Molecular Mechanisms of Plant Stress Memory: Roles of Non-Coding RNAs and Alternative Splicing
Abstract
1. Introduction
2. Non-Coding RNAs and Alternative Splicing
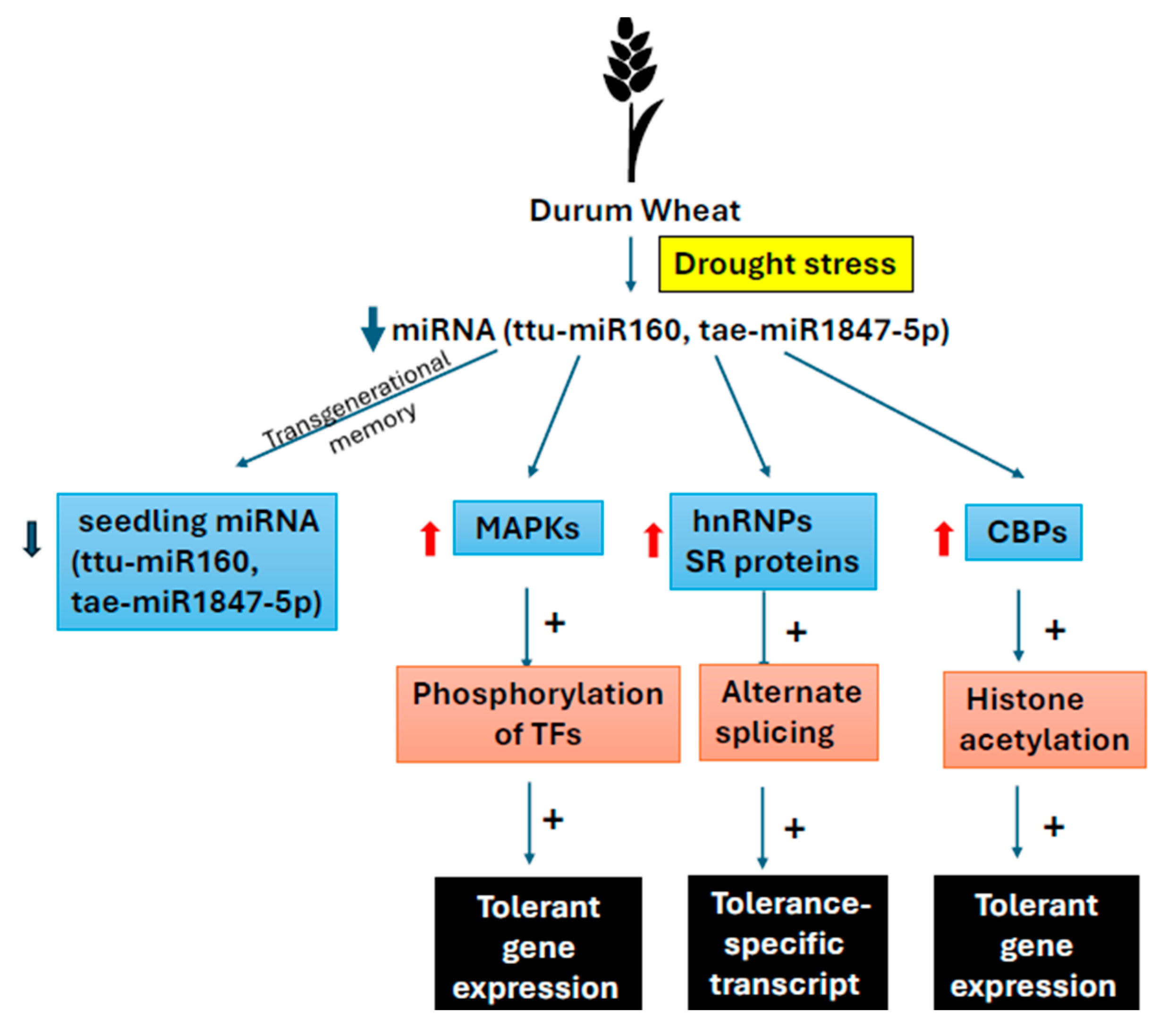
3. Drought Stress Memory
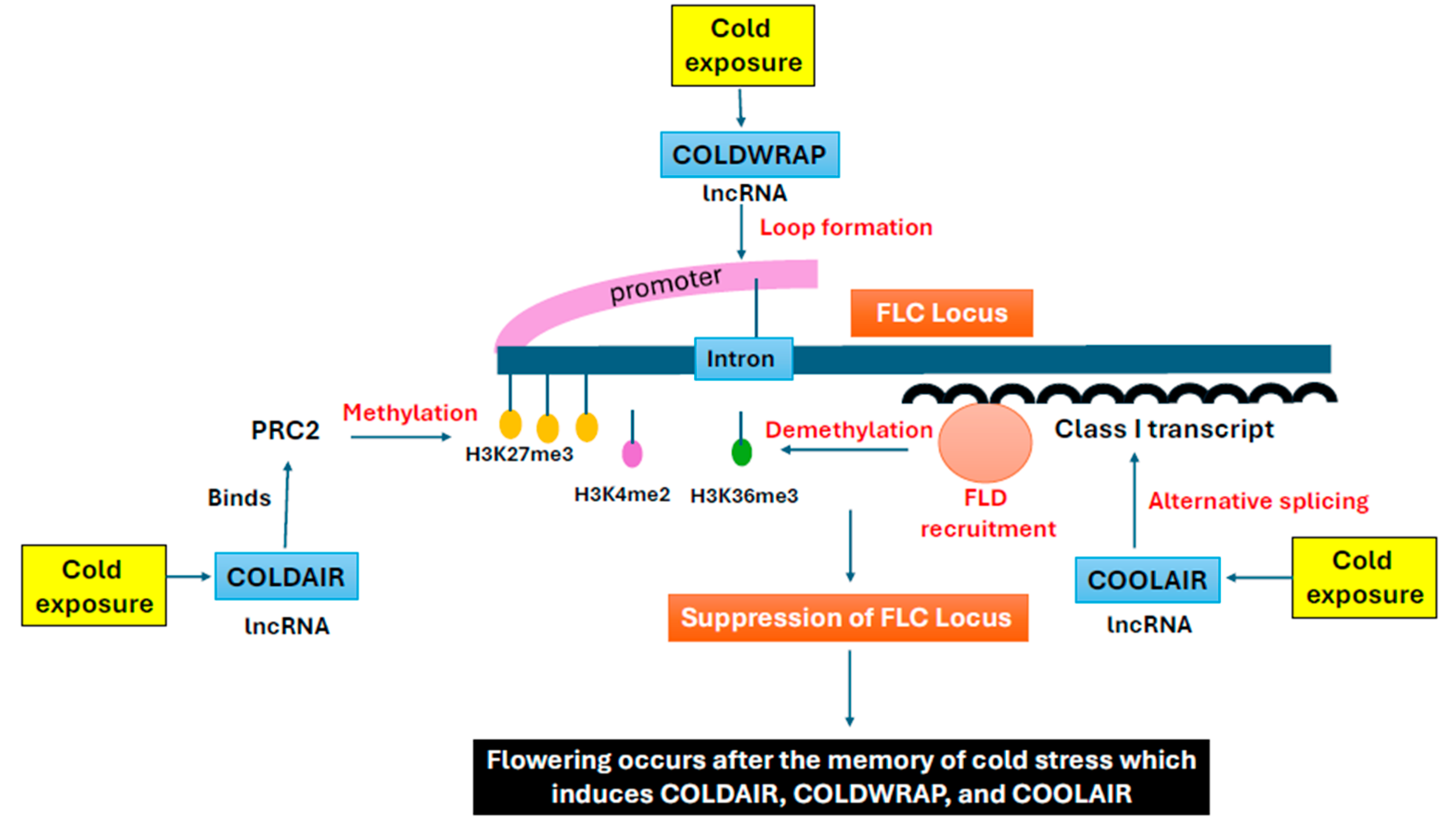
4. Cold Stress Memory
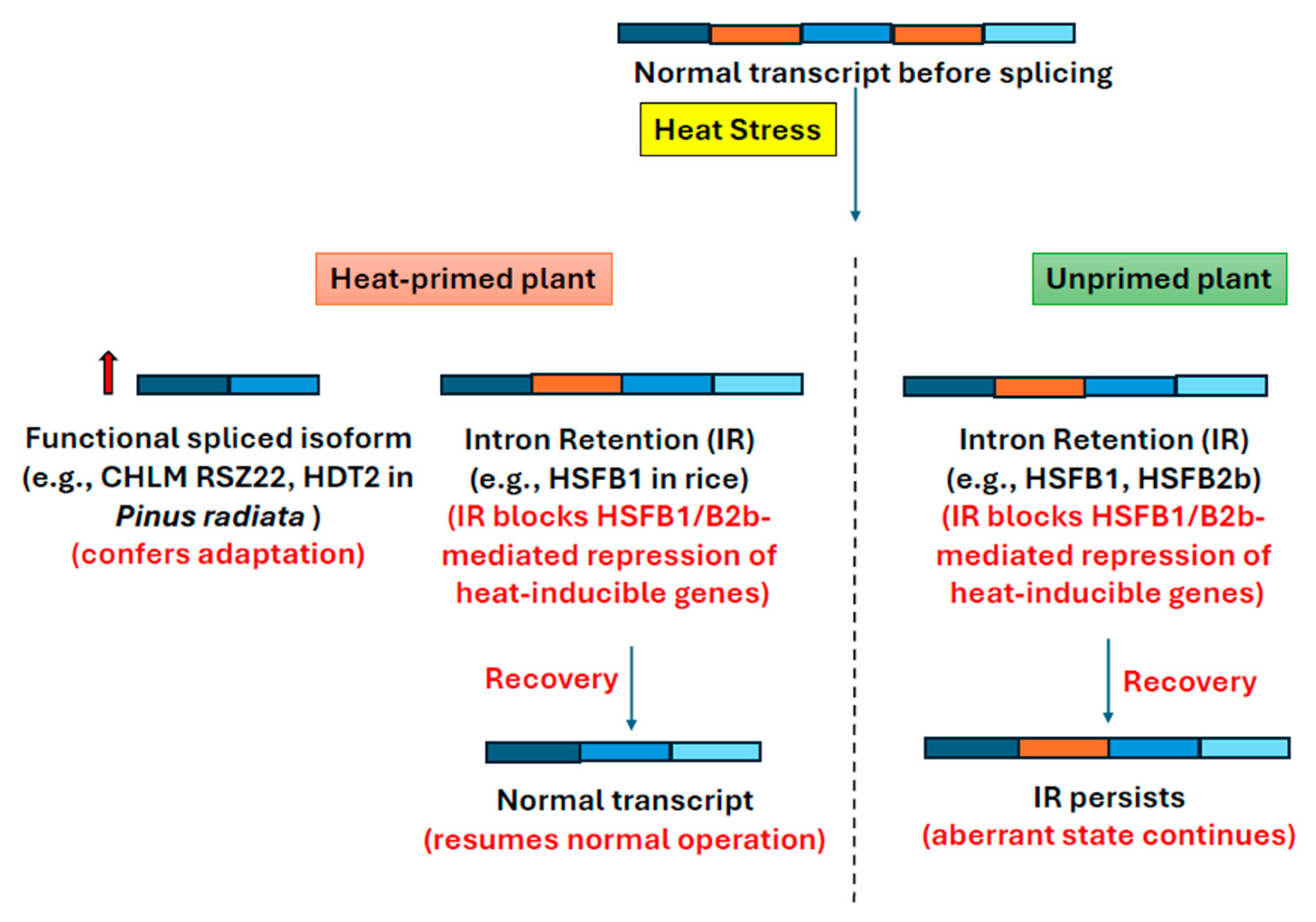
5. Heat Stress Memory
6. Heavy Metal Stress Memory
7. Salt Stress Memory
8. Biotic Stress Memory
9. Conclusions
Funding
Conflicts of Interest
References
- Liu, X.; Quan, W.; Bartels, D. Stress memory responses and seed priming correlate with drought tolerance in plants: An overview. Planta 2022, 255, 45. [Google Scholar] [CrossRef]
- Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant responses to heat stress: Physiology, transcription, noncoding rnas, and epigenetics. Int. J. Mol. Sci. 2020, 22, 117. [Google Scholar] [CrossRef]
- Hu, T.; Jin, Y.; Li, H.; Amombo, E.; Fu, J. Stress memory induced transcriptional and metabolic changes of perennial ryegrass (Lolium perenne) in response to salt stress. Physiol. Plant. 2016, 156, 54–69. [Google Scholar] [CrossRef]
- Di, Q.; Li, Y.; Li, S.; Shi, A.; Zhou, M.; Ren, H.; Yan, Y.; He, C.; Wang, J.; Sun, M.; et al. Photosynthesis Mediated by RBOH-Dependent Signaling Is Essential for Cold Stress Memory. Antioxidants 2022, 11, 969. [Google Scholar] [CrossRef]
- Verma, N.; Giri, S.K.; Singh, G.; Gill, R.; Kumar, A. Epigenetic regulation of heat and cold stress responses in crop plants. Plant Gene 2022, 29, 100351. [Google Scholar] [CrossRef]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N.; et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef]
- Bhatia, P.; Gupta, M. Micronutrient seed priming: New insights in ameliorating heavy metal stress. Environ. Sci. Pollut. Res. Int. 2022, 29, 58590–58606. [Google Scholar] [CrossRef]
- Goh, C.-H.; Nam, H.G.; Park, Y.S. Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 2003, 36, 240–255. [Google Scholar] [CrossRef]
- Virlouvet, L.; Fromm, M. Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 2015, 205, 596–607. [Google Scholar] [CrossRef]
- Pintó-Marijuan, M.; Cotado, A.; Fleta-Soriano, E.; Munné-Bosch, S. Drought stress memory in the photosynthetic mechanisms of an invasive CAM species, Aptenia cordifolia. Photosyn. Res. 2017, 131, 241–253. [Google Scholar] [CrossRef]
