Metabarcoding Analysis Reveals Microbial Diversity and Potential Soilborne Pathogens Associated with Almond Dieback and Decline
Abstract
1. Introduction
2. Materials and Methods
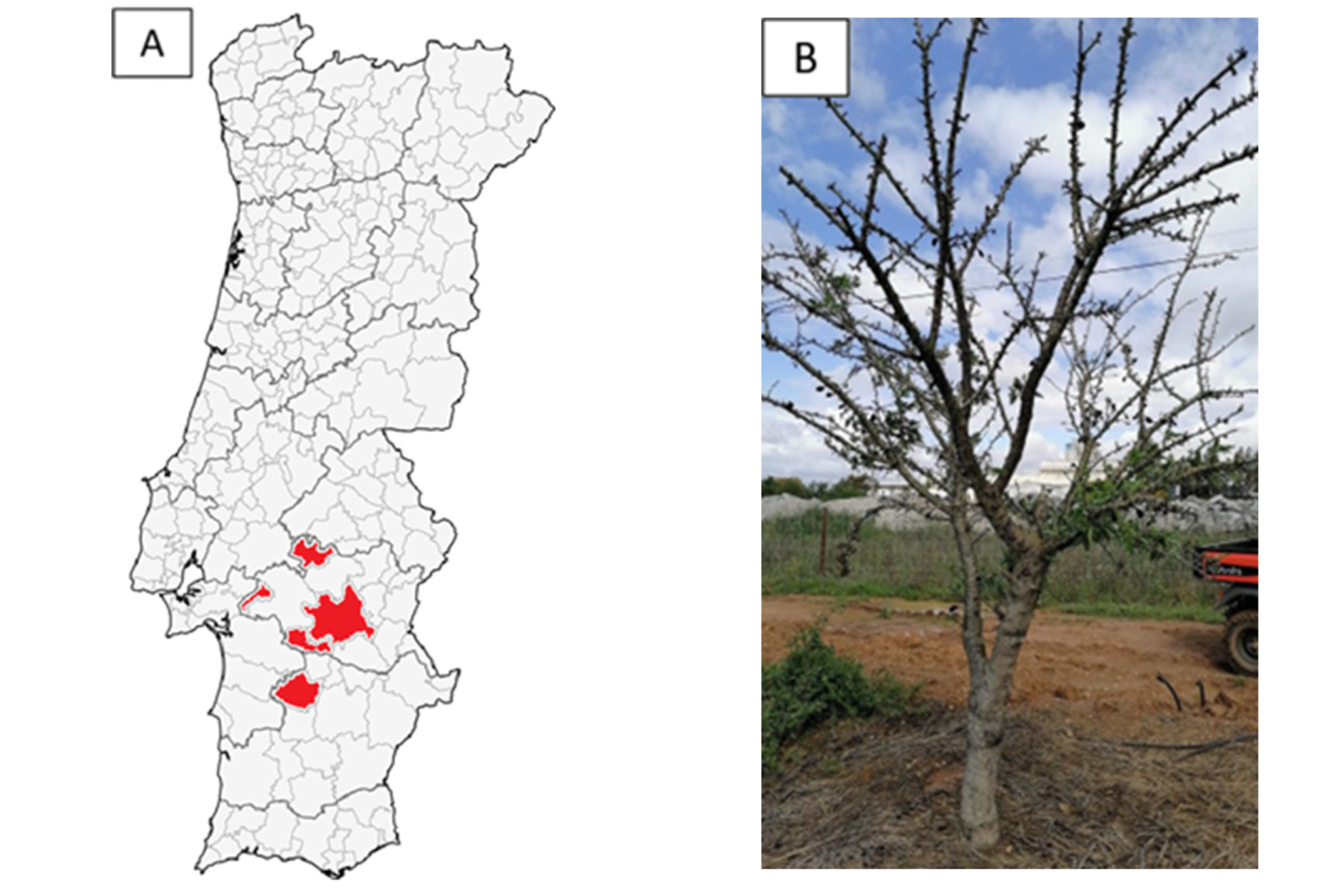
2.1. Sampling Conditions
2.2. Sample Processing and Fungi Isolation
2.3. DNA Extraction, PCR, and Sanger Sequencing of Plate Isolates
2.4. Sample DNA Extraction and Metabarcoding
