Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation
Abstract
1. Introduction
2. Epitranscriptomic RNA Methylation: A Critical Layer in Drought Stress Adaptation of Rice
| Component Type | Gene Name (Rice) | Arabidopsis Homolog | Putative Function | Known/Proposed Role in Drought Stress | Reference |
|---|---|---|---|---|---|
| Writer | OsMTA | AtMTA (METTL3) | Catalytic subunit of the m6A methyltransferase complex | May regulate drought-responsive transcripts via m6A deposition; expression modulated under abiotic stress | [31,32] |
| Writer | OsMTB | AtMTB (METTL14) | Forms heterodimer with OsMTA; provides structural support | Facilitates target specificity of OsMTA; role in drought not yet clarified | [33] |
| Writer | OsFIP37 | FIP37 | Adaptor protein linking MTA-MTB complex to RNA targets | Essential for embryogenesis in Arabidopsis; drought role in rice unknown | [33] |
| Eraser | OsALKBH2 | ALKBH9B/10B | m6A demethylase | Potentially removes m6A from transcripts to modulate gene expression under stress; deregulated under stress | [13,34] |
| Reader | OsECT2 | ECT2 | YTH-domain-containing protein that binds m6A-modified RNAs | May regulate transcript stability and translation during drought | [35] |
| Reader | OsYTHDF1-like | YTHDF1/2/3 | Cytoplasmic reader of m6A marks | Controls translation efficiency and mRNA decay of stress-responsive genes | [35,36,37] |
2.1. Overview of RNA Methylation in Plants: Types, Distribution, and Detection Technologies
2.1.1. Translational Potential of Epitranscriptomics in Crop Improvement
2.1.2. N6-Methyladenosine (m6A): A Key Epitranscriptomic Mark
2.2. Tools for Mapping m6A: MeRIP-Seq, m6A-Seq, miCLIP
Functional Validation of m6A RNA Methylation
2.3. Role of m6A RNA Methylation in Drought Stress Adaptation
3. Dynamic m6A Methylation Landscape Under Drought in Rice
3.1. Cross-Talk Between RNA Methylation and Other Regulatory Pathways
3.1.1. Interplay with Small RNAs (miRNAs, siRNAs)
3.1.2. Interaction with Chromatin Modifiers and Histone Marks
3.1.3. Coordination with Transcriptional and Translational Controls
3.1.4. Potential Feedback Loops Between ABA Signaling and RNA Methylation
4. Critical Issues and Research Gaps
5. Future Research Directions
- ▪
- To fully realize the potential of RNA methylation as a target for improving drought tolerance in rice, several key research directions must be prioritized. First, functional genomics studies are urgently needed to elucidate the roles of RNA methylation regulatory proteins writers, erasers, and readers using CRISPR/Cas9-based gene editing, overexpression, or knockdown approaches. Particular attention should be given to tissue-specific and stress-inducible expression patterns to uncover spatial and temporal regulation under drought conditions.
- ▪
- Second, high-resolution mapping of RNA modifications using techniques such as miCLIP or nanopore-based direct RNA sequencing should be extended to drought-treated rice plants. These studies should include time-course analyses and organ-specific profiling to capture how the modification landscape dynamically changes during drought progression and recovery. Integrating these data with transcriptomic, proteomic, and metabolomic datasets will be essential for identifying functionally significant target genes and pathways.
- ▪
- Third, mechanistic studies are needed to dissect the functional effects of RNA methylation on RNA stability, alternative splicing, nuclear export, and translational efficiency under drought stress. Additionally, the cross-regulatory interactions between RNA methylation, small RNAs, histone modifications, and phytohormone signaling, particularly ABA, require detailed investigation using multi-omic and genetic approaches.
