Genome-Wide Identification and Evolutionary Analysis of m6A-Related Gene Family in Poplar Nanlin895
Abstract
1. Background
2. Materials and Methods
2.1. Identification of m6A-Related Members in NL895
2.2. Characterization of m6A-Related Proteins in NL895
2.3. Chromosomal Location and Synteny Analysis
2.4. Phylogenetic Tree Analysis
2.5. Gene Structure and Motif Analysis
2.6. Cis-Acting Element Identification
2.7. Gene Expression Analysis
2.8. Plant Materials, Growth Conditions, Cd Treatments and Phytohormone Treatments
2.9. Total RNA Extraction and Real-Time qPCR (RT-qPCR)
3. Results
3.1. Identification of m6A-Related Genes
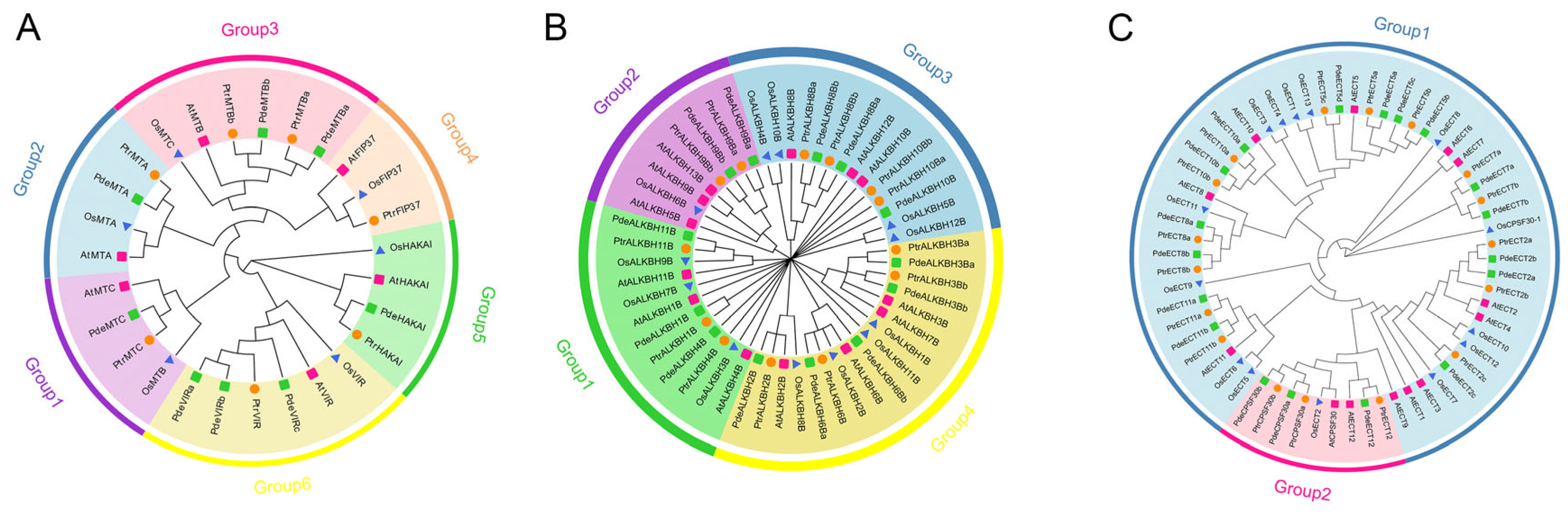
3.2. Phylogenetic Analysis of m6A-Related Gene Families
3.3. Gene Structure, Conserved Domain, and Motif Analysis
3.4. Prediction of Physicochemical Properties and Subcellular Localization of m6A-Related Proteins
3.5. Synteny Analysis of m6A-Related Genes
3.6. Cis-Acting Elements Analysis in m6A-Related Genes
3.7. Gene Expression Pattern Analysis of m6A-Related Genes
3.8. Cadmium Stress Response of m6A Writers
4. Discussion
4.1. Characteristics and Functional Evolution of m6A-Related Genes in NL895
4.2. Genome-Wide Identification of m6A-Related Genes
4.3. Chromosome Localization and Evolutionary Conservation
4.4. Cis-Acting Elements and Their Role in Gene Expression Regulation
4.5. m6A in Plant Stress Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
References
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.-S.; Huang, Y.-C.; Joseph, B.; Lin, C.-J.; Despic, V.; et al. The m6A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; He, C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013, 29, 108–115. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, Y.; Bao, H.; Jiang, Y.; Xu, C.; Wu, J.; Shi, Y. A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res. 2016, 44, 969–982. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Bertero, A.; Brown, S.; Madrigal, P.; Osnato, A.; Ortmann, D.; Yiangou, L.; Kadiwala, J.; Hubner, N.C.; de Los Mozos, I.R.; Sadée, C.; et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 2018, 555, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.-C.; Wei, L.-H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B Is an RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m6A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef]
- Chen, M.; Urs, M.J.; Sánchez-González, I.; Olayioye, M.A.; Herde, M.; Witte, C.P. m6A RNA Degradation Products Are Catabolized by an Evolutionarily Conserved N6-Methyl-AMP Deaminase in Plant and Mammalian Cells. Plant Cell 2018, 30, 1511–1522. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.-C.; Liao, J.-Y.; Zhou, Y.-F.; Feng, Y.-Z.; Yang, Y.-W.; Lei, M.-Q.; Bai, M.; Wu, H.; Chen, Y.-Q.; et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019, 15, e1008120. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, R.; Li, X.; Tian, S.; Li, B.; Qin, G. N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 2021, 22, 168. [Google Scholar] [CrossRef]
