Functional Analysis of Zinc Finger Protein Transcription Factor ZmZFP69 Under Low-Temperature Stress at Maize Seedling Stage
Abstract
1. Introduction
2. Results
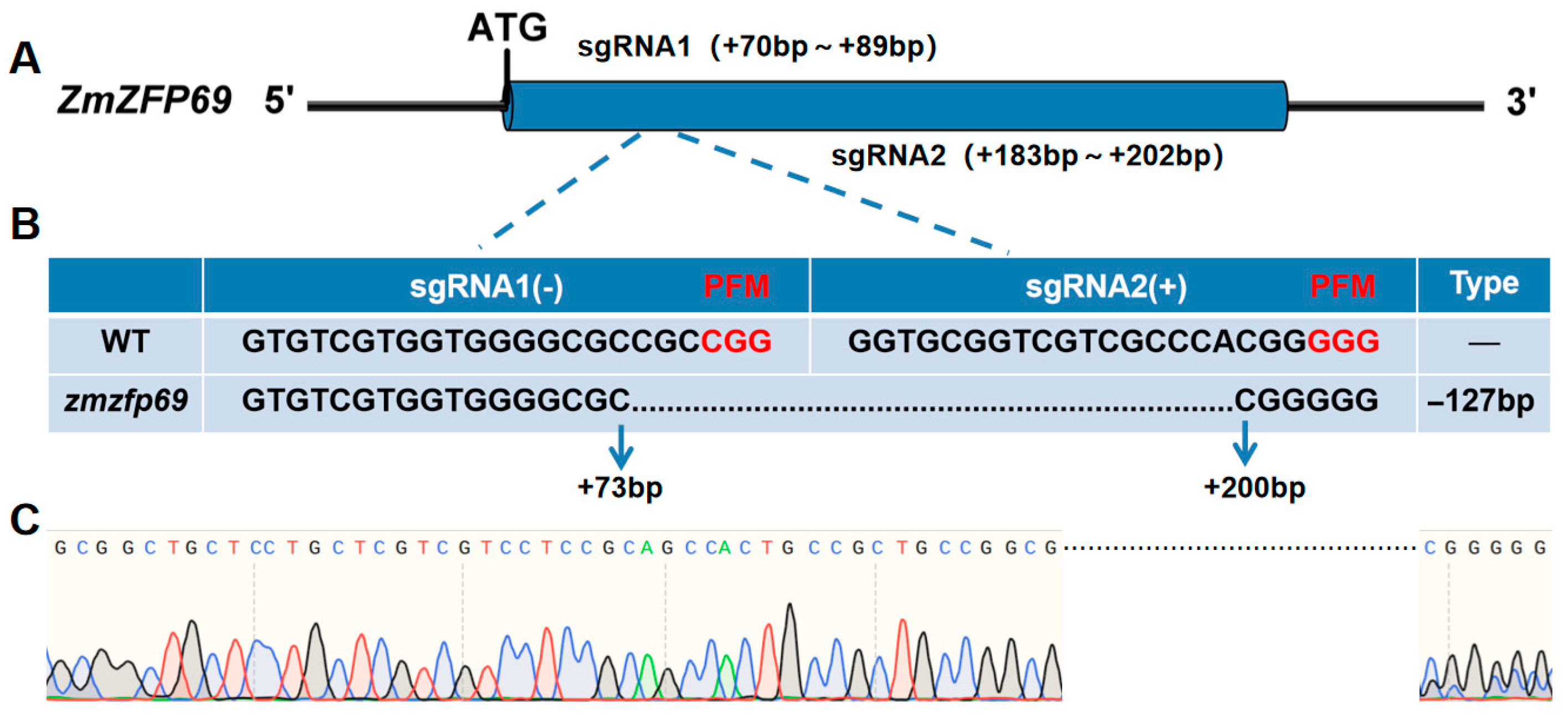
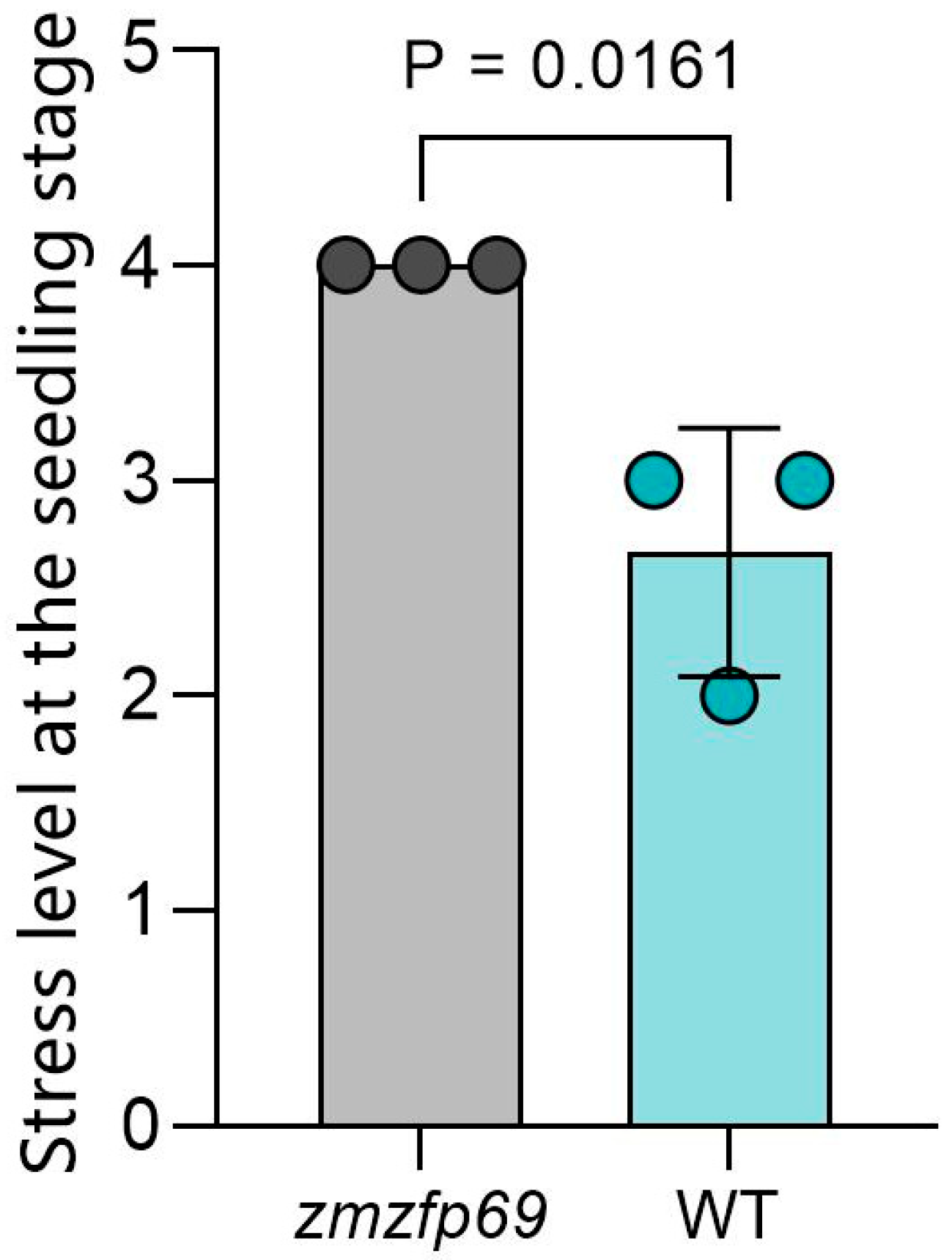
2.1. Identification of Low-Temperature Resistance of Mutant Materials
2.1.1. Phenotypic Identification of Mutant Materials
2.1.2. Determination of Hormone Content in Mutant Materials
2.2. ZmZFP69 Downstream Target Gene Screening Through Multi-Omics Joint Analysis
2.2.1. ZmZFP69 Potential Downstream Target Gene Screening by RNA-Seq
2.2.2. DNA Affinity Purification Sequencing (DAP-Seq)
2.2.3. Combined Analysis of DAP-Seq and RNA-Seq to Screen ZmZFP69 Target Genes
2.3. Luciferase Assay Verified That ZmZFP69 Recognizes the ZmAOX2 Promoter
2.4. Identification of Downstream Interacting Proteins with InterProDesign
2.5. Subcellular Localization
2.6. BiFC Verification of Interacting Proteins
3. Discussion
4. Materials and Methods
4.1. Identification of Low-Temperature Resistance Properties of Mutant Materials
4.1.1. Phenotypic Identification of Mutant Materials
4.1.2. Determination of Hormone Indicators of Maize Mutant Materials
4.2. Multi-Omics Joint Analysis
4.2.1. RNA-Seq
4.2.2. DAP-Seq and Analysis
4.3. Luciferase Reporter Assay
4.4. InterProDesign Identification of Downstream Interacting Proteins
4.5. BiFC and Subcellular Localization Assays
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. Embo J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Wang, L.; Du, H.; Wang, Y.; Wang, W.; Liu, J.; Huang, J.; Huang, W.; Ge, L. Genome-wide study of C2H2 zinc finger gene family in Medicago truncatula. BMC Plant Biol. 2020, 20, 401. [Google Scholar] [CrossRef]
- Liu, X.X.; Fu, C.; Yang, W.W.; Zhang, Q.; Fan, H.; Liu, J. The involvement of TsFtsH8 in Thellungiella salsuginea tolerance to cold and high light stresses. Acta Physiol. Plant. 2016, 38, 62. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Cui, F.; Hou, L.; Zhao, S.; Xia, H.; Qiu, J.; Li, T.; Zhang, Y.; Wang, X.; et al. Genome-Wide Analysis of Gene Expression Provides New Insights into Cold Responses in Thellungiella salsuginea. Front. Plant Sci. 2017, 8, 713. [Google Scholar] [CrossRef]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, M.D.; Park, S.C.; Yang, K.S.; Jeong, J.C.; Lee, H.S.; Kwak, S.S. SCOF-1-expressing transgenic sweet potato plants show enhanced tolerance to low-temperature stress. Plant Physiol. Biochem. 2011, 49, 1436–1441. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, X.; Lei, C.; Cheng, Z.; Lin, Q.; Wang, J.; Wu, F.; Wang, J.; Wan, J. Overexpression of SlCZFP1, a Novel TFIIIA-type Zinc Finger Protein from Tomato, Confers Enhanced Cold Tolerance in Transgenic Arabidopsis and Rice. Plant Mol. Biol. Report. 2011, 29, 185–196. [Google Scholar] [CrossRef]
- Yu, G.H.; Jiang, L.L.; Ma, X.F.; Xu, Z.S.; Liu, M.M.; Shan, S.G.; Cheng, X.G. A soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic Arabidopsis. PLoS ONE 2014, 9, e109399. [Google Scholar] [CrossRef]
- Han, Y.C.; Fu, C.C. Cold-inducible MaC2H2s are associated with cold stress response of banana fruit via regulating MaICE1. Plant Cell Rep. 2019, 38, 673–680. [Google Scholar] [CrossRef]
- Kim, J.C.; Lee, S.H.; Cheong, Y.H.; Yoo, C.M.; Lee, S.I.; Chun, H.J.; Yun, D.J.; Hong, J.C.; Lee, S.Y.; Lim, C.O.; et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001, 25, 247–259. [Google Scholar] [CrossRef]
- Luo, P.; Di, D.W. Precise Regulation of the TAA1/TAR-YUCCA Auxin Biosynthesis Pathway in Plants. Int. J. Mol. Sci. 2023, 24, 8514. [Google Scholar] [CrossRef]
- Gonzales, C.F.; Kelley, D.R. Interplay between auxin and abiotic stresses in maize. J. Exp. Bot. 2025, 76, 1879–1887. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, X.; Gong, D.H.; Huang, Q.Q.; Kandegama, W.; Georgiev, M.I.; Gao, Y.Y.; Liao, P.; Hao, G.F. Sophisticated crosstalk of tryptophan-derived metabolites in plant stress responses. Plant Commun. 2025, 101425. [Google Scholar] [CrossRef]
- El-Bassiouny, H. Physiological responses of wheat to salinity alleviation by nicotinamide and tryptophan. Int. J. Agric. Biol. 2005, 7, 653–659. [Google Scholar]