- ▪
- Fourth, future efforts should focus on translating epitranscriptomic insights into breeding programs by identifying natural allelic variations in RNA methylation regulators across diverse rice germplasms. The development of epigenome-editing tools to manipulate RNA modifications in a site-specific manner could open new avenues for precision crop improvement. Moreover, the identification of RNA modification-based biomarkers associated with drought resilience may facilitate marker-assisted selection in breeding pipelines.
- ▪
- Fifth, multi-omics approaches including epitranscriptomics (e.g., m6A-seq), transcriptomics (RNA-seq), proteomics, metabolomics, and chromatin accessibility assays (ATAC-seq or ChIP-seq) can be integrated to achieve a systems-level understanding of plant drought responses. By correlating m6A modifications with changes in transcript abundance, translation efficiency (assessed via ribosome profiling), and protein or metabolite levels, researchers can identify key regulatory nodes that coordinate stress adaptation. However, the integration of these diverse data types presents significant methodological and analytical challenges, such as ensuring high spatial and temporal resolution, effectively normalizing heterogeneous datasets, and applying advanced machine-learning-based network inference models to unravel complex regulatory interactions.
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| m6A | N6-Methyladenosine |
| ABA | Abscisic Acid |
| MeRIP-seq | Methylated RNA Immunoprecipitation Sequencing |
| miCLIP | Methylation Individual-Nucleotide-Resolution Crosslinking and Immunoprecipitation |
| YTH | YT521-B Homology Domain |
| METTL3 | Methyltransferase-Like 3 |
| METTL14 | Methyltransferase-Like 14 |
| 3′UTR | 3′ Untranslated Region |
| FIP37 | FKBP12 Interacting Protein 37 (WTAP homolog in plants) |
| TF | Transcription Factor |
References
- Birla, D.S.; Malik, K.; Sainger, M.; Chaudhary, D.; Jaiwal, R.; Jaiwal, P.K. Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit. Rev. Food Sci. Nutr. 2017, 57, 2455–2481. [Google Scholar] [CrossRef] [PubMed]
- Mohidem, N.A.; Hashim, N.; Shamsudin, R.; Che Man, H. Rice for food security: Revisiting its production, diversity, rice milling process and nutrient content. Agriculture 2022, 12, 741. [Google Scholar] [CrossRef]
- Kwon, Y.; Kabange, N.R.; Lee, J.-Y.; Seo, B.Y.; Shin, D.; Lee, S.-M.; Cha, J.-K.; Cho, J.-H.; Kang, J.-W.; Park, D.-S. RNA-Seq and electrical penetration graph revealed the role of Grh1-mediated activation of defense mechanisms towards green rice leafhopper (Nephotettix cincticeps Uhler) resistance in rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 10696. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Wang, S.; Wang, Y.; Wei, R.; Liang, Y.; Zuo, L.; Huo, M.; Huang, Z.; Lang, J.; Zhao, X. Impact of abiotic stress on rice and the role of DNA methylation in stress response mechanisms. Plants 2024, 13, 2700. [Google Scholar] [CrossRef]
- Edwards, G.I.; Nelson, K.M.; Le Clec’h, S.; Luu, T.; Coast, O.; Futakuchi, K.; Kok, K. Twenty-five rice research priorities for sustainable rice systems by 2050. Glob. Sustain. 2024, 7, e23. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Satish, L.; Kalendar, R.; Narayanan, M.; Kandasamy, S.; Sharma, A.; Emamverdian, A.; Wei, Q.; Zhou, M. The dynamism of transposon methylation for plant development and stress adaptation. Int. J. Mol. Sci. 2021, 22, 11387. [Google Scholar] [CrossRef]
