Gene Localization and Functional Validation of GmPDH1 in Soybean Against Cyst Nematode Race 4
Abstract
1. Introduction
2. Results
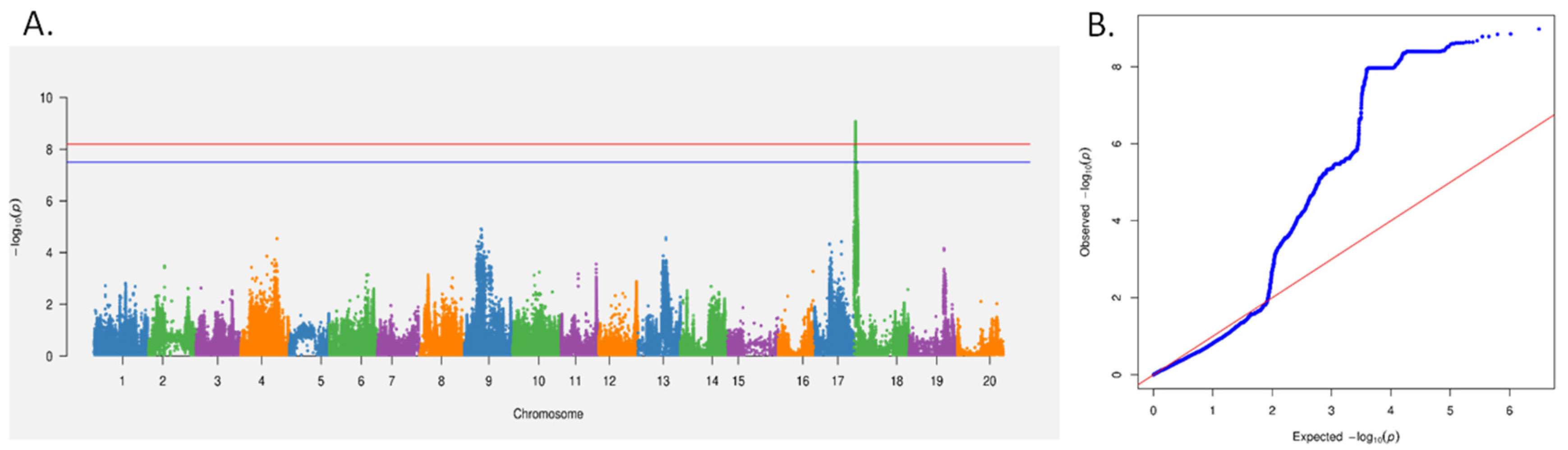
2.1. Phenotypic Profiling and Candidate Gene Identification in Soybean in Response to SCN4 Infestation
2.2. Developmental Stage-Specific Nematode Quantification in Resistant vs. Susceptible Cultivars
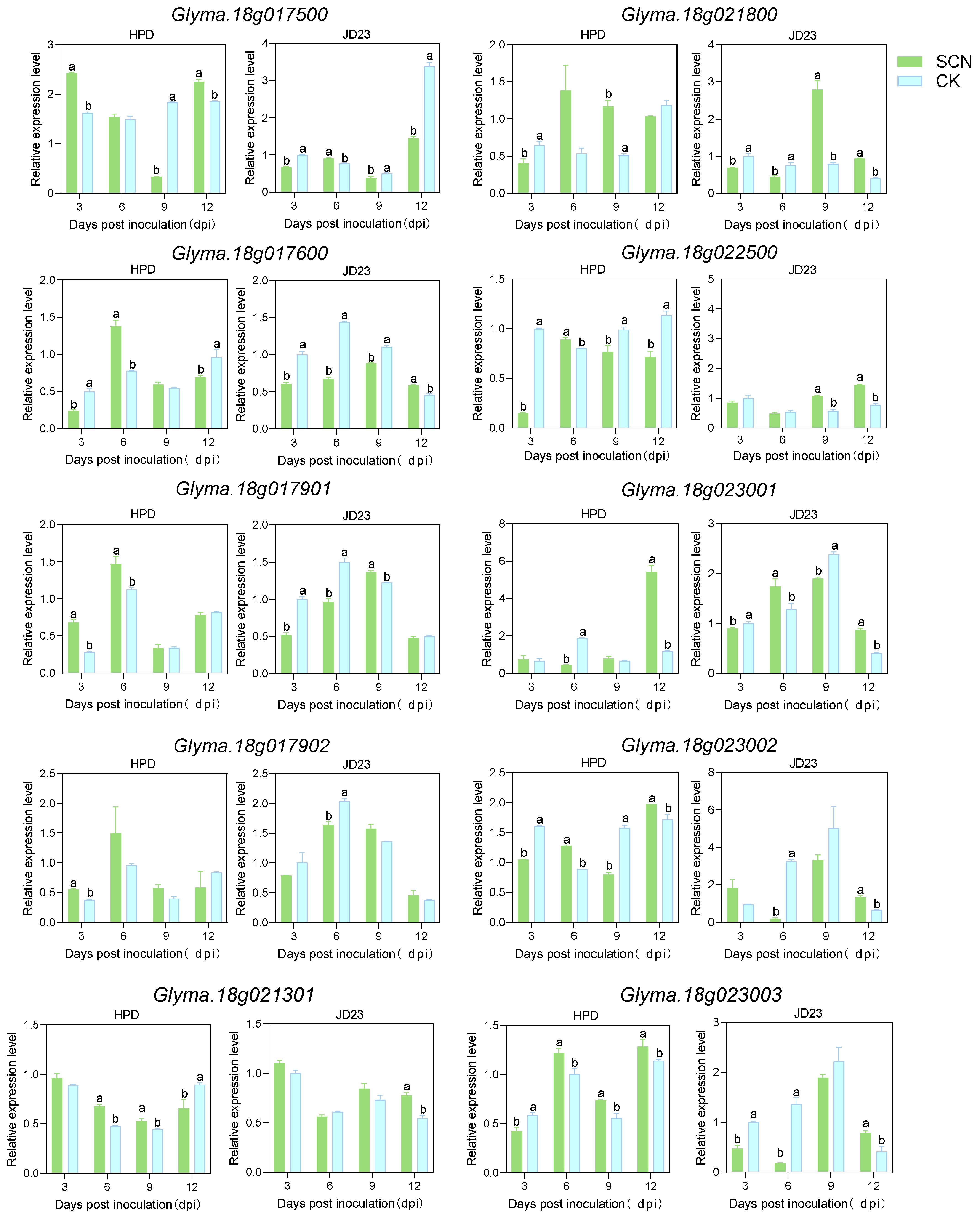
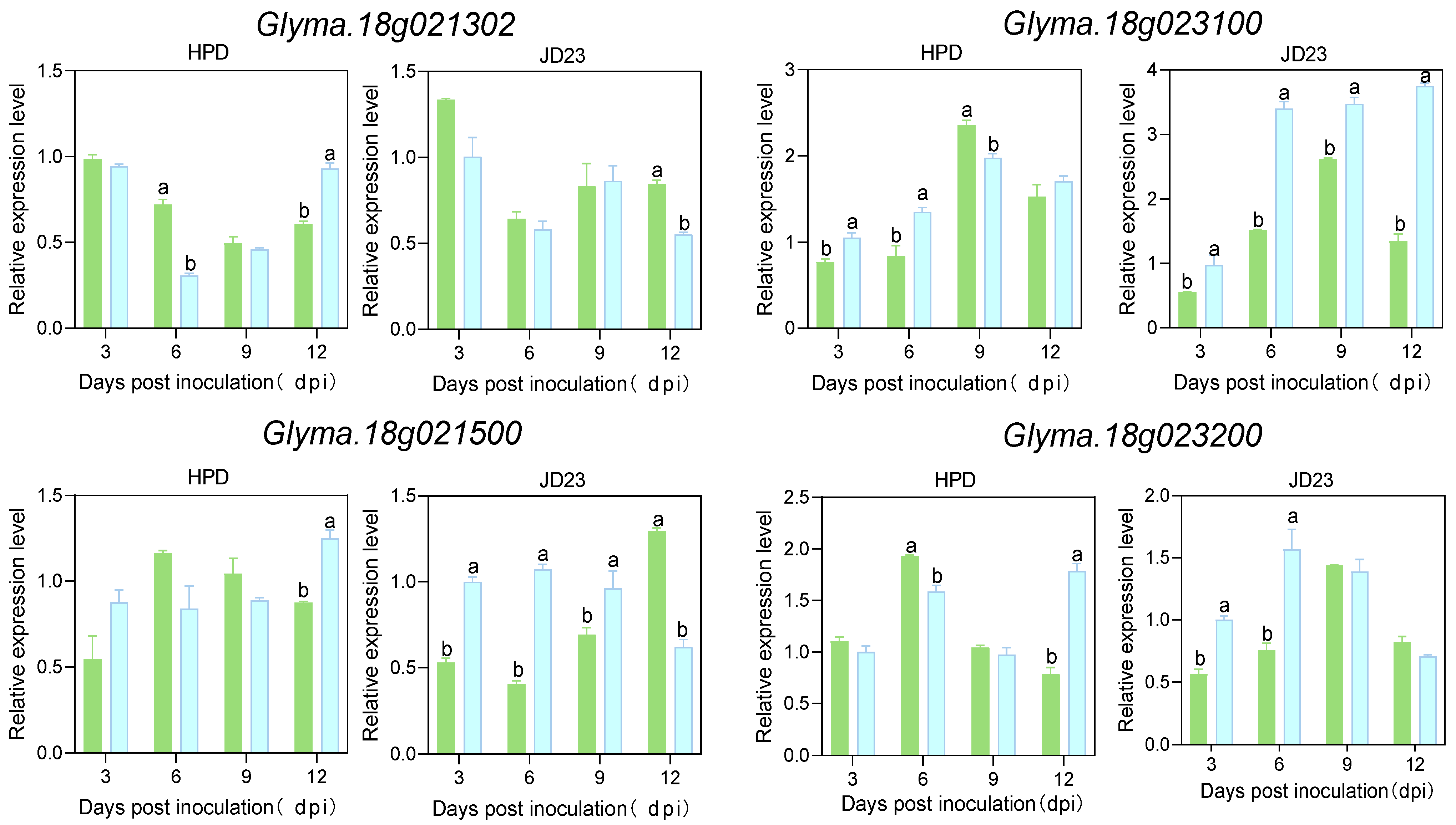
2.3. The Expression Patterns of Candidate Genes During SCN4 Infection
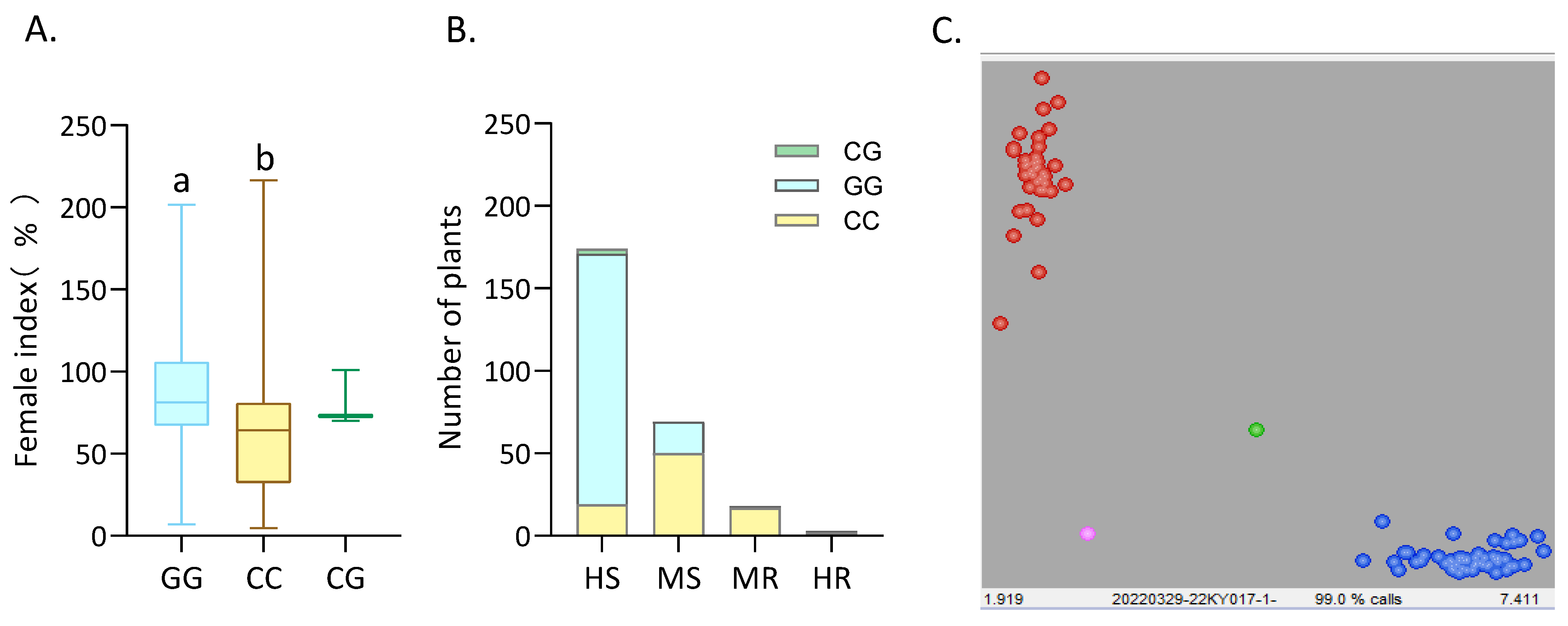
2.4. Kompetitive Allele-Specific PCR (KASP) Marker Development for the SCN Resistance-Associated GmPDH1 Locus
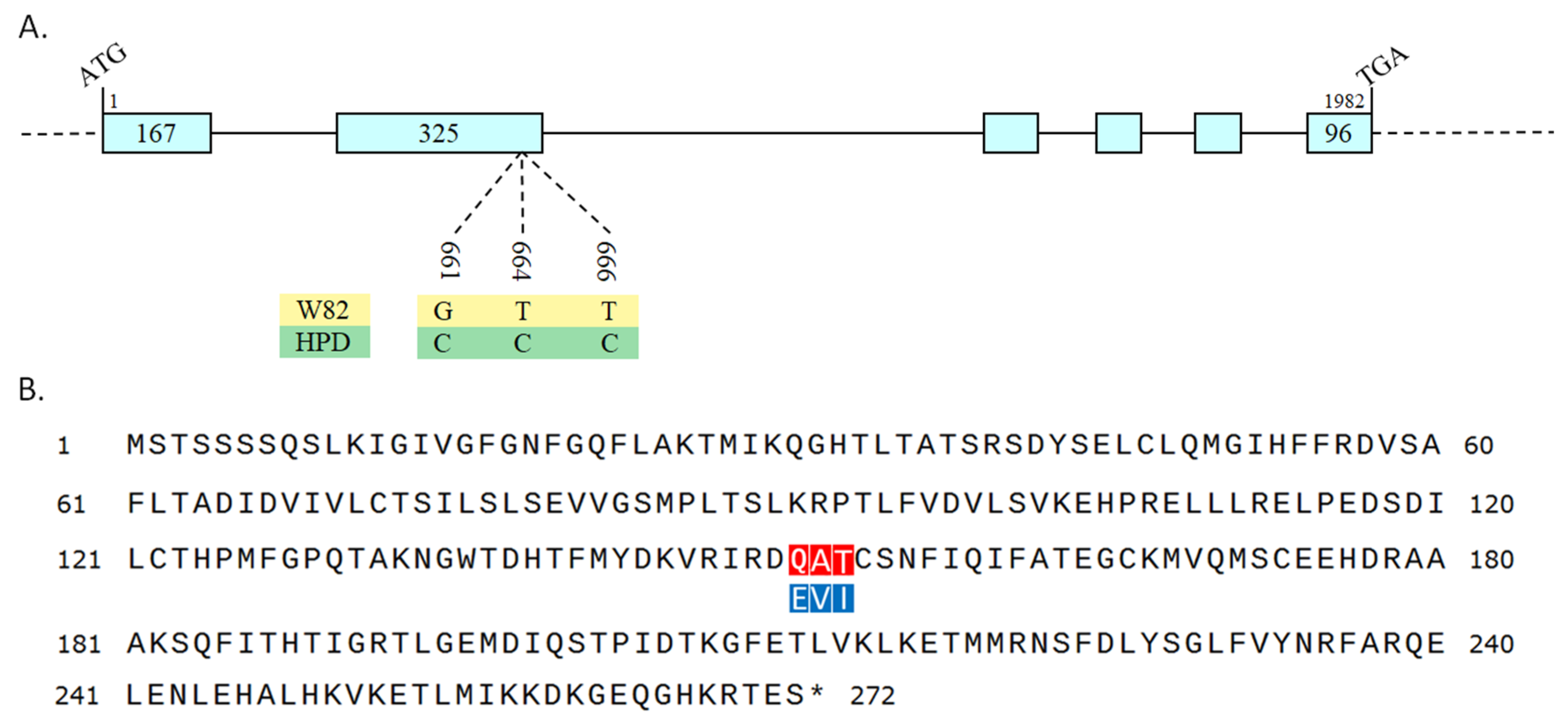
2.5. Structural and Promoter Cis-Element Profiling of GmPDH1
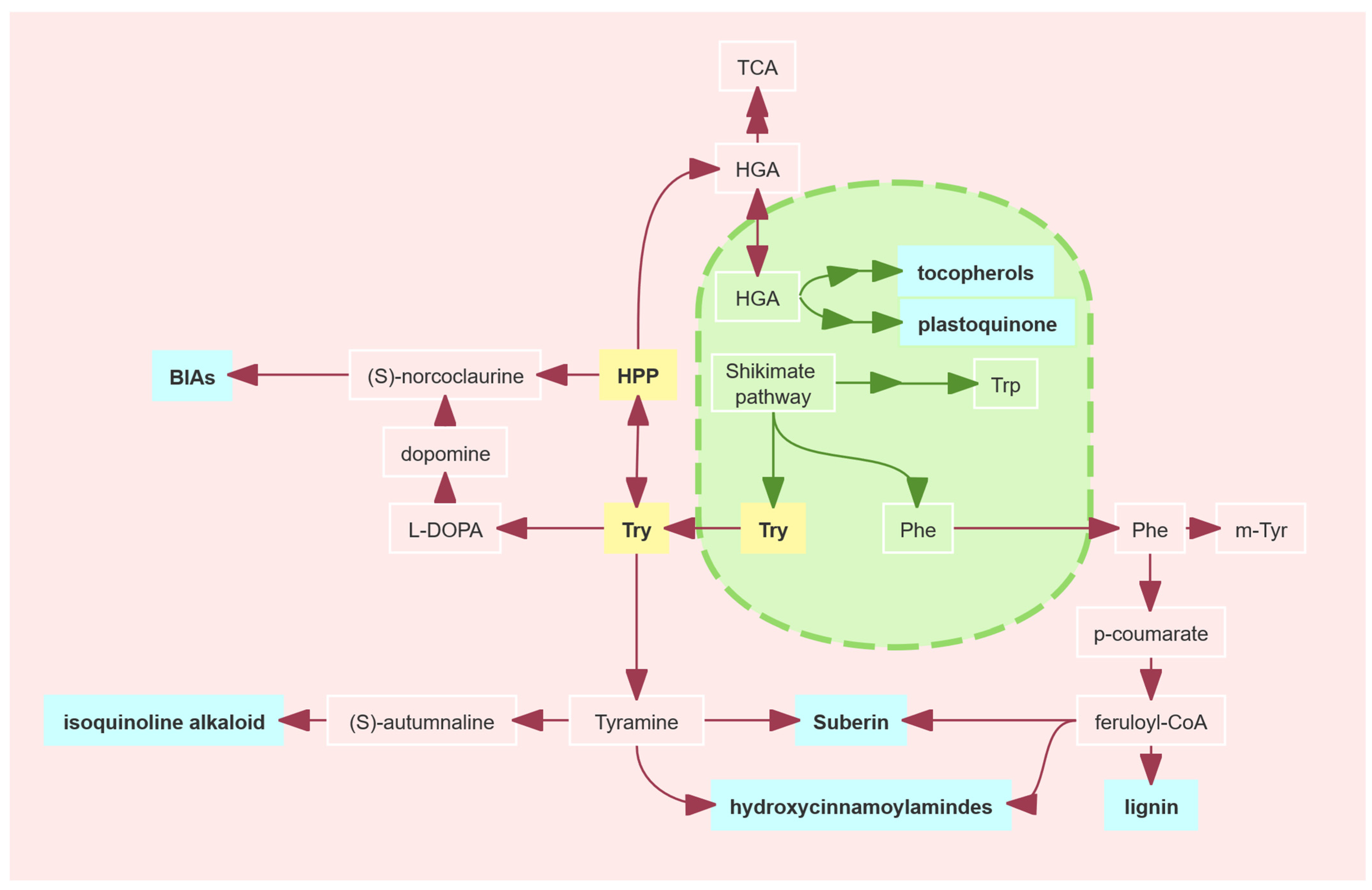
2.6. Physicochemical Characterization and Tertiary Structure Prediction of the GmPDH1 Protein
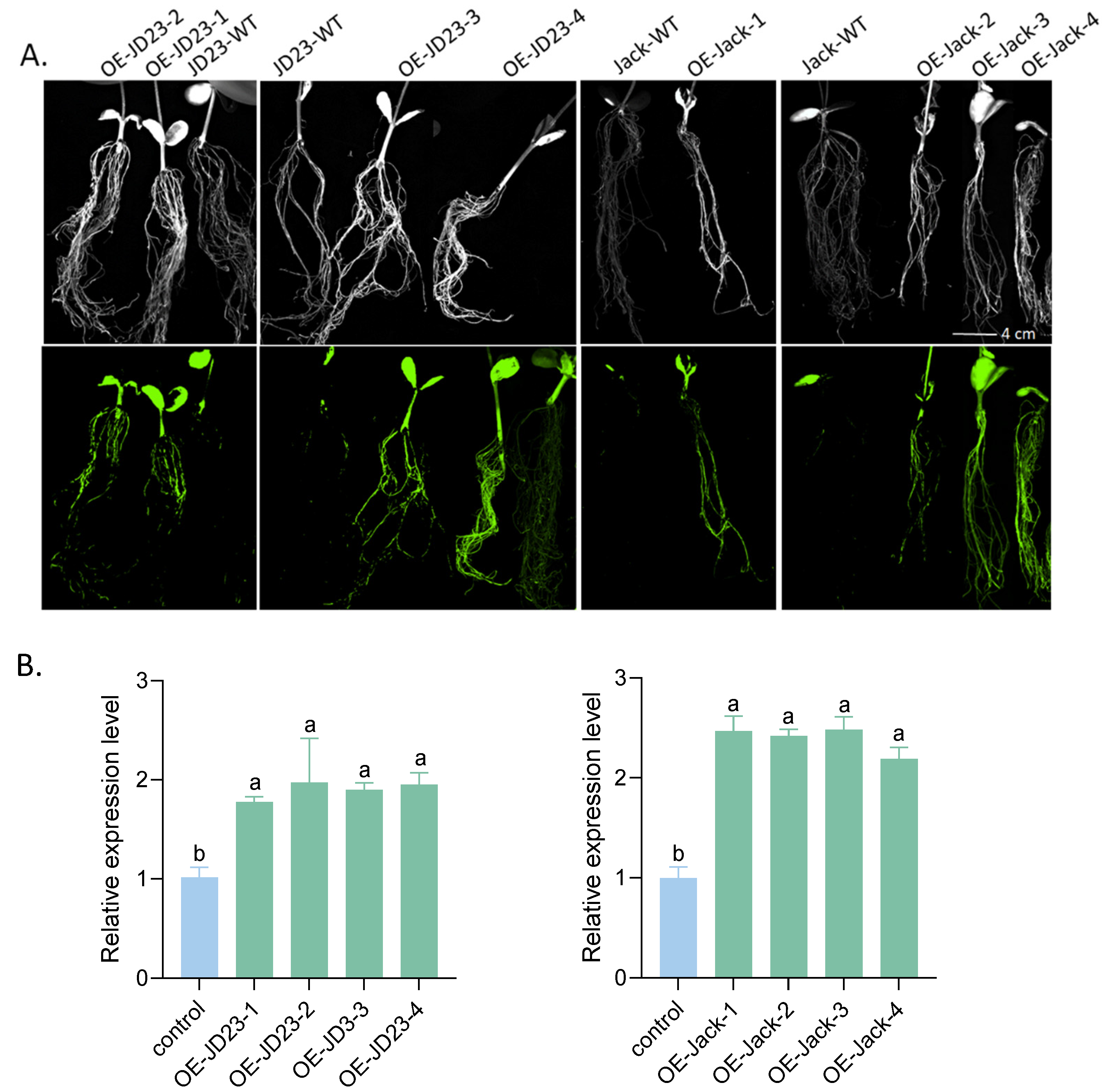
2.7. Transgenic Soybean Overexpressing GmPDH1 Conferred Further Resistance to SCN4
2.8. GmPDH1 Loss of Function Reduces SCN4 Resistance
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phenotype Identification and GWAS
4.3. Development Process of SCN4 and Expression Patterns of Related Genes
4.4. Development and Validation of KASP Markers for the SCN4 Resistance Gene GmPDH1 in Glycine Max
4.5. Cloning and Bioinformatics Analysis of GmPDH1
4.6. GmPDH1 Expression Analysis
4.7. GmPDH1 CRISPR gRNA Design and Off-Target Analysis
4.8. Evaluation of SCN4 Resistance of Transgenic Hairy Roots
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SCN | Soybean cyst nematode |
| RIL | Recombinant inbred line |
| HPD | Huipizhiheidou |
| JD23 | Jindou23 |
| GWAS | Genome-wide association analysis |
| RT-qPCR | Reverse transcription quantitative PCR |
| KASP | Kompetitive Allele Specific PCR |
| NSF | N-ethylmaleimide-sensitive fusion protein |
| Tyr | L-tyrosine |
| CM | Chorismate mutase |
| PPA-AT | Prephenate aminotransferase |
| PDH | Prephenate dehydrogenase |
| ADH | Alcohol dehydrogenase |
| Phe | L-Phenylalanine |
| HPP | 4-hydroxyphenylpyruvic acid |
| DPI | Days post-inoculation |
| CDS | Coding sequence |
| FI | female index |
| gRNA | Guide RNA |