- Wang, X.; Vignjevic, M.; Jiang, D.; Jacobsen, S.; Wollenweber, B. Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J. Exp. Bot. 2014, 65, 6441–6456. [Google Scholar] [CrossRef]
- Ramírez, D.A.; Rolando, J.L.; Yactayo, W.; Monneveux, P.; Mares, V.; Quiroz, R. Improving potato drought tolerance through the induction of long-term water stress memory. Plant Sci. 2015, 238, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Auler, P.A.; Nogueira do Amaral, M.; Bolacel Braga, E.J.; Maserti, B. Drought stress memory in rice guard cells: Proteome changes and genomic stability of DNA. Plant Physiol. Biochem. 2021, 169, 49–62. [Google Scholar] [CrossRef]
- Schulze, W.X.; Altenbuchinger, M.; He, M.; Kränzlein, M.; Zörb, C. Proteome profiling of repeated drought stress reveals genotype-specific responses and memory effects in maize. Plant Physiol. Biochem. 2021, 159, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdallah, M.; Methenni, K.; Nouairi, I.; Zarrouk, M.; Youssef, N.B. Drought priming improves subsequent more severe drought in a drought-sensitive cultivar of olive cv. Chétoui. Sci. Hortic. 2017, 221, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Ellouzi, H.; Ben Hamed, K.; Asensi-Fabado, M.A.; Müller, M.; Abdelly, C.; Munné-Bosch, S. Drought and cadmium may be as effective as salinity in conferring subsequent salt stress tolerance in Cakile maritima. Planta 2013, 237, 1311–1323. [Google Scholar] [CrossRef]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef]
- Rahavi, M.R.; Migicovsky, Z.; Titov, V.; Kovalchuk, I. Transgenerational adaptation to heavy metal salts in Arabidopsis. Front. Plant Sci. 2011, 2, 91. [Google Scholar] [CrossRef]
- Lang-Mladek, C.; Popova, O.; Kiok, K.; Berlinger, M.; Rakic, B.; Aufsatz, W.; Jonak, C.; Hauser, M.-T.; Luschnig, C. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 2010, 3, 594–602. [Google Scholar] [CrossRef]
- Verhoeven, K.J.F.; Jansen, J.J.; van Dijk, P.J.; Biere, A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010, 185, 1108–1118. [Google Scholar] [CrossRef]
