Genome-Wide Characterization of VDAC Gene Family in Soybean (Glycine max L.) and In Silico Expression Profiling in Response to Drought and Salt Stress
Abstract
1. Introduction
2. Results
2.1. Identification, Distribution, and Renaming of VDAC Genes
2.2. Conserved Domain, Localization, and Physico-Chemical Properties of VDAC Proteins
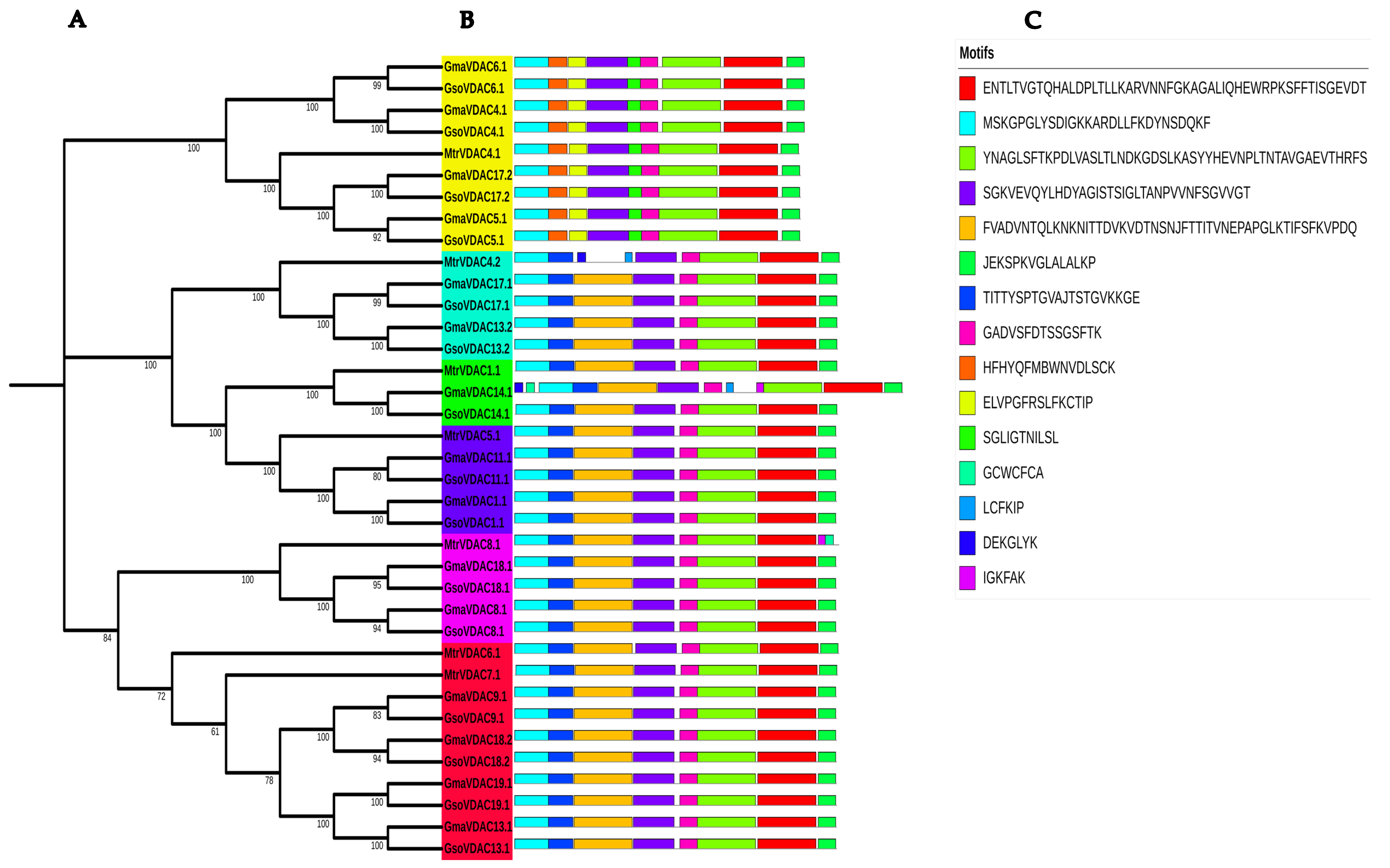
2.3. Classification and Phylogenetic Analysis
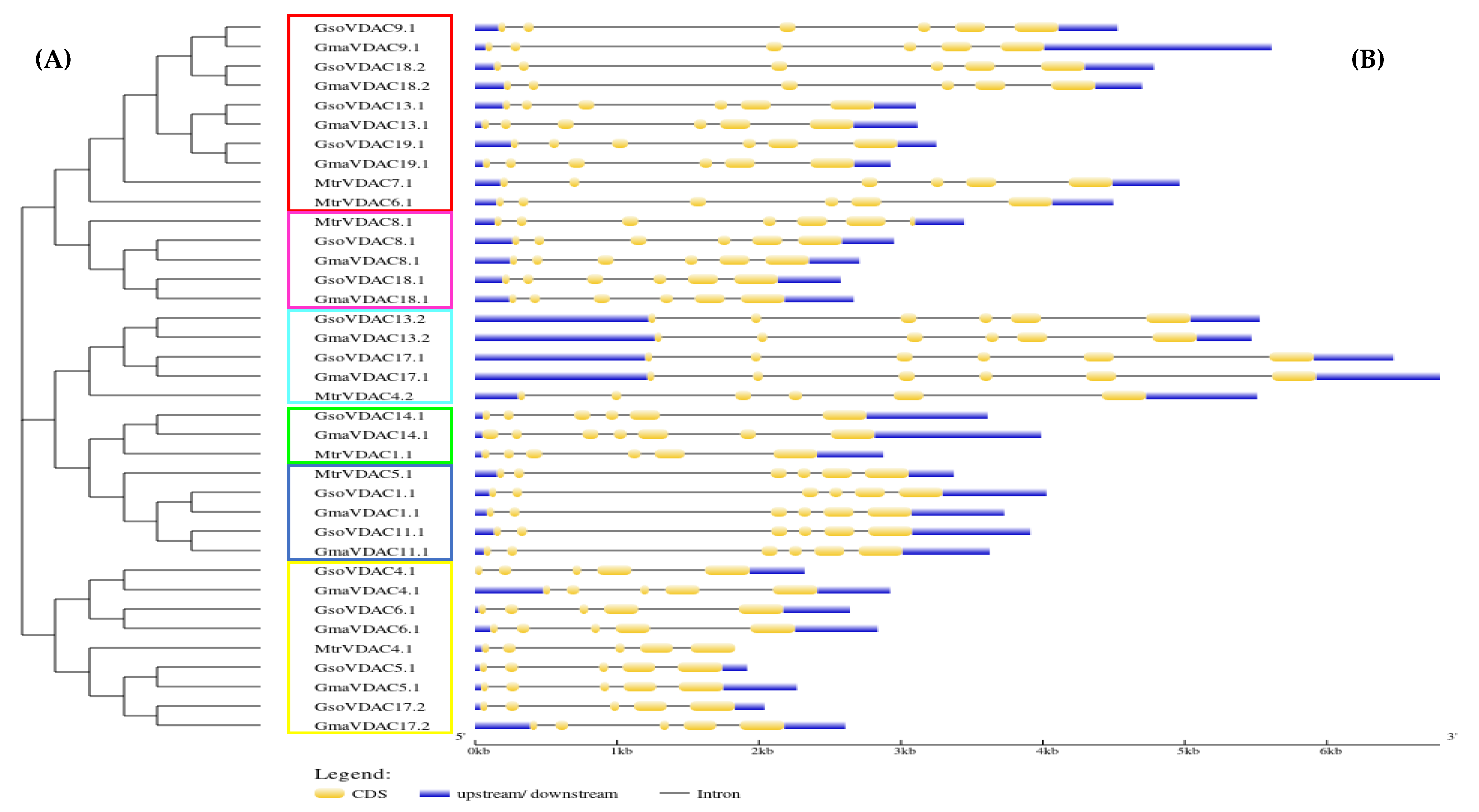
2.4. Conserved Motif and Gene Structure Analysis
2.5. Identification of Cis-Acting Regulatory Elements (CREs) in Promoters of GmaVDAC Genes
2.6. Gene Duplication Events and Divergence Rate
2.7. Gene Positions and Chromosomal Chart
2.8. Synteny Analysis
2.9. Expression Pattern Analysis of GmaVDAC Genes
2.10. Predicted Functions of GmaVDAC Genes
2.11. Protein Interactions and microRNA Targets
3. Discussion
4. Materials and Methods
4.1. Identification of VDAC Genes and Retrieval of Sequences
4.2. Conserved Domain, Subcellular Localization, and Physico-Chemical Properties
4.3. Multiple Sequence Alignment (MSA) and Phylogenetic Relationship
4.4. Gene Structure Prediction and Motif Analysis
4.5. Cis-Regulatory Elements (CREs) and Chromosomal Distribution
4.6. Gene Duplications and Syntenic Analysis
4.7. Protein Interaction Network and microRNA Target Prediction
4.8. Gene Ontology Analysis
4.9. Expression Pattern of GmaVDAC Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, H.; Zhang, L.; Zhang, K.; Ran, Y. Progresses, Challenges, and Prospects of Genome Editing in Soybean (Glycine max). Front. Plant Sci. 2020, 11, 571138. [Google Scholar] [CrossRef] [PubMed]
- Aleem, M.; Aleem, S.; Sharif, I.; Wu, Z.; Aleem, M.; Tahir, A.; Atif, R.M.; Cheema, H.M.N.; Shakeel, A.; Lei, S.; et al. Characterization of SOD and GPX Gene Families in the Soybeans in Response to Drought and Salinity Stresses. Antioxidants 2022, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Rostovtseva, T.K.; Bezrukov, S.M.; Hoogerheide, D.P.; Kmita, H.; Messina, A.A. Regulation of Mitochondrial Respiration by VDAC Is Enhanced by Membrane-Bound Inhibitors with Disordered Polyanionic C-Terminal Domains. Int. J. Mol. Sci. 2021, 22, 7358. [Google Scholar] [CrossRef]