2.5. Bioinformatic and Statistical Analyses
3. Results
3.1. Isolate Sanger Sequencing
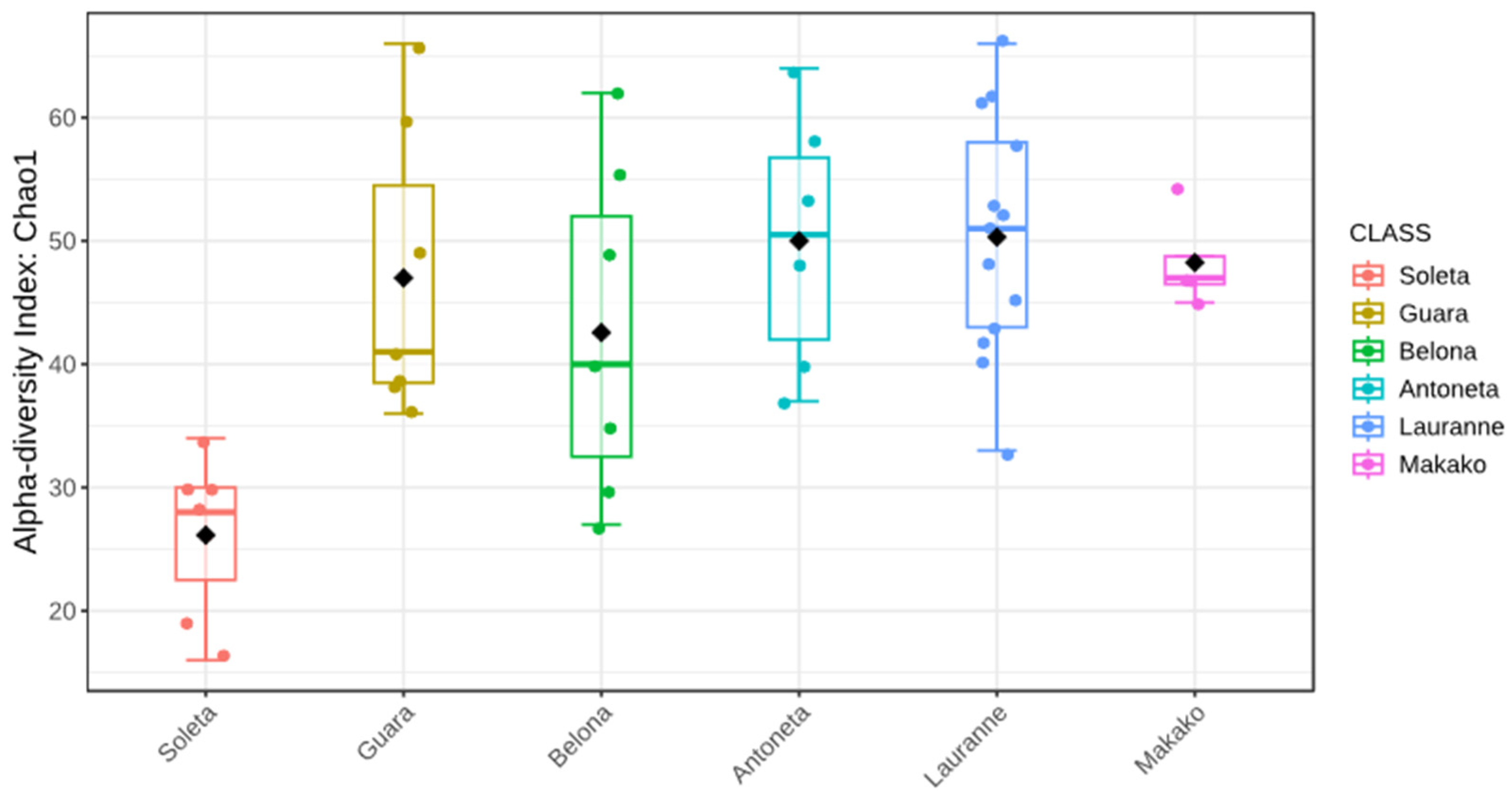
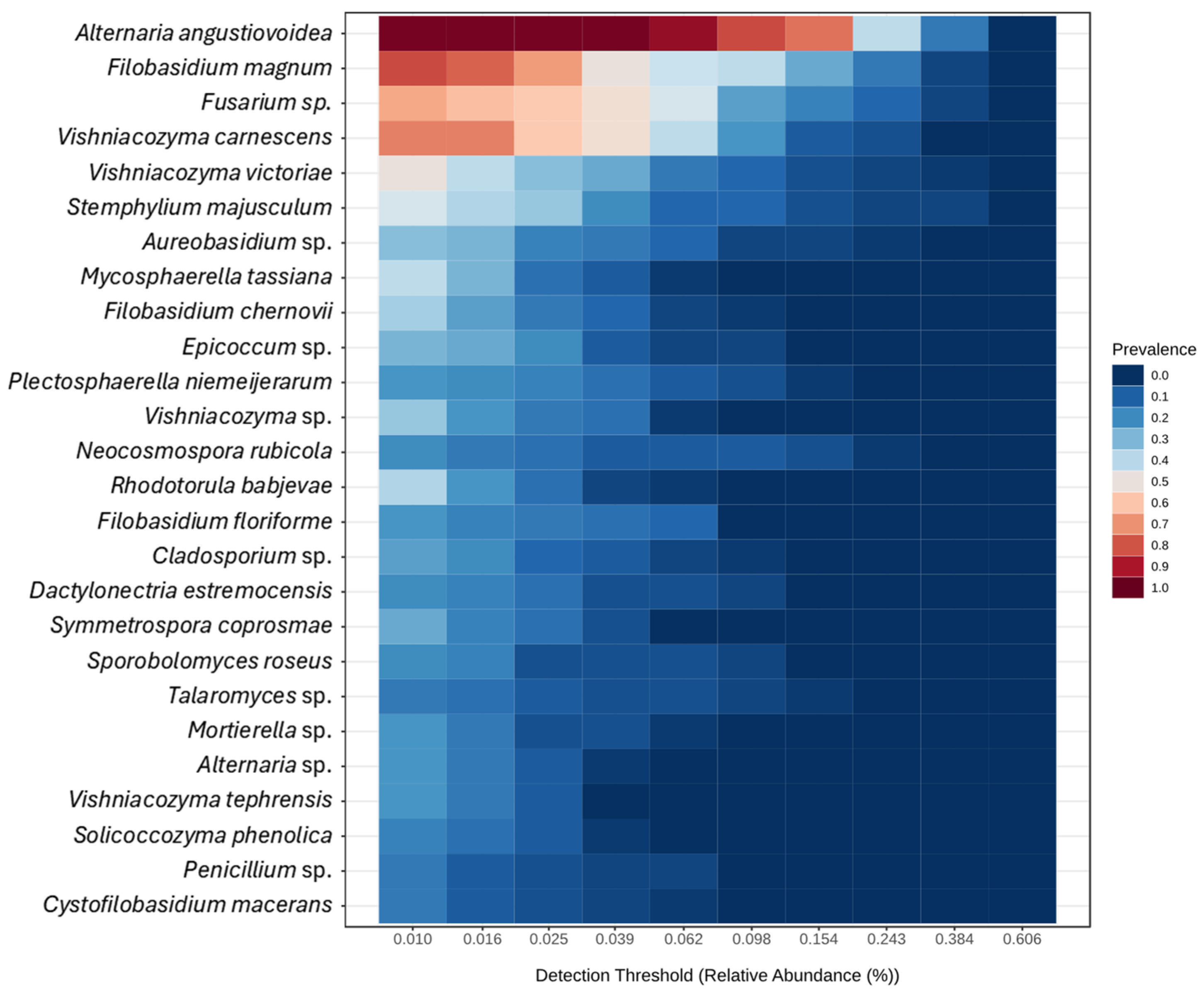
3.2. Metabarcoding of the Fungal Communities in Almond Trees
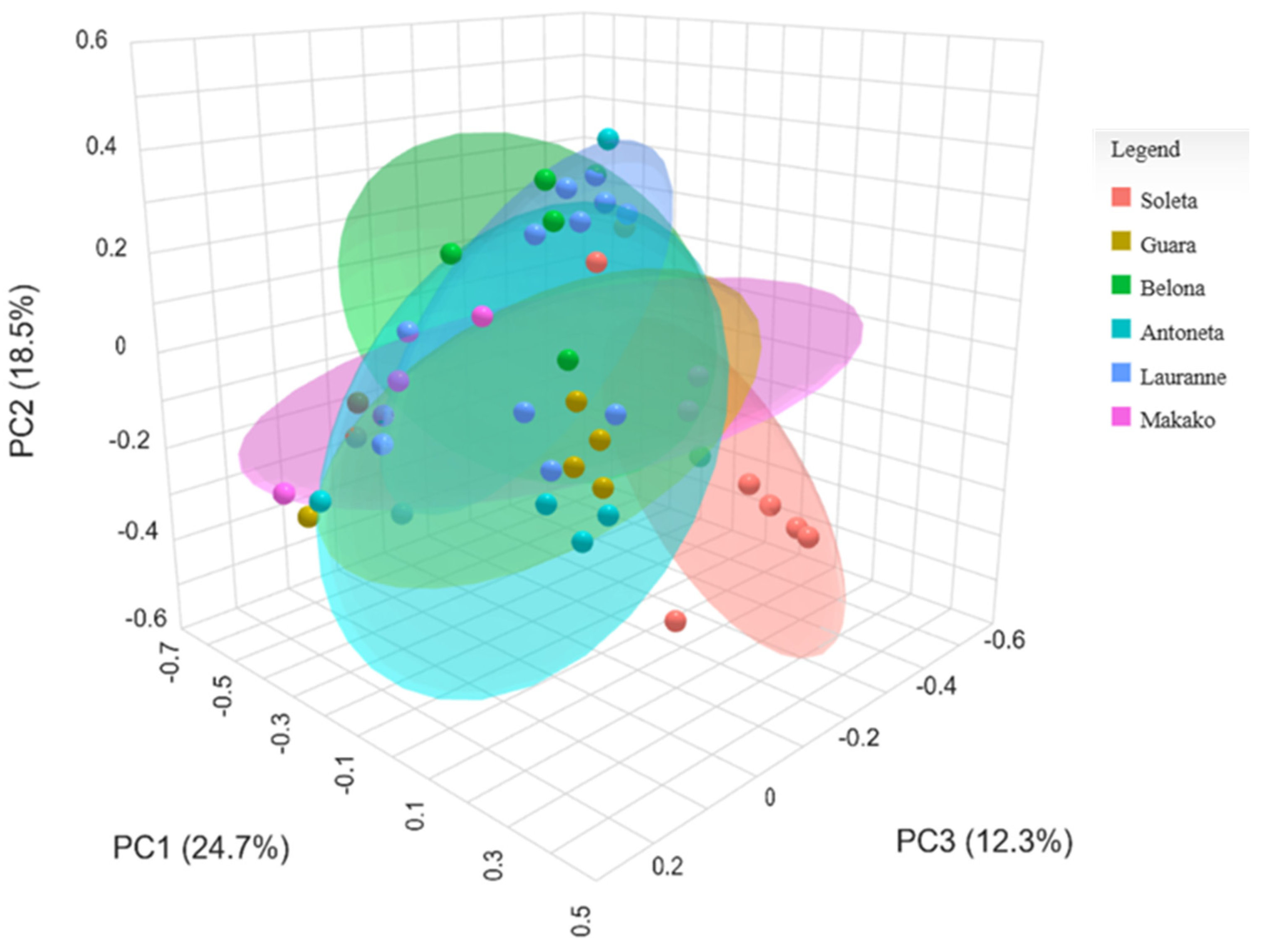
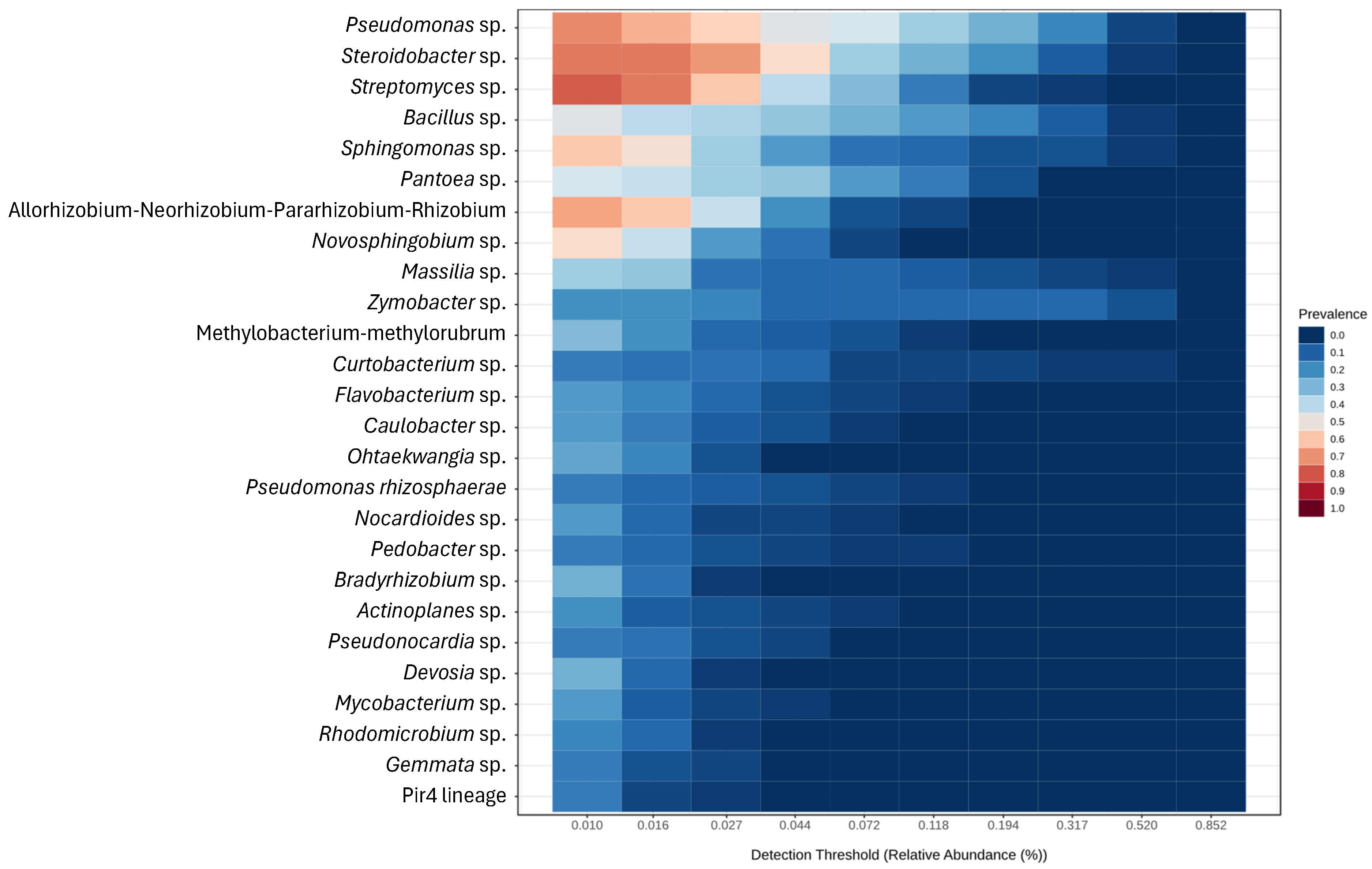
3.3. Metabarcoding of the Bacterial Communities in Almond Trees