- Kathiria, P.; Sidler, C.; Golubov, A.; Kalischuk, M.; Kawchuk, L.M.; Kovalchuk, I. Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 2010, 153, 1859–1870. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Transgenerational Effects of Water-Deficit and Heat Stress on Germination and Seedling Vigour-New Insights from Durum Wheat microRNAs. Plants 2020, 9, 189. [Google Scholar] [CrossRef]
- Rasmann, S.; De Vos, M.; Casteel, C.L.; Tian, D.; Halitschke, R.; Sun, J.Y.; Agrawal, A.A.; Felton, G.W.; Jander, G. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 2012, 158, 854–863. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Chae, S.; Oh, N.-I.; Nguyen, N.H.; Cheong, J.-J. Recurrent drought conditions enhance the induction of drought stress memory genes in Glycine max L. Front. Genet. 2020, 11, 576086. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Ding, Y.; Virlouvet, L.; Liu, N.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 141. [Google Scholar] [CrossRef]
- Yung, W.-S.; Wang, Q.; Huang, M.; Wong, F.-L.; Liu, A.; Ng, M.-S.; Li, K.-P.; Sze, C.-C.; Li, M.-W.; Lam, H.-M. Priming-induced alterations in histone modifications modulate transcriptional responses in soybean under salt stress. Plant J. 2022, 109, 1575–1590. [Google Scholar] [CrossRef]
- Levine, M. Paused RNA polymerase II as a developmental checkpoint. Cell 2011, 145, 502–511. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Cho, J. Transposon-Derived Non-coding RNAs and Their Function in Plants. Front. Plant Sci. 2018, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Matzke, M.A.; Matzke, A.J.; Pruss, G.J.; Vance, V.B. RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 2001, 11, 221–227. [Google Scholar] [CrossRef]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA Methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.-K. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011, 14, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, B. siRNA-directed DNA Methylation in Plants. Curr. Genom. 2015, 16, 23–31. [Google Scholar] [CrossRef]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Timmermans, M.C.P. Small RNAs are on the move. Nature 2010, 467, 415–419. [Google Scholar] [CrossRef]
