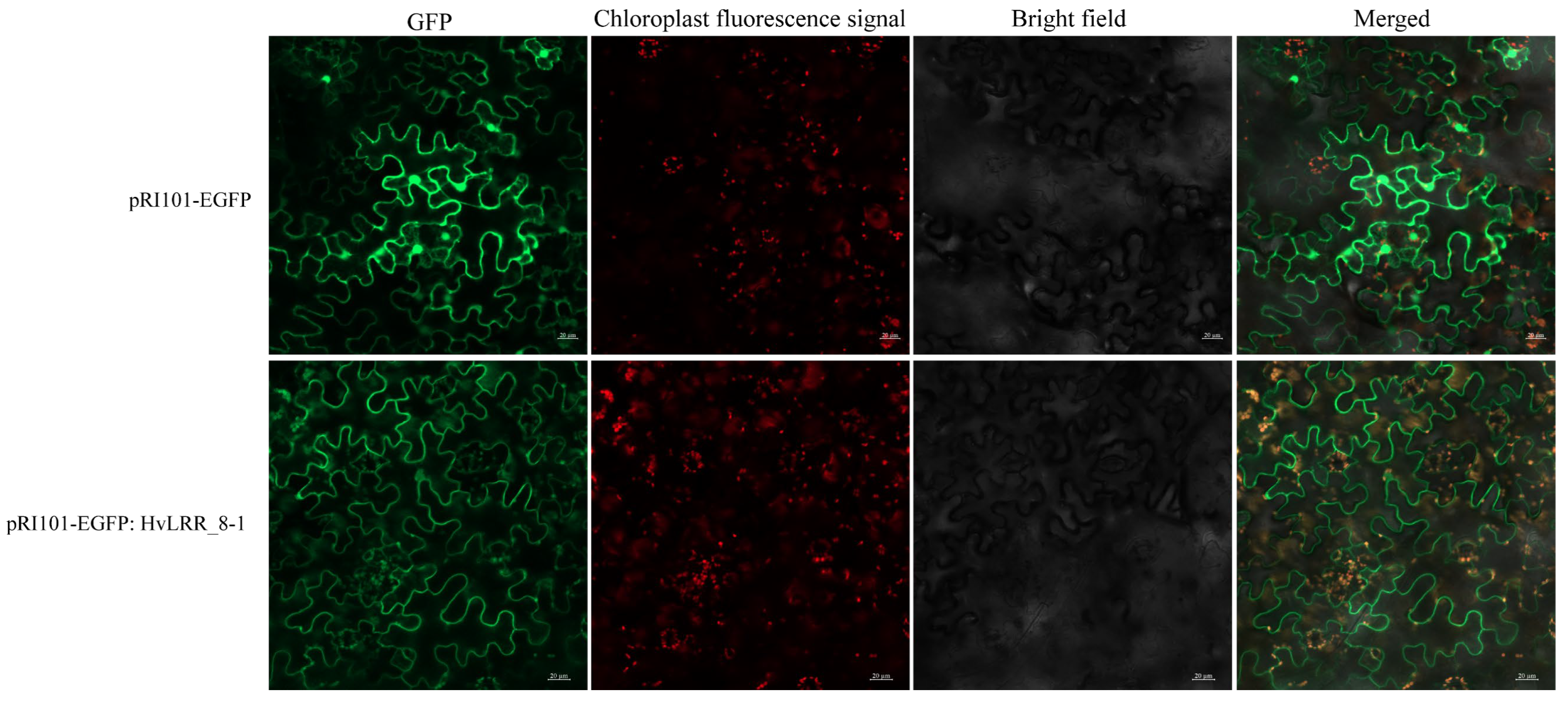
2.7. Subcellular Localization of HvLRR_8-1 Fusion Protein in Tobacco
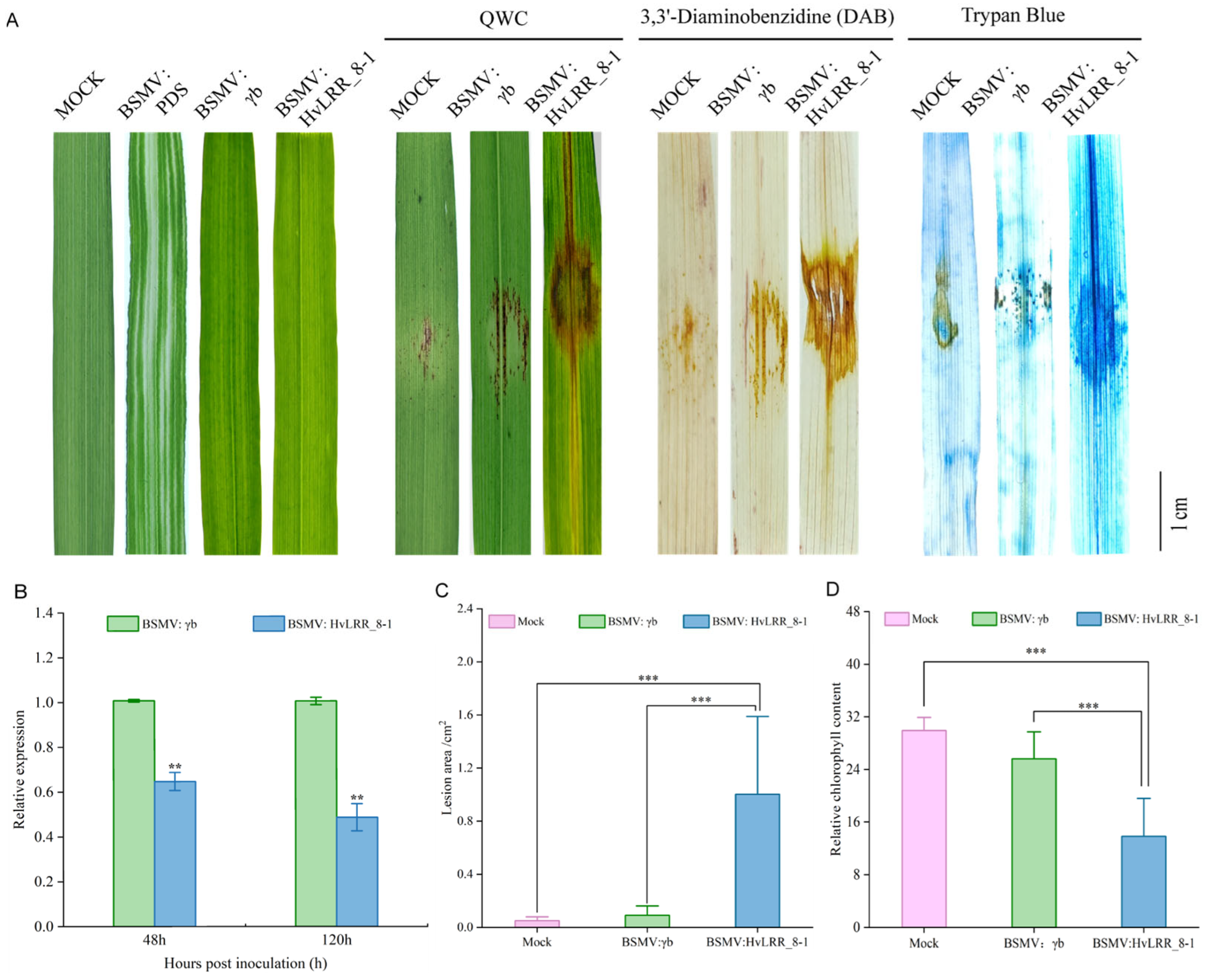
2.8. Barley HvLRR_8-1 Gene Is Crucial for Resistance to Pg
3. Discussion
3.1. Ganpi2 Has Broad-Spectrum Resistance to Pg Collected from China
3.2. Resistance/Susceptibility Temporal Analysis of Barley Embryo Responses to Pyrenophora graminea Infection
3.3. Plant-Pathogen Interaction Related to Defense Responses to Pg
3.4. Resistance Genes Participated in Response to Pg Infection
3.5. HvLRR_8-1 Gene Potentially Involved in Defense Response Against Pg Infection
4. Materials and Methods
4.1. Fungal Strains, Plant Material, Inoculation, and Symptom Observation
4.2. RNA Extraction, mRNA Library Construction, and Sequencing
4.3. RNA Sequencing Data Analysis
4.4. Quantitative PCR (qRT-PCR) Validation of Gene Expression Data
4.5. Cloning and Analysis of HvLRR_8-1
4.6. Subcellular Localization
4.7. Barley Stripe Mosaic Virus-Induced HvLRR_8-1 Gene Silencing and Plant Inoculation
4.8. qRT-PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Valè, G.; Tacconi, G.; Francia, E.; Dall’Aglio, E.; Govoni, C.; Pecchioni, N.; Arru, L.; Delogu, G.; Porta-Puglia, A.; Stanca, A. Advances in understanding barley Pyrenophora graminea interactions. In Proceedings of the 2nd International Workshop on Barley Leaf Blights, Aleppo, Syria, 7–11 April 2002. [Google Scholar]
- Bulgarelli, D.; Boselli, C.; Collins, N.C.; Consonni, G.; Stanca, A.M.; Schulzelefert, P.; Valè, G. The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS ONE 2010, 5, 12599. [Google Scholar] [CrossRef]
- Haegi, A.; Bonardi, V.; Dall’Aglio, E.; Glissant, D.; Tumino, G.; Collins, N.C.; Bulgarelli, D.; Infantino, A.; Stanca, A.M. Histological and molecular analysis of Rdg2a barley resistance to leaf stripe. Mol. Plant Pathol. 2008, 9, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Haegi, A.; Porta-Puglia, A. Purification and partial characterization of a toxic compound produced by Pyrenophora graminea. Physiol. Mol. Plant Pathol. 1995, 46, 429–444. [Google Scholar] [CrossRef]
- Guo, M.; Si, E.J.; Hou, J.J.; Yao, L.R.; Wang, J.C.; Meng, Y.X.; Ma, X.L.; Li, B.C.; Wang, H.J. Pgmiox mediates stress response and plays a critical role for pathogenicity in Pyrenophora graminea, the agent of barley leaf stripe. Plant Sci. 2025, 350, 112308. [Google Scholar] [CrossRef] [PubMed]