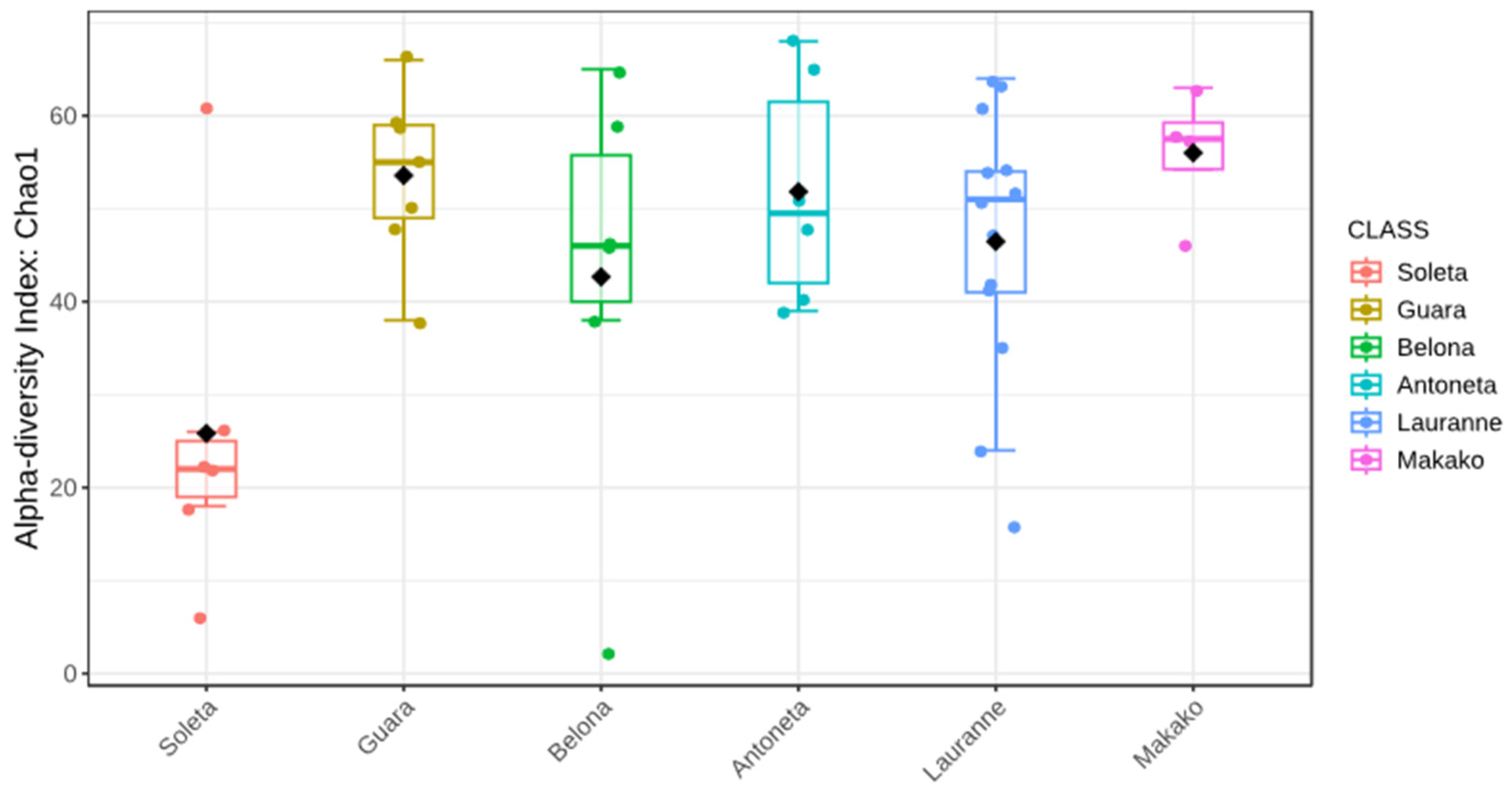
3.4. Metabarcoding of the Soil Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalak, L.F. Almond: Multiple uses of a mediterranean heritage. Acta Hortic. 2014, 1032, 29–36. [Google Scholar] [CrossRef]
- Martins, M.; Tenreiro, R.; Oliveira, M.M. Genetic relatedness of Portuguese almond cultivars assessed by RAPD and ISSR markers. Plant Cell Rep. 2003, 22, 71–78. [Google Scholar] [CrossRef]
- FAO. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 27 March 2024).
- Lusa, A. Almond Production in Portugal Increases, but Algarve Loses Representation. Available online: https://www.sulinformacao.pt/en/2024/03/producao-de-amendoa-em-portugal-aumenta-mas-algarve-perde-representatividade/ (accessed on 18 March 2025).