| SNP | Single nucleotide polymorphism |
| MLM | Mixed linear model |
| AQP | Allele-Specific Quantitative PCR |
| OE | Overexpression |
| EVs | Empty vector controls |
References
- Abad, P.; Gouzy, J.; Aury, J.-M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome Sequence of the Metazoan Plant-Parasitic Nematode Meloidogyne Incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.H.; Wang, J.; Li, Y.G.; Han, Y.P. Soybean Disease Resistance, Yield and Quality Correlation. Sci. Agric. Sin. 2024, 57, 2061–2064. [Google Scholar]
- Bandara, A.Y.; Weerasooriya, D.K.; Bradley, C.A.; Allen, T.W.; Esker, P.D. Dissecting the Economic Impact of Soybean Diseases in the United States over Two Decades. PLoS ONE 2020, 15, e0231141. [Google Scholar] [CrossRef] [PubMed]
- Tylka, G.L.; Marett, C.C. Known Distribution of the Soybean Cyst Nematode, Heterodera glycines, in the United States and Canada in 2020. Plant Health Prog. 2021, 22, 72–74. [Google Scholar] [CrossRef]
- Kim, D.G.; Riggs, R.D.; Robbins, R.T.; Rakes, L. Distribution of Races of Heterodera glycines in the Central United States. J. Nematol. 1997, 29, 173–179. [Google Scholar]
- Klink, V.P.; Hosseini, P.; Matsye, P.; Alkharouf, N.W.; Matthews, B.F. A Gene Expression Analysis of Syncytia Laser Microdissected from the Roots of the Glycine Max (Soybean) Genotype PI 548402 (Peking) Undergoing a Resistant Reaction after Infection by Heterodera glycines (Soybean Cyst Nematode). Plant Mol. Biol. 2009, 71, 525–567. [Google Scholar] [CrossRef]
- Zhao, M.M. Soybean Cyst Nematode Occurrence Characteristics and Green Prevention and Control Measures. Seed Sci. Technol. 2022, 40, 97–99. [Google Scholar]
- Tian, Z.; Nepomuceno, A.L.; Song, Q.; Stupar, R.M.; Liu, B.; Kong, F.; Ma, J.; Lee, S.-H.; Jackson, S.A. Soybean2035: A Decadal Vision for Soybean Functional Genomics and Breeding. Mol. Plant 2025, 18, 245–271. [Google Scholar] [CrossRef]
- Meng, F.L.; Yu, J.Y.; Li, C.J.; Huang, M.H.; Zhao, L.; Wang, X.; Jiang, Y.; Qin, R.F.; Wang, C.L. Research progress on occurrence and management of soybean cyst nematode in Northeast China. J. Northeast. Agric. Univ. 2022, 53, 87–94. [Google Scholar]