- Saffari, Y.; Sadrzadeh, S.M. Green tea metabolite EGCG protects membranes against oxidative damage in vitro. Life Sci. 2004, 74, 1513–1518. [Google Scholar] [CrossRef]
- Li, Q.Z.; Li, C.W.; Yang, T.W. Stress Response and Memory Mediated by DNA Methylation in Plants. Plant Physiol. J. 2014, 50, 725–734. [Google Scholar] [CrossRef]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 2007, 282, 9260–9268. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Mittler, R. The zinc finger network of plants. Cell Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Wang, K.; Ding, Y.; Cai, C.; Chen, Z.; Zhu, C. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. Plant 2019, 165, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.X.; Zhang, F.C.; Zhang, W.Z.; Song, L.F.; Wu, W.H.; Chen, Y.F. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 2013, 6, 1487–1502. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Ye, T.; Chen, F.; Deng, J.; Yang, P.; Zhang, Y.; Chan, Z. The Cysteine2/Histidine2-Type Transcription Factor Zinc Finger Of Arabidopsis Thaliana6 Modulates Biotic and Abiotic Stress Responses by Activating Salicylic Acid-Related Genes and C-Repeat-Binding Factor Genes in Arabidopsis. Plant Physiol. 2014, 165, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006, 580, 6537–6542. [Google Scholar] [CrossRef]
- Chen, J.; Yang, L.; Yan, X.; Liu, Y.; Wang, R.; Fan, T.; Ren, Y.; Tang, X.; Xiao, F.; Liu, Y.; et al. Zinc-Finger Transcription Factor ZAT6 Positively Regulates Cadmium Tolerance through the Glutathione-Dependent Pathway in Arabidopsis. Plant Physiol. 2016, 171, 707–719. [Google Scholar] [CrossRef]
- Liu, X.M.; An, J.; Han, H.J.; Kim, S.H.; Lim, C.O.; Yun, D.J.; Chung, W.S. ZAT11, a zinc finger transcription factor, is a negative regulator of nickel ion tolerance in Arabidopsis. Plant Cell Rep. 2014, 33, 2015–2021. [Google Scholar] [CrossRef]
- Qureshi, M.K.; Sujeeth, N.; Gechev, T.S.; Hille, J. The zinc finger protein ZAT11 modulates paraquat-induced programmed cell death in Arabidopsis thaliana. Acta Physiol. Plant. 2013, 35, 1863–1871. [Google Scholar] [CrossRef]