- Barata, C. Portugal já é auto-suficiente em frutos secos: Amendoal invadiu o sedento Alentejo. Público 2023. [Google Scholar]
- Campos, C.R.; Sousa, B.; Silva, J.; Braga, M.; Araújo, S.D.; Sales, H.; Pontes, R.; Nunes, J. Positioning Portugal in the Context of World Almond Production and Research. Agriculture 2023, 13, 1716. [Google Scholar] [CrossRef]
- Denman, S.; Brown, N.; Vanguelova, E.; Crampton, B. Chapter 14—Temperate Oak Declines: Biotic and abiotic predisposition drivers. In Forest Microbiology; Asiegbu, F.O., Kovalchuk, A., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 239–263. [Google Scholar] [CrossRef]
- Spies, C.F.J.; Mostert, L.; Carlucci, A.; Moyo, P.; van Jaarsveld, W.J.; du Plessis, I.L.; van Dyk, M.; Halleen, F. Dieback and decline pathogens of olive trees in South Africa. Persoonia 2020, 45, 196–220. [Google Scholar] [CrossRef]
- Gramaje, D.; Agustí-Brisach, C.; Pérez-Sierra, A.; Moralejo, E.; Olmo, D.; Mostert, L.; Damm, U.; Armengol, J. Fungal trunk pathogens associated with wood decay of almond trees on Mallorca (Spain). Persoonia 2012, 28, 1–13. [Google Scholar] [CrossRef]
- Agustí-Brisach, C.; Moldero, D.; Raya, M.d.C.; Lorite, I.J.; Orgaz, F.; Trapero, A. Water Stress Enhances the Progression of Branch Dieback and Almond Decline under Field Conditions. Plants 2020, 9, 1213. [Google Scholar] [CrossRef]
- Bettenfeld, P.; Fontaine, F.; Trouvelot, S.; Fernandez, O.; Courty, P.-E. Woody Plant Declines. What’s Wrong with the Microbiome? Trends Plant Sci. 2020, 25, 381–394. [Google Scholar] [CrossRef]
- Eljounaidi, K.; Lee, S.K.; Bae, H. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases—Review and future prospects. Biol. Control 2016, 103, 62–68. [Google Scholar] [CrossRef]
- Scortichini, M.; Manetti, G.; Brunetti, A.; Lumia, V.; Sciarroni, L.; Pilotti, M. Xylella fastidiosa subsp. pauca, Neofusicoccum spp. and the Decline of Olive Trees in Salento (Apulia, Italy): Comparison of Symptoms, Possible Interactions, Certainties and Doubts. Plants 2023, 12, 3593. [Google Scholar] [CrossRef]
- Goura, K.; Lahlali, R.; Bouchane, O.; Baala, M.; Radouane, N.; Kenfaoui, J.; Ezrari, S.; El Hamss, H.; El Alami, N.; Amiri, S.; et al. Identification and Characterization of Fungal Pathogens Causing Trunk and Branch Cankers of Almond Trees in Morocco. Agronomy 2023, 13, 130. [Google Scholar] [CrossRef]
- van der Merwe, R.; Halleen, F.; van Dyk, M.; Jacobs, V.G.; Mostert, L. Occurrence of Canker and Wood Rot Pathogens on Stone Fruit Propagation Material and Nursery Trees in the Western Cape of South Africa. Plant Dis. 2021, 105, 3586–3599. [Google Scholar] [CrossRef]
- Azenzem, R.; Koussa, T.; Alfeddy, M.N. Root and crown rot caused by oomycetes: An emerging threat to olive trees. Trop. Plant Pathol. 2024, 49, 331–345. [Google Scholar] [CrossRef]
- Salgado-Salazar, C.; Rossman, A.Y.; Chaverri, P. The genus Thelonectria (Nectriaceae, Hypocreales, Ascomycota) and closely related species with cylindrocarpon-like asexual states. Fungal Divers. 2016, 80, 411–455. [Google Scholar] [CrossRef]
- Devkota, P.; Hammerschmidt, R. The infection process of Armillaria mellea and Armillaria solidipes. Physiol. Mol. Plant Pathol. 2020, 112, 101543. [Google Scholar] [CrossRef]
- La Porta, N.; Hietala, A.M.; Baldi, P. Chapter 6—Bacterial diseases in forest trees. In Forest Microbiology; Asiegbu, F.O., Kovalchuk, A., Eds.; Academic Press: Cambridge, MA, USA, 2023; Volume 3, pp. 139–166. [Google Scholar]
- Schilling, M.; Farine, S.; Péros, J.-P.; Bertsch, C.; Gelhaye, E. Chapter Six—Wood degradation in grapevine diseases. In Advances in Botanical Research; Morel-Rouhier, M., Sormani, R., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 99, pp. 175–207. [Google Scholar]
- Lamichhane, J.R.; Venturi, V. Synergisms between microbial pathogens in plant disease complexes: A growing trend. Front. Plant Sci. 2015, 6, 385. [Google Scholar] [CrossRef]
- Patel, D.; Shittu, T.A.; Baroncelli, R.; Muthumeenakshi, S.; Osborne, T.H.; Janganan, T.K.; Sreenivasaprasad, S. Genome Sequence of the Biocontrol Agent Coniothyrium minitans Conio (IMI 134523). Mol. Plant-Microbe Interact. 2020, 34, 222–225. [Google Scholar] [CrossRef]
- Whitelaw-Weckert, M.A.; Rahman, L.; Appleby, L.M.; Hall, A.; Clark, A.C.; Waite, H.; Hardie, W.J. Co-infection by Botryosphaeriaceae and lyonectria spp. fungi during propagation causes decline of young grafted grapevines. Plant Pathol. 2013, 62, 1226–1237. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Zhang, W.; Zhang, J.; Wang, H.; Peng, J.; Wang, X.; Yan, J. Belowground microbiota analysis indicates that Fusarium spp. exacerbate grapevine trunk disease. Environ. Microbiome 2023, 18, 29. [Google Scholar] [CrossRef]
- Patanita, M.; Félix, M.d.R.; Ribeiro, J.A.; Varanda, C.M.R.; Albuquerque, A.; Materatski, P.; Garrido, N.; Campos, M.D. Insights into Grapevine Defence Response Against Fungal and Oomycete Diseases Towards a Sustainable Plant Breeding. In Plant Pathogen Interaction; Verma, P.K., Mishra, S., Srivastava, V., Mehrotra, S., Eds.; Springer Nature: Singapore, 2023; pp. 119–160. [Google Scholar]