- Cai, J.; Shen, L.; Kang, H.; Xu, T. RNA modifications in plant adaptation to abiotic stresses. Plant Commun. 2025, 6, 101229. [Google Scholar] [CrossRef]
- Su, T.; Chen, J.; Huo, X.; Kuang, L.; Yan, T.; Gao, F.; Wu, D. Transcriptome-wide m6A methylation and metabolomic analysis reveal regulatory networks in rice roots under manganese stress. Environ. Exp. Bot. 2024, 226, 105906. [Google Scholar] [CrossRef]
- Tang, L.; Risalat, H.; Cao, R.; Hu, Q.; Pan, X.; Hu, Y.; Zhang, G. Food security in China: A brief view of rice production in recent 20 years. Foods 2022, 11, 3324. [Google Scholar] [CrossRef]
- Barbosa Amorim, L.L.; Bezerra-Neto, J.P.; da Fonseca do Santos, R.; Ferreira Neto, J.R.C.; Kido, E.A.; Matos, M.; Benko-Iseppon, A.M. Transcription Factors Involved in Plant Drought Tolerance Regulation. In Drought Stress Tolerance in Plants, Vol 2: Molecular and Genetic Perspectives; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 315–358. [Google Scholar]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef]
- Hou, N.; Li, C. MdMTA-mediated m(6) A modification enhances drought tolerance by promoting mRNA stability and translation efficiency of genes involved in lignin deposition and oxidative stress. New Phytol. 2022, 234, 1294–1314. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Lei, D.; Yang, J.; Chen, S.; Wang, X.; Huang, X.; Zhang, S.; Cai, Z.; Zhu, S.; Wan, J. OsALKBH9-mediated m6A demethylation regulates tapetal PCD and pollen exine accumulation in rice. Plant Biotechnol. J. 2024, 22, 2410–2423. [Google Scholar] [CrossRef] [PubMed]
- Boulias, K.; Greer, E.L. Biological roles of adenine methylation in RNA. Nat. Rev. Genet. 2023, 24, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Sreepathi, N.; Puttegowda, D.; Firdose, N.; Gangadharappa, B.S.; Chandana Kumari, V.; Ramu, R. GABA as a Signaling Molecule in Plants. In GABA in Plants: Biosynthesis, Plant Development, and Food Security; John Wiley & Sons: Hoboken, NJ, USA, 2025; pp. 65–96. [Google Scholar]
- Muhammad, Z.M.; Muhammad, S.; Ghulam, A.; Javaid, A.; Zia, U.-Q. Drought stress impairs grain yield and quality of rice genotypes by impaired photosynthetic attributes and K nutrition. Rice Sci. 2020, 27, 5. [Google Scholar]
- Wilhelm, C.; Selmar, D. Energy dissipation is an essential mechanism to sustain the viability of plants: The physiological limits of improved photosynthesis. J. Plant Physiol. 2011, 168, 79–87. [Google Scholar] [CrossRef]
- Sandhu, D.; Coleman, Z.; Atkinson, T.; Rai, K.M.; Mendu, V. Genetics and physiology of the nuclearly inherited yellow foliar mutants in soybean. Front. Plant Sci. 2018, 9, 471. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Van Breusegem, F. Improving oxidative stress resilience in plants. Plant J. 2022, 109, 359–372. [Google Scholar] [CrossRef]
- Ouyang, W.; Struik, P.C.; Yin, X.; Yang, J. Stomatal conductance, mesophyll conductance, and transpiration efficiency in relation to leaf anatomy in rice and wheat genotypes under drought. J. Exp. Bot. 2017, 68, 5191–5205. [Google Scholar] [CrossRef]
- Aslam, M.M.; Rashid, M.A.R.; Siddiqui, M.A.; Khan, M.T.; Farhat, F.; Yasmeen, S.; Khan, I.A.; Raja, S.; Rasool, F.; Sial, M.A. Recent insights into signaling responses to cope drought stress in rice. Rice Sci. 2022, 29, 105–117. [Google Scholar] [CrossRef]