- Zhang, D.; Ding, X.; Wang, Z.; Li, W.; Li, L.; Liu, L.; Zhou, H.; Yu, J.; Zheng, C.; Wu, H.; et al. A C2H2 zinc finger protein, OsZOS2-19, modulates ABA sensitivity and cold response in rice. Plant Cell Physiol. 2025, 66, 753–765. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.; Ren, H.; Zhang, C.; Xiao, D.; Li, Y.; Hou, X.; Liu, T. Regulatory interaction of BcWRKY33A and BcHSFA4A promotes salt tolerance in non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic. Res. 2022, 9, uhac113. [Google Scholar] [CrossRef]
- Zeng, R.; Shi, Y.; Guo, L.; Fu, D.; Li, M.; Zhang, X.; Li, Z.; Zhuang, J.; Yang, X.; Zuo, J.; et al. A natural variant of COOL1 gene enhances cold tolerance for high-latitude adaptation in maize. Cell 2025, 188, 1315–1329.e1313. [Google Scholar] [CrossRef]
- Yuan, H.; Zhao, K.; Lei, H.; Shen, X.; Liu, Y.; Liao, X.; Li, T. Genome-wide analysis of the GH3 family in apple (Malus × domestica). BMC Genom. 2013, 14, 297. [Google Scholar] [CrossRef]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Hagen, G.; Martin, G.; Li, Y.; Guilfoyle, T.J. Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 1991, 17, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.L.; Shi, Y.T.; Yang, S.H. Molecular mechanisms of plant responses to cold stress. Chin. Bull. Life Sci. 2015, 27, 398–405. [Google Scholar] [CrossRef]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. Plant Cell 2009, 21, 3823–3838. [Google Scholar] [CrossRef]
- Kodaira, K.S.; Qin, F.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011, 157, 742–756. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; García-Caparrós, P.; Rahman, M.A.; Ogwugwa, V.H.; Saeed, F.; Jin, W. Melatonin-mediated temperature stress tolerance in plants. GM Crops Food 2022, 13, 196–217. [Google Scholar] [CrossRef]
- Sun, J.L.; Li, J.Y.; Wang, M.J.; Song, Z.T.; Liu, J.X. Protein Quality Control in Plant Organelles: Current Progress and Future Perspectives. Mol. Plant 2021, 14, 95–114. [Google Scholar] [CrossRef]
- Luo, H.X.; An, G.; Dai, X.; Wang, S.S.; Guo, Z.Q. Research Progress and Future Prospect of Protein Quality Control in Plant Organelles. Agric. Technol. Equip. 2021, 14, 91–92. [Google Scholar]