- Si, E.J.; Guo, M.; Liu, H.Y.; Li, C.D.; Wang, J.C.; Yao, L.R.; Meng, Y.X.; Ma, X.L.; Li, B.C.; Yang, K. The essentials of PgPG1, a polygalacturonase-encoding gene for the invasion of Pyrenophora graminea to Hordeum vulgare. Int. J. Mol. Sci. 2025, 26, 2401. [Google Scholar] [CrossRef]
- Porta-Puglia, A.; Delogu, G.; Vannacci, G. Pyrenophora graminea on winter barley seed: Effect on disease incidence and yield losses. J. Phytopathol. 1986, 117, 26–33. [Google Scholar] [CrossRef]
- Arabi, M.I.E.; Jawhar, M.; Al-Safadi, B.; Mirali, N. Yield responses of barley to leaf stripe (Pyrenophora graminea) under experimental conditions in southern syria. J. Phytopathol. 2004, 152, 519–523. [Google Scholar] [CrossRef]
- Kovacs, A.I. The effect of seed treatment with chlormequat on the control of barley leaf stripe, Drechslera graminea (Rabenh. et Schlecht.) Shoemaker, with fungicides. Crop Prot. 1982, 1, 369–372. [Google Scholar] [CrossRef]
- Delogu, G.; Portapuglia, A.; Stanca, A.M.; Vannacci, G. Interaction between barley and Pyrenophora graminea: An overview of research in Italy. Barley Wheat News 1995, 14, 29–34. [Google Scholar]
- Borgen, A.; Nielsen, B. Effect of seed treatment with acetic acid in control of seed borne diseases. In Proceedings of the BCPC Symposium. Seed Treatment Challenges and Opportunities, Brighton, UK, 12–15 November 2001; p. 76. [Google Scholar]
- Mueller, K.J.; Valè, G.; Enneking, D. Selection of resistant spring barley accessions after natural infection with leaf strips (Pyrenophora graminea) under organic farming conditions in Germany and by Sandwich test. J. Plant Pathol. 2003, 85, 9–14. [Google Scholar]
- Si, E.J.; Meng, Y.X.; Li, B.C.; Ma, X.L.; Zhang, Y.; Wang, H.J. Association analysis between barley resistance to Pyrenophora graminea and SSR markers. J. Plant Prot. 2019, 46, 1073–1085. [Google Scholar]
- Biselli, C.; Urso, S.; Bernardo, L.; Tondelli, A.; Tacconi, G.; Martino, V.; Grando, S.; Valè, G. Identification and mapping of the leaf stripe resistance gene Rdg1a in Hordeum spontaneum. Theor. Appl. Genet. 2010, 120, 1207–1218. [Google Scholar] [CrossRef]
- Tacconi, G.; Cattivelli, L.; Faccini, N.; Pecchioni, N.; Stanca, A.M.; Valè, G. Identification and mapping of a new leaf stripe resistance gene in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2001, 102, 1286–1291. [Google Scholar] [CrossRef]
- Si, E.J.; Zhang, Y.; Meng, Y.X.; Li, B.C.; Ma, X.L.; Wang, H.J. Molecular mapping of a gene for resistance to barley leaf stripe. J. Plant Prot. 2019, 46, 723–729. [Google Scholar]
- Arru, L.; Niks, R.E.; Lindhout, P.; Valé, G.; Francia, E.; Pecchioni, N. Genomic regions determining resistance to leaf stripe (Pyrenophora graminea) in barley. Genome 2002, 45, 460–466. [Google Scholar] [CrossRef]