- Liang, Z.; Riaz, A.; Chachar, S.; Ding, Y.; Du, H.; Gu, X. Epigenetic modifications of mRNA and DNA in plants. Mol. Plant 2020, 13, 14–30. [Google Scholar] [CrossRef]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Song, Z.; Tang, K.; Chen, L.; Wang, L.; Ban, T.; Guo, Z.; Kim, C.; Zhang, H.; Duan, C.-G. A histone H3K4me1-specific binding protein is required for siRNA accumulation and DNA methylation at a subset of loci targeted by RNA-directed DNA methylation. Nat. Commun. 2021, 12, 3367. [Google Scholar] [CrossRef] [PubMed]
- Kuluev, B.; Mikhaylova, E.; Ermoshin, A.; Veselova, S.; Tugbaeva, A.; Gumerova, G.; Gainullina, K.; Zaikina, E. The ARGOS-LIKE genes of Arabidopsis and tobacco as targets for improving plant productivity and stress tolerance. J. Plant Physiol. 2019, 242, 153033. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Madsen, J.G.S.; Rauch, A.; Van Hauwaert, E.L.; Schmidt, S.F.; Winnefeld, M.; Mandrup, S. Integrated analysis of motif activity and gene expression changes of transcription factors. Genome Res. 2018, 28, 243–255. [Google Scholar] [CrossRef]
- Anderson, S.J.; Kramer, M.C.; Gosai, S.J.; Yu, X.; Vandivier, L.E.; Nelson, A.D.L.; Anderson, Z.D.; Beilstein, M.A.; Fray, R.G.; Lyons, E.; et al. N6-Methyladenosine Inhibits Local Ribonucleolytic Cleavage to Stabilize mRNAs in Arabidopsis. Cell Rep. 2018, 25, 1146–1157.e3. [Google Scholar] [CrossRef]
- Hu, J.; Xu, T.; Kang, H. Crosstalk between RNA m6A modification and epigenetic factors for gene regulation in plants. Plant Commun. 2024, 5, 101037. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Xue, F.; Zhang, J. m(6)A demethylase OsALKBH5 is required for double-strand break formation and repair by affecting mRNA stability in rice meiosis. New Phytol. 2024, 244, 2326–2342. [Google Scholar] [CrossRef]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.N.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.K.H.; Kang, H. Reading m6A marks in mRNA: A potent mechanism of gene regulation in plants. J. Integr. Plant Biol. 2024, 66, 2586–2599. [Google Scholar] [CrossRef] [PubMed]
- Scutenaire, J.; Deragon, J.-M.; Jean, V.; Benhamed, M.; Raynaud, C.; Favory, J.-J.; Merret, R.; Bousquet-Antonelli, C. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 2018, 30, 986–1005. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Tang, Q.; Song, P.; Tian, E.; Yang, J.; Jia, G. The m6A reader ECT8 is an abiotic stress sensor that accelerates mRNA decay in Arabidopsis. Plant Cell 2024, 36, 2908–2926. [Google Scholar] [CrossRef]
- Ma, W.; Cui, S.; Lu, Z.; Yan, X.; Cai, L.; Lu, Y.; Cai, K.; Zhou, H.; Ma, R.; Zhou, S.; et al. YTH Domain Proteins Play an Essential Role in Rice Growth and Stress Response. Plants 2022, 11, 2206. [Google Scholar] [CrossRef]
- Yu, F.; Qi, H.; Gao, L.; Luo, S.; Njeri Damaris, R.; Ke, Y.; Wu, W.; Yang, P. Identifying RNA modifications by direct RNA sequencing reveals complexity of epitranscriptomic dynamics in rice. Genom. Proteom. Bioinform. 2023, 21, 788–804. [Google Scholar] [CrossRef]
- Li, Y.; Yin, M.; Wang, J.; Zhao, X.; Xu, J.; Wang, W.; Fu, B. Epitranscriptome profiles reveal participation of the RNA methyltransferase gene OsMTA1 in rice seed germination and salt stress response. BMC Plant Biol. 2025, 25, 115. [Google Scholar] [CrossRef]