- Brucker, E.; Carlson, S.; Wright, E.; Niblack, T.; Diers, B. Rhg1 Alleles from Soybean PI 437654 and PI 88788 Respond Differentially to Isolates of Heterodera glycines in the Greenhouse. Theor. Appl. Genet. 2005, 111, 44–49. [Google Scholar] [CrossRef]
- Kim, M.; Hyten, D.; Niblack, T.; Diers, B. Stacking Resistance Alleles from Wild and Domestic Soybean Sources Improves Soybean Cyst Nematode Resistance. Crop Sci. 2011, 51, 934–943. [Google Scholar] [CrossRef]
- Jiao, Y.; Vuong, T.D.; Liu, Y.; Meinhardt, C.; Liu, Y.; Joshi, T.; Cregan, P.B.; Xu, D.; Shannon, J.G.; Nguyen, H.T. Identification and Evaluation of Quantitative Trait Loci Underlying Resistance to Multiple HG Types of Soybean Cyst Nematode in Soybean PI 437655. Theor. Appl. Genet. 2015, 128, 15–23. [Google Scholar] [CrossRef]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kandoth, P.K.; Lakhssassi, N.; Kang, J.; Colantonio, V.; Heinz, R.; Yeckel, G.; Zhou, Z.; Bekal, S.; Dapprich, J.; et al. The Soybean GmSNAP18 Gene Underlies Two Types of Resistance to Soybean Cyst Nematode. Nat. Commun. 2017, 8, 14822. [Google Scholar] [CrossRef]
- He, L.; Ghani, N.N.U.; Chen, L.; Liu, Q.; Zheng, J.; Han, S. Research Progress on the Functional Study of Host Resistance-Related Genes against Heterodera glycines. Crop Health 2023, 1, 8. [Google Scholar] [CrossRef]
- Bayless, A.M.; Zapotocny, R.W.; Grunwald, D.J.; Amundson, K.K.; Diers, B.W.; Bent, A.F. An Atypical N-Ethylmaleimide Sensitive Factor Enables the Viability of Nematode-Resistant Rhg1 Soybeans. Proc. Natl. Acad. Sci. USA 2018, 115, E4512–E4521. [Google Scholar] [CrossRef]
- Shaibu, A.S.; Zhang, S.; Ma, J.; Feng, Y.; Huai, Y.; Qi, J.; Li, J.; Abdelghany, A.M.; Azam, M.; Htway, H.T.P.; et al. The GmSNAP11 Contributes to Resistance to Soybean Cyst Nematode Race 4 in Glycine Max. Front. Plant Sci. 2022, 13, 939763. [Google Scholar] [CrossRef]
- Patil, G.B.; Lakhssassi, N.; Wan, J.; Song, L.; Zhou, Z.; Klepadlo, M.; Vuong, T.D.; Stec, A.O.; Kahil, S.S.; Colantonio, V.; et al. Whole-genome Re-sequencing Reveals the Impact of the Interaction of Copy Number Variants of the Rhg1 and Rhg4 Genes on Broad-based Resistance to Soybean Cyst Nematode. Plant Biotechnol. J. 2019, 17, 1595–1611. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kandoth, P.K.; Warren, S.D.; Yeckel, G.; Heinz, R.; Alden, J.; Yang, C.; Jamai, A.; El-Mellouki, T.; Juvale, P.S.; et al. A Soybean Cyst Nematode Resistance Gene Points to a New Mechanism of Plant Resistance to Pathogens. Nature 2012, 492, 256–260. [Google Scholar] [CrossRef]