- Yu, M.; Yu, Y.; Song, T.; Zhang, Y.; Wei, F.; Cheng, J.; Zhang, B.; Zhang, X. Characterization of the Voltage-Dependent Anion Channel (VDAC) Gene Family in Wheat (Triticum aestivum L.) and Its Potential Mechanism in Response to Drought and Salinity Stresses. Gene 2022, 809, 146031. [Google Scholar] [CrossRef]
- Ling, H.Q.; Wang, J.; Zhao, S.; Liu, D.; Wang, J.; Sun, H.; Zhang, C.; Fan, H.; Li, D.; Dong, L.; et al. Draft Genome of the Wheat A-Genome Progenitor Triticum urartu. Nature 2013, 496, 87–90. [Google Scholar] [CrossRef]
- Gonçalves, R.P.; Buzhynskyy, N.; Prima, V.; Sturgis, J.N.; Scheuring, S. Supramolecular Assembly of VDAC in Native Mitochondrial Outer Membranes. J. Mol. Biol. 2007, 369, 413–418. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Shteinfer-Kuzmine, A.; Verma, A. VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases. Biomolecules 2020, 10, 1485. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen Peroxide Metabolism and Functions in Plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Meyer, Y.; Buchanan, B.B.; Vignols, F.; Reichheld, J.P. Thioredoxins and Glutaredoxins: Unifying Elements in Redox Biology. Annu. Rev. Genet. 2009, 43, 335–367. [Google Scholar] [CrossRef]
- Kneeshaw, S.; Keyani, R.; Delorme-Hinoux, V.; Imrie, L.; Loake, G.J.; Le Bihan, T.; Reichheld, J.P.; Spoel, S.H. Nucleoredoxin Guards against Oxidative Stress by Protecting Antioxidant Enzymes. Proc. Natl. Acad. Sci. USA 2017, 114, 8414–8419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Takano, T.; Liu, S.; Zhang, X. Arabidopsis Mitochondrial Voltage-Dependent Anion Channel 3 (AtVDAC3) Protein Interacts with Thioredoxin M2. FEBS Lett. 2015, 589, 1207–1213. [Google Scholar] [CrossRef]
- Kusano, T.; Tateda, C.; Takahashi, Y.; Berberich, T. Voltage-Dependent Anion Channels: Their Roles in Plant Defense and Cell Death. Plant Cell Rep. 2009, 28, 1301–1308. [Google Scholar] [CrossRef]
- Saidani, H.; Grobys, D.; Léonetti, M.; Kmita, H.; Homblé, F. Towards Understanding of Plant Mitochondrial VDAC Proteins: An Overview of Bean (Phaseolus vulgaris) VDAC Proteins. AIMS Biophys. 2017, 4, 43–62. [Google Scholar] [CrossRef]
- Yang, M.; Duan, X.; Wang, Z.; Yin, H.; Zang, J.; Zhu, K.; Wang, Y.; Zhang, P. Overexpression of a Voltage-Dependent Anion-Selective Channel (VDAC) Protein-Encoding Gene, MsVDAC, from Medicago sativa Confers Cold and Drought Tolerance to Transgenic Tobacco. Genes 2021, 12, 1706. [Google Scholar] [CrossRef]