- Fang, Y.; Chen, L.; Lin, K.; Feng, Y.; Zhang, P.; Pan, X.; Sanders, J.; Wu, Y.; Wang, X.-e.; Su, Z. Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice. Genome Res. 2019, 29, 1287–1297. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Tang, Y.; Gao, C.-C.; Gao, Y.; Yang, Y.; Shi, B.; Yu, J.-L.; Lyu, C.; Sun, B.-F.; Wang, H.-L.; Xu, Y.; et al. OsNSUN2-Mediated 5-Methylcytosine mRNA Modification Enhances Rice Adaptation to High Temperature. Dev. Cell 2020, 53, 272–286.e7. [Google Scholar] [CrossRef]
- Wang, L.; Zhuang, H.; Fan, W.; Zhang, X.; Dong, H.; Yang, H.; Cho, J. m6A RNA methylation impairs gene expression variability and reproductive thermotolerance in Arabidopsis. Genome Biol. 2022, 23, 244. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Cai, Z. Principles, functions, and biological implications of m(6)A in plants. RNA 2024, 30, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Arribas-Hernández, L.; Brodersen, P. Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 2020, 182, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Jia, Y.; Liu, W.; Zhao, Q.; Pan, W.; Jia, Y.; Lv, S.; Liu, X.; Nie, X. Transcriptome-wide m6A methylation profile reveals its potential role underlying drought response in wheat (Triticum aestivum L.). Planta 2024, 260, 65. [Google Scholar] [CrossRef] [PubMed]
- Boughanem, H.; Pilo, J.; Rego, A.; Garcia-Flores, L.A.; de Vera, T.D.; Tinahones, F.J.; Martin-Nuñez, G.M.; González, M.M. A multi-omic integrative approach combining m6A-epitranscriptomic, transcriptomic, and splicing alternative events reveals potential candidates for colorectal cancer diagnosis. Genes Dis. 2025, 101537. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Y.; Wang, T.; Li, S.; Jiang, J.; Chen, S.; Chen, F.; Wang, L. mRNA m6A regulates gene expression via H3K4me3 shift in 5’UTR. Genome Biol. 2025, 26, 54. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, Y.; Yang, J.; Dong, L.; Zhang, R.; Tian, S.; Yu, Y.; Ren, L.; Hou, W.; Zhu, F.; et al. Multi-omics data integration using ratio-based quantitative profiling with Quartet reference materials. Nat. Biotechnol. 2024, 42, 1133–1149. [Google Scholar] [CrossRef]
- Shen, L.; Yu, H. RNA m6A modification meets plant hormones. Nat. Plants 2025, 11, 686–695. [Google Scholar] [CrossRef]
- Bhat SS, Bielewicz D, Gulanicz T, Bodi Z, Yu X, Anderson SJ, Szewc L, Bajczyk M, Dolata J, Grzelak N, Smolinski DJ, Gregory BD, Fray RG, Jarmolowski A, Szweykowska-Kulinska Z. mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA, 2020; 17, 21785–21795. [CrossRef]
- Wei, L.H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.C.; Jia, G. The m(6)A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef]
- Quadrana, L.; Bortolini Silveira, A.; Mayhew, G.F.; LeBlanc, C.; Martienssen, R.A.; Jeddeloh, J.A.; Colot, V. The Arabidopsis thaliana mobilome and its impact at the species level. elife 2016, 5, e15716. [Google Scholar] [CrossRef]
- Govindan, G.; Sharma, B.; Li, Y.F.; Armstrong, C.D.; Merum, P.; Rohila, J.S.; Gregory, B.D. mRNA N(6)-methyladenosine is critical for cold tolerance in Arabidopsis. Plant J. 2022, 111, 1052–1068. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Shil, S.; Rime, J.; Alice, A.K.; Yumkhaibam, T.; Mounika, V.; Singh, A.P.; Kundu, M.; Lalhmangaihzuali, H.; Hazarika, T.K. Phytohormonal signaling in plant resilience: Advances and strategies for enhancing abiotic stress tolerance. Plant Growth Regul. 2025, 105, 329–360. [Google Scholar] [CrossRef]