- Hu, J.; Cai, J.; Umme, A.; Chen, Y.; Xu, T.; Kang, H. Unique features of mRNA m6A methylomes during expansion of tomato (Solanum lycopersicum) fruits. Plant Physiol. 2022, 188, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Sun, X.; Li, J.; Song, Y.; Song, J.; Wang, F.; Liu, L.; Zhang, X.; Sui, N. Analysis of N6-methyladenosine reveals a new important mechanism regulating the salt tolerance of sweet sorghum. Plant Sci. 2021, 304, 110801. [Google Scholar] [CrossRef] [PubMed]
- Huong, T.T.; Ngoc, L.N.T.; Kang, H. Functional Characterization of a Putative RNA Demethylase ALKBH6 in Arabidopsis Growth and Abiotic Stress Responses. Int. J. Mol. Sci. 2020, 21, 6707. [Google Scholar] [CrossRef]
- Prall, W.; Sheikh, A.H.; Bazin, J.; Bigeard, J.; Almeida-Trapp, M.; Crespi, M.; Hirt, H.; Gregory, B.D. Pathogen-induced m6A dynamics affect plant immunity. Plant Cell 2023, 35, 4155–4172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhu, W.; Diao, S.; Wu, X.; Lu, J.; Ding, C.; Su, X. The poplar pangenome provides insights into the evolutionary history of the genus. Commun. Biol. 2019, 2, 215. [Google Scholar] [CrossRef]
- Gui, J.; Luo, L.; Zhong, Y.; Sun, J.; Umezawa, T.; Li, L. Phosphorylation of LTF1, an MYB Transcription Factor in Populus, Acts as a Sensory Switch Regulating Lignin Biosynthesis in Wood Cells. Mol. Plant. 2019, 12, 1325–1337. [Google Scholar] [CrossRef]
- Li, R.; Wang, Z.; Wang, J.W.; Li, L. Combining single-cell RNA sequencing with spatial transcriptome analysis reveals dynamic molecular maps of cadmium differentiation in the primary and secondary growth of trees. Plant Commun. 2023, 4, 100665. [Google Scholar] [CrossRef]
- Luo, J.; Nvsvrot, T.; Wang, N. Comparative transcriptomic analysis uncovers conserved pathways involved in adventitious root formation in poplar. Physiol. Mol. Biol. Plants 2021, 27, 1903–1918. [Google Scholar] [CrossRef]
- Luo, J.; Wang, Y.; Li, Z.; Wang, Z.; Cao, X.; Wang, N. Haplotype-resolved genome assembly of poplar line NL895 provides a valuable tree genomic resource. For. Res. 2024, 4, e015. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Luo, J.; Xia, W.; Cao, P.; Xiao, Z.; Zhang, Y.; Liu, M.; Zhan, C.; Wang, N. Integrated Transcriptome Analysis Reveals Plant Hormones Jasmonic Acid and Salicylic Acid Coordinate Growth and Defense Responses upon Fungal Infection in Poplar. Biomolecules 2019, 9, 12. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Abdullah, M.; Yu, J.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Expansion and evolutionary patterns of GDSL-type esterases/lipases in Rosaceae genomes. Funct. Integr. Genom. 2018, 18, 673–684. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef]