- Yoshikawa, M. Biogenesis of trans-acting siRNAs, endogenous secondary siRNAs in plants. Genes. Genet. Syst. 2013, 88, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.-L.; Bartel, D.P.; Crété, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes. Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Szachniuk, M. Discovering Structural Motifs in miRNA Precursors from the Viridiplantae Kingdom. Molecules 2018, 23, 1367. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 2004, 14, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Wu, G.; Gonzalez-Sulser, A.; Vaucheret, H.; Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3691–3696. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Bao, N.; Lye, K.-W.; Barton, M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 2004, 7, 653–662. [Google Scholar] [CrossRef]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Roces, V.; Lamelas, L.; Valledor, L.; Carbó, M.; Cañal, M.J.; Meijón, M. Integrative analysis in Pinus revealed long-term heat stress splicing memory. Plant J. 2022, 112, 998–1013. [Google Scholar] [CrossRef]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef]
- Yue, H.; Zhang, H.; Su, N.; Sun, X.; Zhao, Q.; Weining, S.; Nie, X.; Yue, W. Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2022, 23, 5917. [Google Scholar] [CrossRef]
- Guedes, F.A.d.F.; Nobres, P.; Rodrigues Ferreira, D.C.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M.D. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Kantar, M.; Unver, T.; Budak, H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct. Integr. Genom. 2010, 10, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. miR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and Transcriptome Analyses Reveal Short-Term Responses and Formation of Memory Under Drought Stress in Rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef]
- Smékalová, V.; Doskočilová, A.; Komis, G.; Samaj, J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 2014, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Samaj, J.; Ovecka, M.; Hlavacka, A.; Lecourieux, F.; Meskiene, I.; Lichtscheidl, I.; Lenart, P.; Salaj, J.; Volkmann, D.; Bögre, L.; et al. Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth. EMBO J. 2002, 21, 3296–3306. [Google Scholar] [CrossRef]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef]