- Arru, L.; Pecchioni, N. Isolate-specific QTLs of resistance to leaf stripe (Pyrenophora graminea) in the ‘Steptoe’ × ‘Morex’ spring barley cross. Theor. Appl. Genet. 2003, 106, 668–675. [Google Scholar] [CrossRef]
- Pecchioni, N.; Faccioli, P.; Toubia-Rahme, H.; Valè, G.; Terzi, V. Quantitative resistance to barley leaf stripe (Pyrenophora graminea) is dominated by one major locus. Theor. Appl. Genet. 1996, 93, 97–101. [Google Scholar] [CrossRef]
- Faccini, N.; Delbono, S.; Ouz, A.E.; Cattivelli, L.; Tondelli, A. Resistance of European spring 2-row barley cultivars to Pyrenophora graminea and detection of associated loci. Agronomy 2021, 11, 374. [Google Scholar] [CrossRef]
- Ghannam, A.; Alek, H.; Doumani, S.; Mansour, D.; Arabi, M.I.E. Deciphering the transcriptional regulation and spatiotemporal distribution of immunity response in barley to Pyrenophora graminea fungal invasion. BMC Genom. 2016, 17, 256. [Google Scholar] [CrossRef]
- Arabi, M.I.E.; Jawhar, M. Heterogeneity in Pyrenophora graminea as revealed by ITS-RFLP. J. Plant Pathol. 2007, 89, 391–395. [Google Scholar]
- Bayraktar, H.; Akan, K. Genetic characterization of Pyrenophora graminea isolates the reactions of some barley cultivars to leaf stripe disease under greenhouse conditions Turk. J. Agric. For. 2012, 36, 329–339. [Google Scholar]
- Si, E.J.; Meng, Y.X.; Ma, X.L.; Li, B.C.; Wang, J.C.; Ren, P.R.; Yao, L.R.; Yang, K.; Zhang, Y.; Shang, X.W.; et al. Development and characterization of microsatellite markers based on whole-genome sequences and pathogenicity differentiation of Pyrenophora graminea, the causative agent of barley leaf stripe. Eur. J. Plant Pathol. 2019, 154, 227–241. [Google Scholar] [CrossRef]
- Liang, Q.; Li, B.; Wang, J.; Ren, P.; Yao, L.; Meng, Y.; Si, E.; Shang, X.; Wang, H. PGPBS, a mitogen-activated protein kinase kinase, is required for vegetative differentiation, cell wall integrity, and pathogenicity of the barley leaf stripe fungus Pyrenophora graminea. Gene 2019, 696, 95–104. [Google Scholar] [CrossRef]
- Bennett, S. Solexa Ltd. Pharmacogenomics 2004, 5, 433–438. [Google Scholar] [CrossRef]
- Zeng, X.Q.; Luo, X.M.; Wang, Y.L.; Xu, Q.J.; Bai, L.J.; Yuan, H.J.; Tashi, N. Transcriptome sequencing in a tibetan barley landrace with high resistance to powdery mildew. Sci. World J. 2014, 2014, 594579. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Q.; Yao, Y.; Cui, Y.; Bai, Y.; An, L.; Li, X.; Ding, B.; Yao, X.; Wu, K. Transcriptome, miRNA, and degradome sequencing reveal the leaf stripe (Pyrenophora graminea) resistance genes in Tibetan hulless barley. BMC Plant Biol. 2025, 25, 7. [Google Scholar] [CrossRef]