- Desai, M.K.; Mishra, R.N.; Verma, D.; Nair, S.; Sopory, S.K.; Reddy, M.K. Structural and Functional Analysis of a Salt Stress Inducible Gene Encoding Voltage Dependent Anion Channel (VDAC) from Pearl Millet (Pennisetum glaucum). Plant Physiol. Biochem. 2006, 44, 483–493. [Google Scholar] [CrossRef]
- Xu, X.; Tan, Y.P.; Cheng, G.; Liu, X.Q.; Xia, C.J.; Luo, F.Y.; Wang, C.T. Genomic Survey and Gene Expression Analysis of the VDAC Gene Family in Rice. Genet. Mol. Res. 2015, 14, 15683–15696. [Google Scholar] [CrossRef]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J.; et al. Genomic Insights into Salt Adaptation in a Desert Poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef]
- Severin, A.J.; Cannon, S.B.; Graham, M.M.; Grant, D.; Shoemaker, R.C. Changes in Twelve Homoeologous Genomic Regions in Soybean Following Three Rounds of Polyploidy. Plant Cell 2011, 23, 3129–3136. [Google Scholar] [CrossRef]
- Zhu, Y.; Yin, J.; Liang, Y.; Liu, J.; Jia, J.; Huo, H.; Wu, Z.; Yang, R.; Gong, H. Transcriptomic Dynamics Provide an Insight into the Mechanism for Silicon-Mediated Alleviation of Salt Stress in Cucumber Plants. Ecotoxicol. Environ. Saf. 2019, 174, 245–254. [Google Scholar] [CrossRef]
- Wandrey, M.; Trevaskis, B.; Brewin, N.; Udvardi, M.K. Molecular and Cell Biology of a Family of Voltage-Dependent Anion Channel Porins in Lotus japonicus. Plant Physiol. 2004, 134, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Tateda, C.; Watanabe, K.; Kusano, T. Molecular and Genetic Characterization of the Gene Family Encoding the Voltage-Dependent Anion Channel in Arabidopsis. J. Exp. Bot. 2011, 62, 4773–4785. [Google Scholar] [CrossRef]
- Mok, D.W.S.; Peloquin, S.J. Breeding Value of 2n Pollen (Diplandroids) in Tetraploid x Diploid Crosses in Potatoes. Theor. Appl. Genet. 1975, 46, 307–314. [Google Scholar] [CrossRef]
- Singh, N.; Bhatt, V.; Rana, N.; Shivaraj, S.M. Advances of Next-Generation Sequencing (NGS) Technologies to Enhance the Biofortifications in Crops. In Advances in Agri-Food Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 427–450. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene Duplication as a Major Force in Evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic Analysis of Maize Class III Peroxidase Gene Family Reveals a Conserved Subfamily Involved in Abiotic Stress Response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A Conserved Domain Database for Interactive Domain Family Analysis. Nucleic Acids Res. 2006, 35, D237–D240. [Google Scholar] [CrossRef]