- Ling, Y.; Alshareef, S.; Butt, H.; Lozano-Juste, J.; Li, L.; Galal, A.A.; Moustafa, A.; Momin, A.A.; Tashkandi, M.; Richardson, D.N.; et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 2017, 89, 291–309. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Dixon, L.E.; Hepworth, J.; Irwin, J.A. Vernalisation. In Encyclopedia of Life Sciences; Wiley: Hoboken, NJ, USA, 2005; pp. 1–11. ISBN 9780470015902. [Google Scholar]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Nielsen, M.; Menon, G.; Zhao, Y.; Mateo-Bonmati, E.; Wolff, P.; Zhou, S.; Howard, M.; Dean, C. COOLAIR and PRC2 function in parallel to silence FLC during vernalization. Proc. Natl. Acad. Sci. USA 2024, 121, e2311474121. [Google Scholar] [CrossRef] [PubMed]
- Hornyik, C.; Duc, C.; Rataj, K.; Terzi, L.C.; Simpson, G.G. Alternative polyadenylation of antisense RNAs and flowering time control. Biochem. Soc. Trans. 2010, 38, 1077–1081. [Google Scholar] [CrossRef]
- Marquardt, S.; Raitskin, O.; Wu, Z.; Liu, F.; Sun, Q.; Dean, C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 2014, 54, 156–165. [Google Scholar] [CrossRef]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef]
- Yang, H.; Howard, M.; Dean, C. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 2014, 24, 1793–1797. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.; Zhang, Z.; Mullasseri, S.; Kalendar, R.; Ahmad, Z.; Sharma, A.; Liu, G.; Zhou, M.; Wei, Q. Epigenetic stress memory: A new approach to study cold and heat stress responses in plants. Front. Plant Sci. 2022, 13, 1075279. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312.e4. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.-R.; Bäurle, I. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef]
- Cho, S.H.; Coruh, C.; Axtell, M.J. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Lu, Y.; de Ruiter, M.; Cariaso, M.; Prins, M.; van Tunen, A.; He, Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 2012, 63, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Wang, Y.; Yao, Y.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Stief, A.; Brzezinka, K.; Lämke, J.; Bäurle, I. Epigenetic responses to heat stress at different time scales and the involvement of small RNAs. Plant Signal. Behav. 2014, 9, e970430. [Google Scholar] [CrossRef] [PubMed]