- Lemcke, R.; Kamble, M.; Schneider, S.; Lyngkjær, M.F.; Radutoiu, S.; Wienkoop, S. Integrative transcript to proteome analysis of barley during Ramularia collo-cygni leaf spot development identified several proteins that are related to fungal recognition and infection responses. Front. Plant Sci. 2024, 15, 1367271. [Google Scholar] [CrossRef]
- Alkan, M.; Bozoğlu, T.; Yeken, M.; Yeken, M.Z.; Emiralioğlu, O.; Tekin, F.; Lahlali, R.; Türkkan, M.; Derviş, S.; Özer, G. Development of an LSU rRNA-targeted qPCR assay and transcriptional profiling of defense-related genes to elucidate barley resistance to Bipolaris sorokiniana. Physiol. Mol. Plant Pathol. 2025, 139, 102805. [Google Scholar] [CrossRef]
- Daoude, A.; Shehadah, E.; Shoaib, A.; Jawhar, M.; Arabi, M.I.E. Salicylic acid pathway changes in barley plants challenged with either a biotrophic or a necrotrophic pathogen. Cereal Res. Commun. 2019, 47, 324–333. [Google Scholar] [CrossRef]
- Saake, P.; Brands, M.; Endeshaw, A.B.; Stolze, S.C.; Westhoff, P.; Balcke, G.U.; Hensel, G.; Holton, N.; Zipfel, C.; Tissier, A.; et al. Ergosterol-induced immune response in barley involves phosphorylation of phosphatidylinositol phosphate metabolic enzymes and activation of diterpene biosynthesis. New Phytol. 2025, 246, 1236–1255. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kaushik, S.; Nandety, R.S. Evolving disease resistance genes. Curr. Opin. Plant Biol. 2025, 8, 129–134. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Huo, W.; Wang, X.X.; Ren, Z.Y.; Zhao, J.J.; Liu, Y.G.; He, K.L.; Zhang, F.; Li, W.; Jin, S.X.; et al. Origin, evolution, and diversification of the wall-associated kinase gene family in plants. Plant Cell Rep. 2023, 42, 1891–1906. [Google Scholar] [CrossRef]
- Wu, Y.; Xun, Q.; Guo, Y.; Zhang, J.; Cheng, K.; Shi, T.; Li, J. Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant. 2016, 9, 289–300. [Google Scholar] [CrossRef]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Boil. 2016, 16, 58. [Google Scholar] [CrossRef]