- Liang, Z.; Riaz, A.; Chachar, S.; Ding, Y.; Du, H.; Gu, X. Epigenetic Modifications of mRNA and DNA in Plants. Mol. Plant 2020, 13, 14–30. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wu, W.; Yang, Y.; Wilson, I.; Shao, F.; Qiu, D. Genome-Wide Identification of m6A Writers, Erasers and Readers in Poplar 84K. Genes 2022, 13, 1018. [Google Scholar] [CrossRef]
- Shen, H.; Luo, B.; Wang, Y.; Li, J.; Hu, Z.; Xie, Q.; Wu, T.; Chen, G. Genome-Wide Identification, Classification and Expression Analysis of m6A Gene Family in Solanum lycopersicum. Int. J. Mol. Sci. 2022, 23, 4522. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, B.; Gu, L.; Chen, Y.; Mora, M.; Zhu, M.; Noory, E.; Wang, Q.; Lin, C. A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 2021, 7, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Si, Z.; Mei, G.; Cheng, Y.; Zhang, J.; Jiang, T.; Chen, J.; Xiong, H.; Zhang, T.; Hu, Y. N6-methyladenosine RNA modification regulates photoperiod sensitivity in cotton. Plant Physiol. 2024, 196, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Zhao, H.; Zhang, S.; Zong, Z.; Li, C.; Ming, L.; Xie, W.; Yu, H. Feedback regulation of m6A modification creates local auxin maxima essential for rice microsporogenesis. Dev. Cell 2025, 60, 1454–1466.e4. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Su, T.; Zhang, S.; Zhang, Y.; Wong, C.E.; Ma, J.; Shao, Y.; Hua, C.; Shen, L.; Yu, H. N6-methyladenosine-mediated feedback regulation of abscisic acid perception via phase-separated ECT8 condensates in Arabidopsis. Nat. Plants 2024, 10, 469–482. [Google Scholar] [CrossRef]
- Lee, K.P.; Liu, K.; Kim, E.Y.; Medina-Puche, L.; Dong, H.; Di, M.; Singh, R.M.; Li, M.; Qi, S.; Meng, Z.; et al. The m6A reader ECT1 drives mRNA sequestration to dampen salicylic acid-dependent stress responses in Arabidopsis. Plant Cell 2024, 36, 746–763. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, G.; Tang, R.; Wang, W.; Wang, Y.; Tian, S.; Qin, G. m6 A-mediated regulation of crop development and stress responses. Plant Biotechnol. J. 2022, 20, 1447–1455. [Google Scholar] [CrossRef]
- Lu, L.; Zhang, Y.; He, Q.; Qi, Z.; Zhang, G.; Xu, W.; Yi, T.; Wu, G.; Li, R. MTA, an RNA m6A Methyltransferase, Enhances Drought Tolerance by Regulating the Development of Trichomes and Roots in Poplar. Int. J. Mol. Sci. 2020, 21, 2462. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, K.; Tian, Y.; Jia, K.; El-Kassaby, Y.A.; Wu, Y.; Liu, J.; Si, H.; Sun, Y.; Li, Y. N6-methyladenosine mRNA methylation positively regulated the response of poplar to salt stress. Plant Cell Environ. 2024, 47, 1797–1812. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, Q.; Cao, S.; Tian, Y.; Han, K.; Sun, Y.; Li, J.; Yang, Q.; Ji, Q.; Sederoff, R.; et al. Genome-wide identification of the AlkB homologs gene family, PagALKBH9B and PagALKBH10B regulated salt stress response in Populus. Front. Plant Sci. 2022, 13, 994154. [Google Scholar] [CrossRef]