- Charng, Y.-Y.; Liu, H.-C.; Liu, N.-Y.; Chi, W.-T.; Wang, C.-N.; Chang, S.-H.; Wang, T.-T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef]
- Szaker, H.M.; Darkó, É.; Medzihradszky, A.; Janda, T.; Liu, H.-C.; Charng, Y.-Y.; Csorba, T. miR824/AGAMOUS-LIKE16 Module Integrates Recurring Environmental Heat Stress Changes to Fine-Tune Poststress Development. Front. Plant Sci. 2019, 10, 1454. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef]
- Matsunaga, W.; Ohama, N.; Tanabe, N.; Masuta, Y.; Masuda, S.; Mitani, N.; Yamaguchi-Shinozaki, K.; Ma, J.F.; Kato, A.; Ito, H. A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Front. Plant Sci. 2015, 6, 48. [Google Scholar] [CrossRef]
- Matsunaga, W.; Kobayashi, A.; Kato, A.; Ito, H. The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 824–833. [Google Scholar] [CrossRef]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Gu, X.; Deng, X.; Qiu, Q.; Li, Q.; Zhang, Y.; Wang, M.; Deng, Y.; Wang, E.; et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019, 29, 379–390. [Google Scholar] [CrossRef]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; von Wettstein, D.; Liu, B. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 2016, 5, e13546. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Maekawa, S.; Yasuda, S.; Sonoda, Y.; Katoh, E.; Ichikawa, T.; Nakazawa, M.; Seki, M.; Shinozaki, K.; Matsui, M.; et al. CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 2009, 60, 852–864. [Google Scholar] [CrossRef]
- Maekawa, S.; Sato, T.; Asada, Y.; Yasuda, S.; Yoshida, M.; Chiba, Y.; Yamaguchi, J. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol. Biol. 2012, 79, 217–227. [Google Scholar] [CrossRef]
- Yasuda, S.; Sato, T.; Maekawa, S.; Aoyama, S.; Fukao, Y.; Yamaguchi, J. Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins. J. Biol. Chem. 2014, 289, 15179–15193. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Du, M.; Liang, X.; Fan, F.; Huang, G.; Zou, Y.; Bai, J.; Lu, D. The calcium-dependent protein kinase CPK28 is targeted by the ubiquitin ligases ATL31 and ATL6 for proteasome-mediated degradation to fine-tune immune signaling in Arabidopsis. Plant Cell 2022, 34, 679–697. [Google Scholar] [CrossRef]