- Cao, Y.R.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lawrence, G.J.; Luck, J.E.; Dodds, P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 1999, 11, 495–506. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Park, S.J.; Moon, J.C.; Park, Y.C.; Kim, J.H.; Kim, D.S.; Jang, C.S. Molecular dissection of the response of a rice leucine-rich repeat receptor-like kinase (LRR-RLK) gene to abiotic stresses. J. Plant Physiol. 2014, 171, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Hao, Z.Q.; Wang, T.Y.; Chen, D.D.; Shen, L.; Zhang, G.H.; Qian, Q.; Zhu, L. Leucine-rich repeat protein family regulates stress tolerance and development in plants. Rice Sci. 2025, 32, 32–43. [Google Scholar]
- Matsushima, N.; Tanaka, T.; Enkhbayar, P.; Mikami, T.; Taga, M.; Yamada, K.; Kuroki, Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genom. 2007, 8, 124. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Angell, S.M.; Baulcombe, D.C. Technical advance: Potato virus X amplicon-mediated silencing of nuclear genes. Plant J. 1999, 20, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Liang, S.; Wang, H.Y.; Han, L.B.; Wang, F.X.; Cheng, H.Q.; Wu, X.M.; Qu, Z.L.; Wu, J.H.; Xia, G.X. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant 2015, 8, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Wang, F.; Cui, Z.; Yuan, S.T.; Meng, L.S.; Liu, D.Q.; Ma, L.R.; Wang, H.Y. Puccinia triticina avirulence protein AvrLr21 directly interacts with wheat resistance protein Lr21 to activate wheat immune response. Commun. Biol. 2024, 7, 1170. [Google Scholar] [CrossRef]
- Wan, B.Q.; Cao, A.Z.; Chen, W.; Xing, L.P.; Du, C.; Gao, X.Q.; Zhang, S.Z.; Zhang, R.Q.; Shen, W.B.; Wang, H.Y. Two members of TaRLK family confer powdery mildew resistance in common wheat. BMC Plant Biol. 2016, 16, 27. [Google Scholar] [CrossRef]
- Hein, I.; Barciszewska-Pacak, M.; Hrubikova, K.; Williamson, S.; Dinesen, M.; Soenderby, I.E.; Sundar, S.; Jarmolowski, A.; Shirasu, K.; Lacomme, C. Virus-Induced Gene Silencing-based functional characterization of genes associated with Powdery Mildew resistance in barley. Plant Physiol. 2005, 138, 2155–2164. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, L.; Feng, S.; Zhao, Z.; Wang, X.; Gao, H. Transcriptome analysis of apple leaves in response to Powdery Mildew (Podosphaera leucotricha) infection. Int. J. Mol. Sci. 2019, 20, 2326. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new END script server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, J.; Voegele, R.T.; Zhang, J.; Duan, Y.H.; Luo, H.Y.; Kang, Z.S. Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in Puccinia striiformis f. sp. tritici using a host-induced RNAi system. PLoS ONE 2012, 7, 49262. [Google Scholar] [CrossRef]
- Lehmann, S.; Serrano, M.; L’Haridon, F.; Tjamos, S.E.; Metraux, J.P. Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 2015, 112, 54–62. [Google Scholar] [CrossRef]