- Almeida, A.B.; Concas, J.; Campos, M.D.; Materatski, P.; Varanda, C.; Patanita, M.; Murolo, S.; Romanazzi, G.; Félix, M.D.R. Endophytic Fungi as Potential Biological Control Agents against Grapevine Trunk Diseases in Alentejo Region. Biology 2020, 9, 420. [Google Scholar] [CrossRef]
- Aragona, M.; Haegi, A.; Valente, M.T.; Riccioni, L.; Orzali, L.; Vitale, S.; Luongo, L.; Infantino, A. New-Generation Sequencing Technology in Diagnosis of Fungal Plant Pathogens: A Dream Comes True? J. Fungi 2022, 8, 737. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. TIG 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. MicrobiomeAnalyst 2.0: Comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef]
- Patanita, M.; Albuquerque, A.; Campos, M.D.; Materatski, P.; Varanda, C.M.R.; Ribeiro, J.A.; Félix, M.d.R. Metagenomic Assessment Unravels Fungal Microbiota Associated to Grapevine Trunk Diseases. Horticulturae 2022, 8, 288. [Google Scholar] [CrossRef]
- Bekris, F.; Vasileiadis, S.; Papadopoulou, E.; Samaras, A.; Testempasis, S.; Gkizi, D.; Tavlaki, G.; Tzima, A.; Paplomatas, E.; Markakis, E.; et al. Grapevine wood microbiome analysis identifies key fungal pathogens and potential interactions with the bacterial community implicated in grapevine trunk disease appearance. Environ. Microbiome 2021, 16, 23. [Google Scholar] [CrossRef]
- Belair, M.; Pensec, F.; Jany, J.-L.; Le Floch, G.; Picot, A. Profiling Walnut Fungal Pathobiome Associated with Walnut Dieback Using Community-Targeted DNA Metabarcoding. Plants 2023, 12, 2383. [Google Scholar] [CrossRef]
- López-Moral, A.; Antón-Domínguez, B.I.; Lovera, M.; Arquero, O.; Trapero, A.; Agustí-Brisach, C. Identification and pathogenicity of Fusarium species associated with wilting and crown rot in almond (Prunus dulcis). Sci. Rep. 2024, 14, 5720. [Google Scholar] [CrossRef]
- Antón-Domínguez, B.I.; López-Moral, A.; Raya, M.C.; Lovera, M.; Melgar, S.; Roca, L.F.; Arquero, O.; Trapero, A.; Agustí-Brisach, C. Fungal Pathogens Associated with Almond Decline Syndrome, an Emerging Disease Complex in Intensive Almond Crops in Southern Spain. Plant Dis. 2023, 107, 3737–3753. [Google Scholar] [CrossRef]
- Markakis, E.A.; Soultatos, S.K.; Kanetis, L.; Goumas, D.E. First Report of Stem Canker of Almond Trees Caused by Fusarium solani in Greece. Plant Dis. 2021, 105, 2724. [Google Scholar] [CrossRef]
- Holland, L.A.; Trouillas, F.P.; Nouri, M.T.; Lawrence, D.P.; Crespo, M.; Doll, D.A.; Duncan, R.A.; Holtz, B.A.; Culumber, C.M.; Yaghmour, M.A.; et al. Fungal Pathogens Associated With Canker Diseases of Almond in California. Plant Dis. 2021, 105, 346–360. [Google Scholar] [CrossRef]
- Ören, E.; Bayraktar, H. Identifying fungi responsible for trunk and scaffold diseases in almonds in Türkiye. Physiol. Mol. Plant Pathol. 2025, 138, 102729. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118. [Google Scholar] [CrossRef]
- Riaz, M.; Akhtar, N.; Msimbira, L.A.; Antar, M.; Ashraf, S.; Khan, S.N.; Smith, D.L. Neocosmospora rubicola, a stem rot disease in potato: Characterization, distribution and management. Front. Microbiol. 2022, 13, 953097. [Google Scholar] [CrossRef]
- Zheng, F.; Xu, G.; Zheng, F.Q.; Ding, X.F.; Xie, C.P. Neocosmospora rubicola Causing Stem Rot of Pitaya (Hylocereus costaricensis) in China. Plant Dis. 2018, 102, 2653. [Google Scholar] [CrossRef]
- Nalim, F.A.; Samuels, G.J.; Wijesundera, R.L.; Geiser, D.M. New species from the Fusarium solani species complex derived from perithecia and soil in the old World tropics. Mycologia 2011, 103, 1302–1330. [Google Scholar] [CrossRef]
- Gramaje, D.; Berlanas, C.; Martínez-Diz, M.D.P.; Diaz-Losada, E.; Antonielli, L.; Beier, S.; Gorfer, M.; Schmoll, M.; Compant, S. Comparative Genomic Analysis of Dactylonectria torresensis Strains from Grapevine, Soil and Weed Highlights Potential Mechanisms in Pathogenicity and Endophytic Lifestyle. J. Fungi 2020, 6, 255. [Google Scholar] [CrossRef]
- Berlanas, C.; Ojeda, S.; López-Manzanares, B.; Andrés-Sodupe, M.; Bujanda, R.; Del Pilar Martínez-Diz, M.; Díaz-Losada, E.; Gramaje, D. Occurrence and Diversity of Black-Foot Disease Fungi in Symptomless Grapevine Nursery Stock in Spain. Plant Dis. 2020, 104, 94–104. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Santos, J.; Phillips, A.J. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 2012, 28, 34–48. [Google Scholar] [CrossRef]
- Raimondo, M.; Carlucci, A. Characterization and pathogenicity of Plectosphaerella spp. collected from basil and parsley in Italy. Phytopathol. Mediterr. 2018, 57, 284–295. [Google Scholar] [CrossRef]
- Heyman, F.; Blair, J.E.; Persson, L.; Wikström, M. Root Rot of Pea and Faba Bean in Southern Sweden Caused by Phytophthora pisi sp. nov. Plant Dis. 2013, 97, 461–471. [Google Scholar] [CrossRef]
- Browne, G.T.; Schmidt, L.S.; Brar, G. First Report of Phytophthora niederhauserii Causing Crown Rot of Almond (Prunus dulcis) in California. Plant Dis. 2015, 99, 1863. [Google Scholar] [CrossRef]
- Browne, G.T.; Viveros, M.A. Lethal Cankers Caused by Phytophthora spp. in Almond Scions: Specific Etiology and Potential Inoculum Sources. Plant Dis. 1999, 83, 739–745. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Nouri, M.T.; Bourret, T.B. Identification and Characterization of Phytophthora Species Associated with Crown and Root Rot of Pistachio Trees in California. Plant Dis. 2021, 106, 197–206. [Google Scholar] [CrossRef]