- Maekawa, S.; Inada, N.; Yauda, S.; Fukao, Y.; Fujiwara, M.; Sato, T.; Yamaguchi, J. The carbon/nitrogen regulator ARABIDOPSIS TOXICOS EN LEVADURA31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis. Plant Physiol. 2014, 164, 879–887. [Google Scholar] [CrossRef]
- Aoyama, S.; Huarancca Reyes, T.; Guglielminetti, L.; Lu, Y.; Morita, Y.; Sato, T.; Yamaguchi, J. Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol. 2014, 55, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Aoyama, S.; Hasegawa, Y.; Sato, T.; Yamaguchi, J. Arabidopsis CBL-Interacting Protein Kinases Regulate Carbon/Nitrogen-Nutrient Response by Phosphorylating Ubiquitin Ligase ATL31. Mol. Plant 2017, 10, 605–618. [Google Scholar] [CrossRef]
- Slaughter, A.; Daniel, X.; Flors, V.; Luna, E.; Hohn, B.; Mauch-Mani, B. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012, 158, 835–843. [Google Scholar] [CrossRef]
- Luna, E.; Bruce, T.J.A.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Maizel, A.; Markmann, K.; Timmermans, M.; Wachter, A. To move or not to move: Roles and specificity of plant RNA mobility. Curr. Opin. Plant Biol. 2020, 57, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Gismondi, A.; Di Marco, G.; Camoni, L.; Montesano, C.; Braglia, R.; Marra, M.; Canini, A. MicroRNA Expression Profiles in Moringa oleifera Lam. Seedlings at Different Growth Conditions. J. Plant Growth Regul. 2023, 42, 2115–2123. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sintaha, M. Molecular Mechanisms of Plant Stress Memory: Roles of Non-Coding RNAs and Alternative Splicing. Plants 2025, 14, 2021. https://doi.org/10.3390/plants14132021
Sintaha M. Molecular Mechanisms of Plant Stress Memory: Roles of Non-Coding RNAs and Alternative Splicing. Plants. 2025; 14(13):2021. https://doi.org/10.3390/plants14132021
Chicago/Turabian StyleSintaha, Mariz. 2025. "Molecular Mechanisms of Plant Stress Memory: Roles of Non-Coding RNAs and Alternative Splicing" Plants 14, no. 13: 2021. https://doi.org/10.3390/plants14132021
APA StyleSintaha, M. (2025). Molecular Mechanisms of Plant Stress Memory: Roles of Non-Coding RNAs and Alternative Splicing. Plants, 14(13), 2021. https://doi.org/10.3390/plants14132021