- Si, E.J.; Meng, Y.X.; Ma, X.L.; Li, B.C.; Wang, J.C.; Yao, L.R.; Yang, K.; Zhang, Y.; Shang, X.W.; Wang, H.J. Genome resource for barley leaf stripe pathogen Pyrenophora graminea. Plant Dis. 2020, 104, 320–322. [Google Scholar] [CrossRef]
- Si, E.J.; Yang, S.; Li, B.C.; Ma, X.L.; Wang, S.; Wang, H.J. Pathogenic analysis, rDNA-ITS and genetic diversity of Pyrenophora garminea in Gansu Province. J. Plant Prot. 2017, 44, 84–92. [Google Scholar]
- Hou, S.; Zhang, C.; Yang, Y.; Wu, D. Recent advances in plant immunity: Recognition, signaling, response, and evolution. Biol. Plant. 2013, 57, 11–25. [Google Scholar] [CrossRef]
- Zhang, D.; Qiu, J. Interactions and co-evolution of plants and pathogens. Chin. Sci. Bull. 2017, 62, 1214–1220. [Google Scholar] [CrossRef][Green Version]
- Moeder, W.; Urquhart, W.; Ung, H.; Yoshioka, K. The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant. 2011, 4, 442–452. [Google Scholar] [CrossRef]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and–susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Fang, Z.; Li, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. Comparative transcriptome analysis between broccoli (Brassica oleracea var. italica) and wild cabbage (Brassica macrocarpa Guss.) in response to Plasmodiophora brassicae during different infection stages. Front. Plant Sci. 2016, 7, 1929. [Google Scholar]
- Do Heo, W.; Lee, S.H.; Kim, J.C. Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. USA 1999, 96, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Mitsuhara, I.; Ito, N.; Seo, S.; Kamada, H.; Ohashi, Y. Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur. J. Biochem. 2001, 268, 3916–3929. [Google Scholar] [CrossRef]
- Yang, X.; Deng, F.; Ramonell, K.M. Receptor-like kinases and receptor-like proteins: Keys to pathogen recognition and defense signaling in plant innate immunity. Front. Biol. 2012, 2, 155–166. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A. Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci. 2008, 13, 145–150. [Google Scholar] [CrossRef]
- Llorente, F.; Alonso, B.C.; Sánchez, R.C.; Jorda, L.; Molina, A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 2005, 43, 165–180. [Google Scholar] [CrossRef]
- Lourdes, G.G.; Thomas, B. FLS2: An LRR receptor-like kinase Involved in the perception of the bacterial elicitor llagellin in Arabidopsis. Mol. Cell 2000, 6, 1003–1011. [Google Scholar]