- Chi, S.M.; Nam, D. WegoLoc: Accurate Prediction of Protein Subcellular Localization Using Weighted Gene Ontology Terms. Bioinformatics 2012, 28, 1028–1030. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. Proteom. Protoc. Handb. 2005, 1, 571–607. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and Analyzing DNA and Protein Sequence Motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE: A Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Korber, B. HIV Signature and Sequence Variation Analysis. In Computational Analysis of HIV Molecular Sequences; Springer: Berlin/Heidelberg, Germany, 2000; Volume 4, pp. 55–72. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 8, 8.13.1–8.13.24. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Shen, Y.; Du, H.; Liu, Y.; Ni, L.; Wang, Z.; Liang, C.; Tian, Z. Update Soybean Zhonghuang 13 Genome to a Golden Reference. Sci. China Life Sci. 2019, 62, 1257–1260. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive Characterization and RNA-Seq Profiling of the HD-Zip Transcription Factor Family in Soybean (Glycine max) during Dehydration and Salt Stress. BMC Genom. 2014, 15, 950. [Google Scholar] [CrossRef]








| Duplicated Pair | Ka | Ks | Ka/Ks | Duplicate Type | Selection Type | T (Mya) |
|---|---|---|---|---|---|---|
| GmaVDAC1.1/GmaVDAC11.1 | 0.01 | 0.07 | 0.18 | Segmental | Purifying | 6.10 |
| GmaVDAC4.1/GmaVDAC6.1 | 0.03 | 0.14 | 0.21 | Segmental | Purifying | 11.39 |
| GmaVDAC5.1/GmaVDAC17.2 | 0.02 | 0.09 | 0.23 | Segmental | Purifying | 7.02 |
| GmaVDAC8.1/GmaVDAC18.1 | 0.01 | 0.14 | 0.06 | Segmental | Purifying | 11.31 |
| GmaVDAC9.1/GmaVDAC18.2 | 0.005 | 0.06 | 0.08 | Segmental | Purifying | 5.16 |
| GmaVDAC13.1/GmaVDAC19.1 | 0.02 | 0.15 | 0.12 | Segmental | Purifying | 12.51 |
| GmaVDAC13.2/GmaVDAC17.1 | 0.02 | 0.11 | 0.18 | Segmental | Purifying | 8.75 |
| Sr. No. | Crop | Botanical name | Database link |
|---|---|---|---|
| 1. | Adzuki bean | Vigna angularis | https://legacy.legumeinfo.org/ (accessed on 17 June 2022) |
| 2. | Arabidopsis | Arabidopsis thaliana | https://www.arabidopsis.org/ (accessed on 17 June, 2022) |
| 3. | Barrel clover | Medicago truncatula | https://phytozome-next.jgi.doe.gov/ (accessed on 20 June 2022) |