- Song, Z.; Yang, Q.; Dong, B.; Wang, S.; Xue, J.; Liu, N.; Zhou, X.; Li, N.; Dandekar, A.M.; Cheng, L.; et al. Nanopore RNA direct sequencing identifies that m6A modification is essential for sorbitol-controlled resistance to Alternaria alternata in apple. Dev. Cell 2025, 60, 1439–1453.e5. [Google Scholar] [CrossRef]
- Cheng, Q.; Wang, P.; Wu, G.; Wang, Y.; Tan, J.; Li, C.; Zhang, X.; Liu, S.; Huang, S.; Huang, T.; et al. Coordination of m6A mRNA methylation and gene transcriptome in rice response to cadmium stress. Rice 2021, 14, 62. [Google Scholar] [CrossRef] [PubMed]






| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplication Type | Selection |
|---|---|---|---|---|---|
| PdeMTBa/PdeMTBb | 0.066449 | 0.243759 | 0.272603 | WGD | Purifying |
| PdeALKBH6Ba/PdeALKBH6Bb | 0.295386 | 0.615411 | 0.479981 | WGD | Purifying |
| PdeALKBH8Ba/PdeALKBH8Bb | 0.078067 | 0.265596 | 0.293931 | WGD | Purifying |
| PdeALKBH9Ba/PdeALKBH9Bb | 0.102159 | 0.282231 | 0.36197 | WGD | Purifying |
| PdeECT2a/PdeECT2b | 0.119733 | 0.387067 | 0.309334 | WGD | Purifying |
| PdeECT2b/PdeECT2c | 0.354481 | 2.112958 | 0.167765 | WGD | Purifying |
| PdeECT2c/PdeECT2a | 0.290612 | 1.626234 | 0.178703 | WGD | Purifying |
| PdeECT5c/PdeECT5b | 0 | 0.022474 | 0 | WGD | Purifying |
| PdeECT7a/PdeECT7b | 0.098606 | 0.249897 | 0.394586 | WGD | Purifying |
| PdeECT10a/PdeECT10b | 0.085928 | 0.311314 | 0.276016 | WGD | Purifying |
| PdeECT11a/PdeECT11b | 0.073604 | 0.221573 | 0.33219 | WGD | Purifying |
| PdeCPSF30a/PdeCPSF30b | 0.053137 | 0.309897 | 0.171467 | WGD | Purifying |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Liu, R.; Zhu, M.; Zhang, J.; Li, Z.; Huang, K.; Ren, Z.; Zhao, Y.; Luo, K.; Song, Q. Genome-Wide Identification and Evolutionary Analysis of m6A-Related Gene Family in Poplar Nanlin895. Plants 2025, 14, 2017. https://doi.org/10.3390/plants14132017
Li Z, Liu R, Zhu M, Zhang J, Li Z, Huang K, Ren Z, Zhao Y, Luo K, Song Q. Genome-Wide Identification and Evolutionary Analysis of m6A-Related Gene Family in Poplar Nanlin895. Plants. 2025; 14(13):2017. https://doi.org/10.3390/plants14132017
Chicago/Turabian StyleLi, Zeyu, Rongxia Liu, Mingqiang Zhu, Jinye Zhang, Zhoujin Li, Kaixin Huang, Zehua Ren, Yan Zhao, Keming Luo, and Qin Song. 2025. "Genome-Wide Identification and Evolutionary Analysis of m6A-Related Gene Family in Poplar Nanlin895" Plants 14, no. 13: 2017. https://doi.org/10.3390/plants14132017
APA StyleLi, Z., Liu, R., Zhu, M., Zhang, J., Li, Z., Huang, K., Ren, Z., Zhao, Y., Luo, K., & Song, Q. (2025). Genome-Wide Identification and Evolutionary Analysis of m6A-Related Gene Family in Poplar Nanlin895. Plants, 14(13), 2017. https://doi.org/10.3390/plants14132017