- Hossain, M.M.; Sultana, F.; Li, W.; Tran, L.P.; Mostofa, M.G. Sclerotinia sclerotiorum (Lib.) de Bary: Insights into the Pathogenomic Features of a Global Pathogen. Cells 2023, 12, 1063. [Google Scholar] [CrossRef]
- Xia, S.; Xu, Y.; Hoy, R.; Zhang, J.; Qin, L.; Li, X. The Notorious Soilborne Pathogenic Fungus Sclerotinia sclerotiorum: An Update on Genes Studied with Mutant Analysis. Pathogens 2019, 9, 27. [Google Scholar] [CrossRef]
- Liang, X.; Rollins, J.A. Mechanisms of Broad Host Range Necrotrophic Pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140. [Google Scholar] [CrossRef]
- Hay, F.; Stricker, S.; Gossen, B.D.; McDonald, M.R.; Heck, D.; Hoepting, C.; Sharma, S.; Pethybridge, S. Stemphylium Leaf Blight: A Re-Emerging Threat to Onion Production in Eastern North America. Plant Dis. 2021, 105, 3780–3794. [Google Scholar] [CrossRef]
- Kowalski, T.; Bartnik, C. Cristulariella depraedans as causal agent of leaf spots of maple and other trees and shrubs. Acta Mycol. 2008, 43, 5–12. [Google Scholar] [CrossRef]
- Johnson, D.A.; Pimentel, G.; Dugan, F.M. Cladosporium herbarum Causes a Leaf Spot on Marshmarigold in Western North America. Plant Health Prog. 2008, 9, 2. [Google Scholar] [CrossRef]
- Romero-Cuadrado, L.; Picos, M.C.; Camacho, M.; Ollero, F.J.; Capote, N. Biocontrol of almond canker diseases caused by fungi. Pest Manag. Sci. 2024, 80, 1839–1848. [Google Scholar] [CrossRef]
- Kamilova, F.; Lamers, G.; Lugtenberg, B. Biocontrol strain Pseudomonas fluorescens WCS365 inhibits germination of Fusarium oxysporum spores in tomato root exudate as well as subsequent formation of new spores. Environ. Microbiol. 2008, 10, 2455–2461. [Google Scholar] [CrossRef]
- Abbasi, V.; Rahimian, H.; Tajick-Ghanbari, M.A. Genetic variability of Iranian strains of Pseudomonas syringae pv. syringae causing bacterial canker disease of stone fruits. Eur. J. Plant Pathol. 2013, 135, 225–235. [Google Scholar] [CrossRef]
- Lipps, S.M.; Samac, D.A. Pseudomonas viridiflava: An internal outsider of the Pseudomonas syringae species complex. Mol. Plant Pathol. 2022, 23, 3–15. [Google Scholar] [CrossRef]
- Kozhar, O.; Sitz, R.A.; Woyda, R.; Legg, L.; Ibarra Caballero, J.R.; Pearse, I.S.; Abdo, Z.; Stewart, J.E. Population genomic analysis of an emerging pathogen Lonsdalea quercina affecting various species of oaks in western North America. Sci. Rep. 2023, 13, 14852. [Google Scholar] [CrossRef]
- Zamorano, A.; Zuñiga, T.; Córdova, P.; Higuera, G.; Bertaccini, A.; Fiore, N. Pantoea agglomerans-Induced Dieback in Pistachio in Chile. Horticulturae 2022, 8, 1052. [Google Scholar] [CrossRef]
- Weller-Stuart, T.; De Maayer, P.; Coutinho, T. Pantoea ananatis: Genomic insights into a versatile pathogen. Mol Plant Pathol 2017, 18, 1191–1198. [Google Scholar] [CrossRef]
- Kovács, T.; Schneider, G.; Nagy, I.K.; Ravasz, S.L.; Rákhely, G.; Kovács, K.; Bali, D. First Report of Erwinia rhapontici Causing Bacterial Rot on Peach, Detected in Hungary. Plant Dis. 2020, 104, 3248. [Google Scholar] [CrossRef]
- Caires, N.P.; Guimarães, L.M.S.; Hermenegildo, P.S.; Rodrigues, F.A.; Badel, J.L.; Alfenas, A.C. Bidirectional colonization and biofilm formation by Erwinia psidii in eucalypt plants. Plant Pathol. 2020, 69, 549–558. [Google Scholar] [CrossRef]
- Beluzán, F.; Olmo, D.; León, M.; Abad-Campos, P.; Armengol, J. First Report of Diaporthe amygdali Associated with Twig Canker and Shoot Blight of Nectarine in Spain. Plant Dis. 2021, 105, 3300. [Google Scholar] [CrossRef]
- Makris, G.; Solonos, S.; Christodoulou, M.; Kanetis, L.I. First Report of Diaporthe foeniculina Associated with Grapevine Trunk Diseases on Vitis vinifera in Cyprus. Plant Dis. 2022, 106, 1294. [Google Scholar] [CrossRef]
- Laval, V.; Kerdraon, L.; Barret, M.; Liabot, A.-L.; Marais, C.; Boudier, B.; Balesdent, M.-H.; Fischer-Le Saux, M.; Suffert, F. Assessing the Cultivability of Bacteria and Fungi from Arable Crop Residues Using Metabarcoding Data as a Reference. Diversity 2021, 13, 404. [Google Scholar] [CrossRef]


| Family | Genus/Species |
|---|---|
| Dothioraceae (2) | Aureobasidium pullulans (2) |
| Diaporthaceae (5) | Diaporthe amygdali (4) |
| Diaporthe foeniculina (1) | |
| Trichocomaceae (6) | Aspergillus sp. (1) |
| Penicillium sp. (1) | |
| Penicillium citrinum (4) | |
| Pleosporaceae (15) | Alternaria sp. (10) |
| Alternaria alternata (4) | |
| Alternaria tenuissima (1) | |
| Nectriaceae (13) | Fusarium brachygibbosum (5) |
| Fusarium equiseti (2) | |
| Fusarium oxysporum (3) | |
| Fusarium solani (3) | |
| Hypocreaceae (1) | Trichoderma virens (1) |
| Didymellaceae (1) | Stagonosporopsis sp. (1) |
| Pythiaceae (1) | Phytophthora sp. (1) |
| Apiosporaceae (1) | Arthrinium (1) |
| Botryosphaeriaceae (1) | Diplodia corticola (1) |
| Cystobasidiaceae (1) | Cystobasidium sp. (1) |
| Comparison Pair | Statistic | p-Value | FDR | |||
|---|---|---|---|---|---|---|
| Chao1 | Shannon | Chao1 | Shannon | Chao1 | Shannon | |
| Soleta vs. Guara | −4.104 | −0.961 | 0.002 | 0.361 | 0.009 | 0.563 |
| Soleta vs. Belona | −2.968 | −1.497 | 0.016 | 0.165 | 0.049 | 0.496 |
| Soleta vs. Antoñeta | −4.871 | −2.967 | 0.001 | 0.017 | 0.006 | 0.138 |