| 4. | Birdsfoot trefoil | Lotus japonicus | https://phytozome-next.jgi.doe.gov/ (accessed on 20 June 2022) |
| 5. | Chickpea | Cicer arietinum | https://phytozome-next.jgi.doe.gov/ (accessed on 21 June 2022) |
| 6. | Common bean | Phaseolus vulgaris | https://phytozome-next.jgi.doe.gov/ (accessed on 21 June 2022) |
| 7. | Cowpea | Vigna unguiculata | https://phytozome-next.jgi.doe.gov/ (accessed on 21 June 2022) |
| 8. | Cultivated soybean | Glycine max | https://www.soybase.org/ (accessed on 21 June 2022) |
| 9. | Mungbean | Vigna radiata | https://legacy.legumeinfo.org/ (accessed on 23 June 2022) |
| 10. | Narrow leaf lupin | Lupinus angustifolius | https://legacy.legumeinfo.org/ (accessed on 23 June 2022) |
| 11. | Pea | Pisum sativum | https://legacy.legumeinfo.org/ (accessed on 23 June 2022) |
| 12. | Peanut | Arachis hypogaea | https://phytozome-next.jgi.doe.gov/ (accessed on 24 June 2022) |
| 13. | Pigeon pea | Cajanus cajan | https://legacy.legumeinfo.org/ (accessed on 24 June 2022) |
| 14. | Wild soybean | Glycine soja | https://www.soybase.org/ (accessed on 24 June 2022) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, M.M.; Aleem, M.; Iqbal, M.M.; Riaz, A.; Shi, A. Genome-Wide Characterization of VDAC Gene Family in Soybean (Glycine max L.) and In Silico Expression Profiling in Response to Drought and Salt Stress. Plants 2025, 14, 2101. https://doi.org/10.3390/plants14142101
Ullah MM, Aleem M, Iqbal MM, Riaz A, Shi A. Genome-Wide Characterization of VDAC Gene Family in Soybean (Glycine max L.) and In Silico Expression Profiling in Response to Drought and Salt Stress. Plants. 2025; 14(14):2101. https://doi.org/10.3390/plants14142101
Chicago/Turabian StyleUllah, Muhammad Muneeb, Muqadas Aleem, Muhammad Mudassar Iqbal, Awais Riaz, and Ainong Shi. 2025. "Genome-Wide Characterization of VDAC Gene Family in Soybean (Glycine max L.) and In Silico Expression Profiling in Response to Drought and Salt Stress" Plants 14, no. 14: 2101. https://doi.org/10.3390/plants14142101
APA StyleUllah, M. M., Aleem, M., Iqbal, M. M., Riaz, A., & Shi, A. (2025). Genome-Wide Characterization of VDAC Gene Family in Soybean (Glycine max L.) and In Silico Expression Profiling in Response to Drought and Salt Stress. Plants, 14(14), 2101. https://doi.org/10.3390/plants14142101