- Yang, X.; Liu, T.; Yang, R.; Fan, H.; Liu, X.; Xuan, Y.; Wang, Y.; Chen, L.; Duan, Y.; Zhu, X. Overexpression of GmPAL Genes Enhances Soybean Resistance Against Heterodera glycines. Mol. Plant-Microbe Interact. 2024, 37, 416–423. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. New Insights into the Shikimate and Aromatic Amino Acids Biosynthesis Pathways in Plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Bentley, R. The Shikimate Pathway—A Metabolic Tree with Many Branches. Crit. Rev. Biochem. Mol. Biol. 1990, 25, 307–384. [Google Scholar] [CrossRef]
- Christendat, D.; Turnbull, J.L. Identifying Groups Involved in the Binding of Prephenate to Prephenate Dehydrogenase from Escherichia Coli. Biochemistry 1999, 38, 4782–4793. [Google Scholar] [CrossRef]
- Rippert, P.; Puyaubert, J.; Grisollet, D.; Derrier, L.; Matringe, M. Tyrosine and Phenylalanine Are Synthesized within the Plastids in Arabidopsis. Plant Physiol. 2009, 149, 1251–1260. [Google Scholar] [CrossRef]
- Šindelář, L.; Singh, B.K. Plant Amino Acids. Biochemistry and Biotechnology. Biol. Plant. 2000, 43, 358. [Google Scholar] [CrossRef]
- Schenck, C.A.; Chen, S.; Siehl, D.L.; Maeda, H.A. Non-Plastidic, Tyrosine-Insensitive Prephenate Dehydrogenases from Legumes. Nat. Chem. Biol. 2015, 11, 52–57. [Google Scholar] [CrossRef]
- Rippert, P.; Matringe, M. Purification and Kinetic Analysis of the Two Recombinant Arogenate Dehydrogenase Isoforms of Arabidopsis Thaliana. Eur. J. Biochem. 2002, 269, 4753–4761. [Google Scholar] [CrossRef]
- Connelly, J.A.; Conn, E.E. Tyrosine Biosynthesis in Sorghum Bicolor: Isolation and Regulatory Properties of Arogenate Dehydrogenase. Z. Für Naturforschung. C 1986, 41, 69–78. [Google Scholar] [CrossRef]
- Byng, G.; Whitaker, R.; Flick, C.; Jensen, R.A. Enzymology of L-Tyrosine Biosynthesis in Corn (Zea Ma Ys). Phytochemistry 1981, 20, 1289–1292. [Google Scholar] [CrossRef]