- Nishmitha, K.; Singh, R.; Akhtar, J.; Bashyal, B.M.; Dubey, S.C.; Deeba Kamil, A.T. Expression profiling and characterization of key RGA involved in lentil Fusarium wilt race 5 resistance. World J. Microbiol. Biotechnol. 2023, 39, 306. [Google Scholar] [CrossRef] [PubMed]
- Mubassir, M.H.M.; Naser, M.A.; Abdul-Wahab, M.F.; Jawad, T.; Alvy, R.I.; Hamdan, S. Comprehensive in silico modeling of the rice plant PRR Xa21 and its interaction with RaxX21-sY and OsSERK2. RSC Adv. 2020, 27, 15656–16220. [Google Scholar]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Bolger, A.M.; Marc, L.; Bjoern, U. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. String Tie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.V.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, 121. [Google Scholar] [CrossRef]
- Sanseverino, W.; Roma, G.; De Simone, M.; Faino, L.; Melito, S.; Stupka, E.; Frusciante, L.; Ercolano, M.R. PRGdb: A bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2010, 38, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Lüpken, T.; Habekuss, A.; Hensel, G.; Steuernagel, B.; Kilian, B.; Ariyadasa, R.; Himmelbach, A.; Kumlehn, J.; Scholz, U. PROTEIN DISULFIDE ISOMERASE LIKE 5-1 is a susceptibility factor to plant viruses. Proc. Natl. Acad. Sci. USA 2014, 111, 2104–2109. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, B.; Liu, W.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef] [PubMed]



| Isolate Name | Geographic Origin | Disease Incidence (%) | Reaction Types | Isolate Name | Geographic Origin | Disease Incidence (%) | Reaction Types |
|---|---|---|---|---|---|---|---|
| BS-14095-2-1 | Hefei, Anhui | 0.00 | I | BS-13007-7-1 | Eryuan, Dali, Yunnan | 0.00 | I |
| BS-13032-2 | Hohhot, Inner Mongolia | 0.00 | I | BS-13009-8-1 | Eryuan, Dali, Yunnan | 0.00 | I |
| BS-13032-3 | Hohhot, Inner Mongolia | 0.00 | I | BS-13003-1 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13020-1 | Huzhu, Haidong, Qinghai | 0.00 | I | BS-13003-2 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13019-4 | Huangzhogn, Xining, Qinghai | 0.00 | I | BS-13002-3 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13014-5 | Menyuan, Haibei, Qinghai | 0.00 | I | BS-13002-2 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13014-2 | Menyuan, Haibei, Qinghai | 0.00 | I | BS-13004-2-1 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13015-1-2 | Menyuan, Haibei, Qinghai | 0.00 | I | BS-13004-2-3 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13016-3 | Menyuan, Haibei, Qinghai | 0.00 | I | BS-13004-4-2 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13018-3-4 | Haiyan, Haibei, Qinghai | 0.00 | I | BS-13004-3-1 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13018-1-5 | Haiyan, Haibei, Qinghai | 0.00 | I | BS-13004-1-1 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13001-2 | Haiyan, Haibei, Qinghai | 2.56 | HR | BS-13001-2 | Jianchuan, Dali, Yunnan | 0.00 | I |