- Jiang, H.; Ji, Y.; Sheng, J.; Wang, Y.; Liu, X.; Xiao, P.; Ding, H. Genome-Wide Identification of the Bcl-2 Associated Athanogene (BAG) Gene Family in Solanum lycopersicum and the Functional Role of SlBAG9 in Response to Osmotic Stress. Antioxidants 2022, 11, 598. [Google Scholar] [CrossRef]
- Irfan, M.; Kumar, P.; Ahmad, I.; Datta, A. Unraveling the role of tomato Bcl-2-associated athanogene (BAG) proteins during abiotic stress response and fruit ripening. Sci. Rep. 2021, 11, 21734. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Salmon-Divon, M.; Dvinge, H.; Tammoja, K.; Bertone, P. PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci. BMC Bioinform. 2010, 11, 415. [Google Scholar] [CrossRef]
- Hull, R.P.; Srivastava, P.K.; D’Souza, Z.; Atanur, S.S.; Mechta-Grigoriou, F.; Game, L.; Petretto, E.; Cook, H.T.; Aitman, T.J.; Behmoaras, J. Combined ChIP-Seq and transcriptome analysis identifies AP-1/JunD as a primary regulator of oxidative stress and IL-1β synthesis in macrophages. BMC Genom. 2013, 14, 92. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De novo design of protein structure and function with RFdiffusion. Nature 2023, 620, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Haley, O.C.; Harding, S.; Sen, T.Z.; Woodhouse, M.R.; Kim, H.-S.; Andorf, C. Application of RFdiffusion to predict interspecies protein-protein interactions between fungal pathogens and cereal crops. bioRxiv 2024. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Howe, E.S.; Clemente, T.E.; Bass, H.W. Maize histone H2B-mCherry: A new fluorescent chromatin marker for somatic and meiotic chromosome research. DNA Cell Biol. 2012, 31, 925–938. [Google Scholar] [CrossRef]












| No. | Gene ID | Name | Pathway | Notification |
|---|---|---|---|---|
| 1 | Zm00001d034298 | ZmPIF4 | Plant hormone signal transduction | phytochrome-interacting factor 4 |
| 2 | Zm00001d029577 | ZmZH3 | Plant hormone signal transduction | INDOLE-3-ACETIC ACID-AMIDO SYNTHETASE GH3.2-RELATED |
| 3 | Zm00001d016294 | ZmPYL2 | MAPK signaling pathway-plant | ABSCISIC ACID RECEPTOR PYL2-RELATED |
| 4 | Zm00001d013098 | ZmAOX2 | Indole acetaldehyde oxidase | Indoleacetal dehyde oxidase |