- Schenck, C.A.; Maeda, H.A. Tyrosine Biosynthesis, Metabolism, and Catabolism in Plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef] [PubMed]
- Coley, P.D.; Endara, M.-J.; Ghabash, G.; Kidner, C.A.; Nicholls, J.A.; Pennington, R.T.; Mills, A.G.; Soule, A.J.; Lemes, M.R.; Stone, G.N.; et al. Macroevolutionary Patterns in Overexpression of Tyrosine: An Anti-Herbivore Defence in a Speciose Tropical Tree Genus, Inga (Fabaceae). J. Ecol. 2019, 107, 1620–1632. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chapple, C. The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Ithal, N.; Recknor, J.; Nettleton, D.; Hearne, L.; Maier, T.; Baum, T.J.; Mitchum, M.G. Parallel Genome-Wide Expression Profiling of Host and Pathogen During Soybean Cyst Nematode Infection of Soybean. Mol. Plant-Microbe Interact. 2007, 20, 293–305. [Google Scholar] [CrossRef]
- Schenck, C.A.; Westphal, J.; Jayaraman, D.; Garcia, K.; Wen, J.; Mysore, K.S.; Ané, J.-M.; Sumner, L.W.; Maeda, H.A. Role of Cytosolic, Tyrosine-Insensitive Prephenate Dehydrogenase in Medicago Truncatula. Plant Direct 2020, 4, e00218. [Google Scholar] [CrossRef]
- Stacey, M.G.; Cahoon, R.E.; Nguyen, H.T.; Cui, Y.; Sato, S.; Nguyen, C.T.; Phoka, N.; Clark, K.M.; Liang, Y.; Forrester, J.; et al. Identification of Homogentisate Dioxygenase as a Target for Vitamin E Biofortification in Oilseeds. Plant Physiol. 2016, 172, 1506–1518. [Google Scholar] [CrossRef]
- Wang, M.; Toda, K.; Maeda, H.A. Biochemical Properties and Subcellular Localization of Tyrosine Aminotransferases in Arabidopsis Thaliana. Phytochemistry 2016, 132, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Rippert, P.; Scimemi, C.; Dubald, M.; Matringe, M. Engineering Plant Shikimate Pathway for Production of Tocotrienol and Improving Herbicide Resistance. Plant Physiol. 2004, 134, 92–100. [Google Scholar] [CrossRef]
- Karunanandaa, B.; Qi, Q.; Hao, M.; Baszis, S.R.; Jensen, P.K.; Wong, Y.-H.H.; Jiang, J.; Venkatramesh, M.; Gruys, K.J.; Moshiri, F.; et al. Metabolically Engineered Oilseed Crops with Enhanced Seed Tocopherol. Metab. Eng. 2005, 7, 384–400. [Google Scholar] [CrossRef]
- Soares, A.R.; Ferrarese, M.d.L.L.; Siqueira, R.d.C.; Böhm, F.M.L.Z.; Ferrarese-Filho, O. L-DOPA Increases Lignification Associated with Glycine Max Root Growth-Inhibition. J. Chem. Ecol. 2007, 33, 265–275. [Google Scholar] [CrossRef]