| BS-13055-2-3 | Dechang, Liangshan, Sichuan | 0.00 | I | BS-13012-6-5 | Panlong, Kunming, Yunnan | 0.00 | I |
| BS-13055-2-1 | Dechang, Liangshan, Sichuan | 0.00 | I | BS-13012-6-1 | Panlong, Kunming, Yunnan | 0.00 | I |
| Code | Primer | Sequence (5′–3′) | Purpose |
|---|---|---|---|
| 1 | HvLRR_8-1-F | ATGTGTGATAGAAAGCACATG | Cloning HvLRR_8-1 gene |
| 2 | HvLRR_8-1-R | GCTGTGCAACGCTGCAAATG | |
| 3 | 35S-F | GACGCACAATCCCACTATCC | Clone HvLRR_8-1 to EGFP for subcellular localization assay in N. benthamiana |
| 4 | GFP-JR | GGGTCAGCTTGCCGTAGGTG | |
| 5 | γ-HvLRR-F | TTGCTCCTAACATCAATACC | Clone HvLRR_8-1 gene silencing fragment was used for the construction of the BSMV-VIGS clone. (The red font represents the homologous recombination arm of the connection carrier) |
| 6 | γ-HvLRR-R | CCCAGCAAGTATTGGTAAGC | |
| 7 | BSMV: HvLRR-F | CGCTTCCAAGCATCCAAACTT | The primers for the HvLRR_8-1 gene were used for qRT-PCR detection. |
| 8 | BSMV: HvLRR-R | GGCAAGCGAAGTGGGAATTT | |
| 9 | Actin-F | GCTGACCGTATGAGCAAGGA | Expression level analysis of internal control gene Actin in barley |
| 10 | Actin-R | GGAAAGTGCTGAGTGAGGCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Guo, M.; Li, Y.; Yang, Q.; Zhang, H.; Li, C.; Wang, J.; Meng, Y.; Ma, X.; Li, B.; et al. Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea. Plants 2025, 14, 2350. https://doi.org/10.3390/plants14152350
Yang W, Guo M, Li Y, Yang Q, Zhang H, Li C, Wang J, Meng Y, Ma X, Li B, et al. Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea. Plants. 2025; 14(15):2350. https://doi.org/10.3390/plants14152350
Chicago/Turabian StyleYang, Wenjuan, Ming Guo, Yan Li, Qinglan Yang, Huaizhi Zhang, Chengdao Li, Juncheng Wang, Yaxiong Meng, Xiaole Ma, Baochun Li, and et al. 2025. "Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea" Plants 14, no. 15: 2350. https://doi.org/10.3390/plants14152350
APA StyleYang, W., Guo, M., Li, Y., Yang, Q., Zhang, H., Li, C., Wang, J., Meng, Y., Ma, X., Li, B., Yao, L., Zhang, H., Yang, K., Shang, X., Si, E., & Wang, H. (2025). Transcriptome Analysis and Functional Characterization of the HvLRR_8-1 Gene Involved in Barley Resistance to Pyrenophora graminea. Plants, 14(15), 2350. https://doi.org/10.3390/plants14152350