| No. | Protein Name | Hdock Docking Score (kcal/mol) | Protein Description | Gene ID |
|---|---|---|---|---|
| 1 | A0A1D6PKZ7 | −356.74 | PTHR33322:SF4-BAG FAMILY MOLECULAR CHAPERONE REGULATOR 6 | Zm00001d048489 (ZmBG6) |
| 2 | K7UPU9 | −349.24 | PF00931//PF01582-NB-ARC domain (NB-ARC)//TIR domain (TIR) | Zm00001d053244 (ZmTIR) |
| 3 | A0A1D6K803 | −342.77 | 2.7.11.1-Non-specific serine/threonine protein kinase/Threonine-specific protein kinase | Zm00001d029829 (ZmTSP) |
| 4 | B6TCP0 | −342.09 | PTHR13856//PTHR13856:SF81-VHS DOMAIN CONTAINING PROTEIN FAMILY//ENTH/VHS/GAT FAMILY PROTEI | Zm00001d035029 (ZmGAT) |
| 5 | B6UIN4 | −320.92 | Tetratricopeptide repeat (TPR)-like superfamily protein | Zm00001d052177 (ZmTPR) |
| 6 | A0A1D6KFQ1 | −304.85 | KOG0978-E3 ubiquitin ligase involved in syntaxin degradation | Zm00001d031005 (ZmUBL) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.-N.; Sun, Y.; Li, Y.-L.; Sun, M.-H.; Li, S.-J.; Yin, Y.; Yu, T.; Li, X.; Cai, Q.; Zhang, J.-G. Functional Analysis of Zinc Finger Protein Transcription Factor ZmZFP69 Under Low-Temperature Stress at Maize Seedling Stage. Plants 2025, 14, 2114. https://doi.org/10.3390/plants14142114
Li S-N, Sun Y, Li Y-L, Sun M-H, Li S-J, Yin Y, Yu T, Li X, Cai Q, Zhang J-G. Functional Analysis of Zinc Finger Protein Transcription Factor ZmZFP69 Under Low-Temperature Stress at Maize Seedling Stage. Plants. 2025; 14(14):2114. https://doi.org/10.3390/plants14142114
Chicago/Turabian StyleLi, Si-Nan, Yan Sun, Yun-Long Li, Ming-Hao Sun, Shu-Jun Li, Yue Yin, Tao Yu, Xin Li, Quan Cai, and Jian-Guo Zhang. 2025. "Functional Analysis of Zinc Finger Protein Transcription Factor ZmZFP69 Under Low-Temperature Stress at Maize Seedling Stage" Plants 14, no. 14: 2114. https://doi.org/10.3390/plants14142114
APA StyleLi, S.-N., Sun, Y., Li, Y.-L., Sun, M.-H., Li, S.-J., Yin, Y., Yu, T., Li, X., Cai, Q., & Zhang, J.-G. (2025). Functional Analysis of Zinc Finger Protein Transcription Factor ZmZFP69 Under Low-Temperature Stress at Maize Seedling Stage. Plants, 14(14), 2114. https://doi.org/10.3390/plants14142114