| Soleta vs. Lauranne | −6.661 | −2.628 | 4.065 × 10−6 | 0.018 | 6.099 × 10−5 | 0.138 |
| Soleta vs. Makako | −7.054 | −2.377 | 6.518 × 10−5 | 0.068 | 4.888 × 10−4 | 0.339 |
| Guara vs. Belona | 0.663 | −0.355 | 0.52 | 0.729 | 0.747 | 0.781 |
| Guara vs. Antoñeta | −0.487 | −1.507 | 0.636 | 0.16 | 0.795 | 0.496 |
| Guara vs. Lauranne | −0.635 | −0.83 | 0.539 | 0.427 | 0.747 | 0.582 |
| Guara vs. Makako | −0.256 | −1.226 | 0.804 | 0.257 | 0.862 | 0.552 |
| Belona vs. Antoñeta | −1.136 | −1.221 | 0.28 | 0.248 | 0.601 | 0.552 |
| Belona vs. Lauranne | −1.368 | −0.447 | 0.202 | 0.664 | 0.505 | 0.766 |
| Belona vs. Makako | −1.062 | −0.948 | 0.321 | 0.376 | 0.601 | 0.563 |
| Antoñeta vs. Lauranne | −0.061 | 1.028 | 0.952 | 0.331 | 0.953 | 0.563 |
| Antoñeta vs. Makako | 0.373 | 0.144 | 0.72 | 0.89 | 0.831 | 0.89 |
| Lauranne vs. Makako | 0.617 | −0.713 | 0.548 | 0.51 | 0.747 | 0.638 |
| Comparison Pair | F-Value | R-Squared | p-Value | FDR |
|---|---|---|---|---|
| Soleta vs. Guara | 2.298 | 0.161 | 0.028 | 0.19 |
| Soleta vs. Belona | 1.856 | 0.134 | 0.073 | 0.274 |
| Soleta vs. Antoñeta | 1.789 | 0.14 | 0.038 | 0.19 |
| Soleta vs. Lauranne | 2.611 | 0.127 | 0.004 | 0.06 |
| Soleta vs. Makako | 1.663 | 0.156 | 0.111 | 0.333 |
| Guara vs. Belona | 0.514 | 0.041 | 0.887 | 0.943 |
| Guara vs. Antoñeta | 0.502 | 0.044 | 0.869 | 0.943 |
| Guara vs. Lauranne | 0.733 | 0.039 | 0.701 | 0.943 |
| Guara vs. Makako | 0.319 | 0.034 | 0.936 | 0.943 |
| Belona vs. Antoñeta | 0.611 | 0.053 | 0.844 | 0.943 |
| Belona vs. Lauranne | 0.55 | 0.03 | 0.943 | 0.943 |
| Belona vs. Makako | 0.443 | 0.047 | 0.92 | 0.943 |
| Antoñeta vs. Lauranne | 0.812 | 0.046 | 0.675 | 0.943 |
| Antoñeta vs. Makako | 0.457 | 0.054 | 0.924 | 0.943 |
| Lauranne vs. Makako | 0.763 | 0.048 | 0.737 | 0.943 |
| Comparison Pair | Statistic | p-Value | FDR | |||
|---|---|---|---|---|---|---|
| Chao1 | Shannon | Chao1 | Shannon | Chao1 | Shannon | |
| Soleta vs. Guara | −3.332 | −2.232 | 0.012 | 0.048 | 0.093 | 0.226 |
| Soleta vs. Belona | −1.425 | −0.8 | 0.185 | 0.446 | 0.407 | 0.652 |
| Soleta vs. Antoñeta | −2.862 | −2.138 | 0.019 | 0.06 | 0.097 | 0.226 |
| Soleta vs. Lauranne | −2.401 | −2.372 | 0.043 | 0.038 | 0.162 | 0.226 |
| Soleta vs. Makako | −3.601 | −2.684 | 0.009 | 0.033 | 0.093 | 0.226 |
| Guara vs. Belona | 1.124 | 0.868 | 0.301 | 0.409 | 0.502 | 0.652 |
| Guara vs. Antoñeta | 0.285 | 0.349 | 0.782 | 0.734 | 0.782 | 0.847 |
| Guara vs. Lauranne | 1.334 | 0.152 | 0.2 | 0.882 | 0.407 | 0.882 |
| Guara vs. Makako | −0.488 | −0.729 | 0.639 | 0.491 | 0.737 | 0.652 |
| Belona vs. Antoñeta | −0.885 | −0.674 | 0.403 | 0.521 | 0.575 | 0.652 |
| Belona vs. Lauranne | −0.382 | −0.821 | 0.714 | 0.438 | 0.764 | 0.652 |
| Belona vs. Makako | −1.368 | −1.375 | 0.217 | 0.207 | 0.407 | 0.62 |
| Antoñeta vs. Lauranne | 0.832 | −0.235 | 0.421 | 0.818 | 0.575 | 0.876 |
| Antoñeta vs. Makako | −0.676 | −1.089 | 0.518 | 0.323 | 0.648 | 0.652 |
| Lauranne vs. Makako | −1.762 | −0.93 | 0.106 | 0.393 | 0.317 | 0.652 |
| Comparison Pair | F-Value | R-Squared | p-Value | FDR |
|---|---|---|---|---|
| Soleta vs. Guara | 2.61 | 0.192 | 0.013 | 0.065 |
| Soleta vs. Belona | 3.324 | 0.249 | 0.006 | 0.045 |
| Soleta vs. Antoñeta | 2.596 | 0.206 | 0.021 | 0.079 |
| Soleta vs. Lauranne | 4.628 | 0.214 | 0.001 | 0.015 |
| Soleta vs. Makako | 2.075 | 0.206 | 0.045 | 0.135 |
| Guara vs. Belona | 1.143 | 0.094 | 0.29 8 | 0.528 |
| Guara vs. Antoñeta | 0.315 | 0.028 | 0.918 | 0.918 |
| Guara vs. Lauranne | 1.15 | 0.06 | 0.315 | 0.528 |
| Guara vs. Makako | 0.695 | 0.071 | 0.579 | 0.62 |
| Belona vs. Antoñeta | 1.235 | 0.11 | 0.237 | 0.528 |
| Belona vs. Lauranne | 0.935 | 0.052 | 0.519 | 0.62 |
| Belona vs. Makako | 0.89 | 0.1 | 0.519 | 0.62 |
| Antoñeta vs. Lauranne | 1.114 | 0.061 | 0.317 | 0.528 |
| Antoñeta vs. Makako | 0.798 | 0.091 | 0.555 | 0.62 |
| Lauranne vs. Makako | 0.969 | 0.061 | 0.475 | 0.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albuquerque, A.; Patanita, M.; Ribeiro, J.A.; Campos, M.D.; Santos, F.; Monteiro, T.; Basaloco, M.; Félix, M.d.R. Metabarcoding Analysis Reveals Microbial Diversity and Potential Soilborne Pathogens Associated with Almond Dieback and Decline. Plants 2025, 14, 2309. https://doi.org/10.3390/plants14152309
Albuquerque A, Patanita M, Ribeiro JA, Campos MD, Santos F, Monteiro T, Basaloco M, Félix MdR. Metabarcoding Analysis Reveals Microbial Diversity and Potential Soilborne Pathogens Associated with Almond Dieback and Decline. Plants. 2025; 14(15):2309. https://doi.org/10.3390/plants14152309
Chicago/Turabian StyleAlbuquerque, André, Mariana Patanita, Joana Amaro Ribeiro, Maria Doroteia Campos, Filipa Santos, Tomás Monteiro, Margarida Basaloco, and Maria do Rosário Félix. 2025. "Metabarcoding Analysis Reveals Microbial Diversity and Potential Soilborne Pathogens Associated with Almond Dieback and Decline" Plants 14, no. 15: 2309. https://doi.org/10.3390/plants14152309
APA StyleAlbuquerque, A., Patanita, M., Ribeiro, J. A., Campos, M. D., Santos, F., Monteiro, T., Basaloco, M., & Félix, M. d. R. (2025). Metabarcoding Analysis Reveals Microbial Diversity and Potential Soilborne Pathogens Associated with Almond Dieback and Decline. Plants, 14(15), 2309. https://doi.org/10.3390/plants14152309