- Nishihara, E.; Parvez, M.M.; Araya, H.; Kawashima, S.; Fujii, Y. L-3-(3,4-Dihydroxyphenyl)Alanine (L-DOPA), an Allelochemical Exudedfrom Velvetbean (Mucuna pruriens) Roots. Plant Growth Regul. 2005, 45, 113–120. [Google Scholar] [CrossRef]
- Vargas-Ayala, R.; Rodríguez-Kábana, R.; Morgan-Jones, G.; McInroy, J.A.; Kloepper, J.W. Shifts in Soil Microflora Induced by Velvetbean (Mucuna deeringiana) in Cropping Systems to Control Root-Knot Nematodes. Biol. Control 2000, 17, 11–22. [Google Scholar] [CrossRef]
- Fujii, Y. Allelopathy in the Natural and Agricultural Ecosystems and Isolation of Potent Allelochemicals from Velvet Bean (Mucuna Pruriens) and Hairy Vetch (Vicia villosa). Biol. Sci. Space 2003, 17, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Hahlbrock, K.; Scheel, D. Physiology and Molecular Biology of Phenylpropanoid Metabolism. Annu. Rev. Plant Biol. 1989, 40, 347–369. [Google Scholar] [CrossRef]
- Bido, G.d.S.; da Silva, H.A.; Bortolo, T.d.S.C.; Maldonado, M.R.; Marchiosi, R.; dos Santos, W.D.; Ferrarese-Filho, O. Comparative Effects of L-DOPA and Velvet Bean Seed Extract on Soybean Lignification. Plant Signal. Behav. 2018, 13, e1451705. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; de Cássia Siqueira-Soares, R.; Salvador, V.H.; de Lourdes Lucio Ferrarese, M.; Ferrarese-Filho, O. The Effects of L-DOPA on Root Growth, Lignification and Enzyme Activity in Soybean Seedlings. Acta Physiol. Plant. 2012, 34, 1811–1817. [Google Scholar] [CrossRef]
- Bayless, A.M.; Smith, J.M.; Song, J.; McMinn, P.H.; Teillet, A.; August, B.K.; Bent, A.F. Disease Resistance through Impairment of α-SNAP–NSF Interaction and Vesicular Trafficking by Soybean Rhg1. Proc. Natl. Acad. Sci. USA 2016, 113, E7375–E7382. [Google Scholar] [CrossRef]
- Grunwald, D.J.; Bent, A.F.; Zapotocny, R.W.; Ozer, S.; Diers, B.W. Detection of Rare Nematode Resistance Rhg1 Haplotypes in Glycine Soja and a Novel Rhg1 α-SNAP. The Plant Genome. 2021, 15, e20152. [Google Scholar] [CrossRef]
- Li, W.C.; Liu, X.; Qi, Z.Z.; Yu, L.; Wang, F. Bioinformatics of Huipizhi Black soybean GmPUB24 and Expression under Heterodera glycines Infection. Acta Agric. Zhejiangensis. 2022, 34, 1124–1132. [Google Scholar]
- Liu, T.; Yang, R.W.; Wang, X.M.; Yu, B.S.; Wang, H.; Duan, Y.X. Resistance candidate gene GmMIOX4 gene expression analysis, cloning and over expression vector constructing. Soybean Sci. 2016, 35, 730–735. [Google Scholar]
- Liu, S.; Zhang, S.B.; Wang, X.D.; Yu, B.S.; Chen, J.S.; Wang, H.; Duan, Y.X. Expression Analysis of GmCHS Gene and Isoflavone Content Changes Under Soybean Cyst Nematode Stress. Soybean Sci. 2021, 1, 721–727. [Google Scholar]
- Wang, X.M.; Yang, R.W.; Wang, C.; Chen, J.S.; Wang, H.; Duan, Y.X. Expression analysis of GmMIOX gene response to SCN stress. Chin. J. Oil Crop Sci. 2017, 39, 778–784. [Google Scholar]
- Xu, G.H. Structure and function of protein molecules. Bull. Biol. 2010, 45, 24–25. [Google Scholar]
- Sun, W.; Shahinas, D.; Bonvin, J.; Hou, W.; Kimber, M.S.; Turnbull, J.; Christendat, D. The Crystal Structure of Aquifex Aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition. J. Biol. Chem. 2009, 284, 13223–13232. [Google Scholar] [CrossRef] [PubMed]
- Ku, H.-K.; Park, S.-R.; Yang, I.; Kim, S.-K. Expression and Functional Characterization of Prephenate Dehydrogenase from Streptococcus Mutans. Process Biochem. 2010, 45, 607–612. [Google Scholar] [CrossRef]
- Lütke-Eversloh, T.; Stephanopoulos, G. Feedback Inhibition of Chorismate Mutase/Prephenate Dehydrogenase (TyrA) of Escherichia coli: Generation and Characterization of Tyrosine-insensitive Mutants. Appl. Environ. Microbiol. 2005, 71, 7224–7228. [Google Scholar] [CrossRef]
- Sun, W.; Singh, S.; Zhang, R.; Turnbull, J.L.; Christendat, D. Crystal Structure of Prephenate Dehydrogenase from Aquifex Aeolicus. J. Biol. Chem. 2006, 281, 12919–12928. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Y.J.; Jiang, T.B.; Zhou, B.R.; Wang, H.Z. Progress in the regulation of post-translational modifications in lignin biosynthesis. Curr. Biotechnol. 2024, 14, 519–528. [Google Scholar] [CrossRef]
- Millner, P.A.; Barber, J. Plastoquinone as a Mobile Redox Carrier in the Photosynthetic Membrane. FEBS Lett. 1984, 169, 1–6. [Google Scholar] [CrossRef]
- Strack, D.; Vogt, T.; Schliemann, W. Recent Advances in Betalain Research. Phytochemistry 2003, 62, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.C.; Cahoon, E.B. Enhancing Vitamin E in Oilseeds: Unraveling Tocopherol and Tocotrienol Biosynthesis. Lipids 2007, 42, 97–108. [Google Scholar] [CrossRef] [PubMed]
- The Occurrence of Simple Isoquinolines in Plants. In Proceedings in Life Sciences; Springer: Berlin/Heidelberg, Germany, 1985; pp. 47–61. ISBN 978-3-642-70130-6.
- Sato, F.; Inui, T.; Takemura, T. Metabolic Engineering in Isoquinoline Alkaloid Biosynthesis. Curr. Pharm. Biotechnol. 2007, 8, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Marchiosi, R.; Siqueira-Soares, R.; de Lima, C.; Barbosa de Lima, R.; Dantas dos Santos, W.; Ferrarese-Filho, O. The Role of L-DOPA in Plants. Plant Signal. Behav. 2014, 9, e28275. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-DDCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Raman, M.; Martin, K. One Solution for Cloning and Mutagenesis: In-Fusion® HD Cloning Plus. Nat. Methods 2014, 11, iii–v. [Google Scholar] [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, X.; Zhong, L.; Wang, X.; Jin, L.; Lyu, S. One-Step Generation of Composite Soybean Plants with Transgenic Roots by Agrobacterium Rhizogenes-Mediated Transformation. BMC Plant Biol. 2020, 20, 208. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Y.; Zhang, Y.; Li, C.; Wan, K.; Chen, Y.; Nie, M.; Zhang, H. Gene Localization and Functional Validation of GmPDH1 in Soybean Against Cyst Nematode Race 4. Plants 2025, 14, 1877. https://doi.org/10.3390/plants14121877
Dai Y, Zhang Y, Li C, Wan K, Chen Y, Nie M, Zhang H. Gene Localization and Functional Validation of GmPDH1 in Soybean Against Cyst Nematode Race 4. Plants. 2025; 14(12):1877. https://doi.org/10.3390/plants14121877
Chicago/Turabian StyleDai, Yuehua, Yue Zhang, Chuhui Li, Kun Wan, Yan Chen, Mengen Nie, and Haiping Zhang. 2025. "Gene Localization and Functional Validation of GmPDH1 in Soybean Against Cyst Nematode Race 4" Plants 14, no. 12: 1877. https://doi.org/10.3390/plants14121877
APA StyleDai, Y., Zhang, Y., Li, C., Wan, K., Chen, Y., Nie, M., & Zhang, H. (2025). Gene Localization and Functional Validation of GmPDH1 in Soybean Against Cyst Nematode Race 4. Plants, 14(12), 1877. https://doi.org/10.3390/plants14121877