- Ganguly, D.R.; Li, Y.; Bhat, S.S.; Tiwari, S.; Ng, P.J.; Gregory, B.D.; Sunkar, R. mRNA ADENOSINE METHYLASE promotes drought tolerance through N6-methyladenosine-dependent and independent impacts on mRNA regulation in Arabidopsis. New Phytol. 2025, 245, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Fidler, J.; Graska, J.; Gietler, M. PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli. Cells 2022, 11, 1352. [Google Scholar] [CrossRef]
- Bimpong, D.; Zhao, L.; Ran, M.; Zhao, X.; Wu, C.; Li, Z.; Wang, X.; Cheng, L.; Fang, Z.; Hu, Z. Transcriptomic analysis reveals the regulatory mechanisms of messenger RNA (mRNA) and long non-coding RNA (lncRNA) in response to waterlogging stress in rye (Secale cereale L.). BMC Plant Biol. 2024; 24, 534. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, W.; Wei, Y.; Shi, H. MeCIPK23 interacts with Whirly transcription factors to activate abscisic acid biosynthesis and regulate drought resistance in cassava. Plant Biotechnol. J. 2020, 18, 1504. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Shu, X.E.; Mao, Y.; Liu, X.-M.; Yuan, X.; Zhang, X.; Hess, M.E.; Brüning, J.C.; Qian, S.-B. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol. Cell 2018, 69, 636–647.e7. [Google Scholar] [CrossRef]
- Wang, Y.N.; Yu, C.Y.; Jin, H.Z. RNA N(6)-Methyladenosine Modifications and the Immune Response. J. Immunol. Res. 2020, 2020, 6327614. [Google Scholar] [CrossRef]
- Cui, X.; Liang, Z.; Shen, L.; Zhang, Q.; Bao, S.; Geng, Y.; Zhang, B.; Leo, V.; Vardy, L.A.; Lu, T. 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Mol. Plant 2017, 10, 1387–1399. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef]
- Wu, Q.; Bazzini, A.A. Translation and mRNA Stability Control. Annu. Rev. Biochem. 2023, 92, 227–245. [Google Scholar] [CrossRef]
- Hanson, G.; Alhusaini, N.; Morris, N.; Sweet, T.; Coller, J. Translation elongation and mRNA stability are coupled through the ribosomal A-site. RNA 2018, 24, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, Y.; Usman, B.; Kang, H.; Jung, K.-H. Epitranscriptomics: An additional regulatory layer in plants’ development and stress response. Plants 2022, 11, 1033. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Patil, S.; Joshi, S.; Jamla, M.; Kumar, V. Exploring epitranscriptomics for crop improvement and environmental stress tolerance. Plant Physiol. Biochem. 2022, 183, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Röh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M. The role of m6A/m-RNA methylation in stress response regulation. Neuron 2018, 99, 389–403.e9. [Google Scholar] [CrossRef]
- Sun, C.; Ali, K.; Yan, K.; Fiaz, S.; Dormatey, R.; Bi, Z.; Bai, J. Exploration of epigenetics for improvement of drought and other stress resistance in crops: A review. Plants 2021, 10, 1226. [Google Scholar] [CrossRef]
- Nasr, A.; Copeland, N.; Munir, M. Structural Analysis of Virus Regulatory N6-Methyladenosine (m6A) Machinery of the Black Flying Fox (Pteropus alecto) and the Egyptian Fruit Bat (Rousettus aegyptiacus) Shows Evolutionary Conservation Amongst Mammals. Genes 2024, 15, 1361. [Google Scholar] [CrossRef]
- Fan, Y.; Sun, C.; Yan, K.; Li, P.; Hein, I.; Gilroy, E.M.; Kear, P.; Bi, Z.; Yao, P.; Liu, Z. Recent advances in studies of genomic DNA methylation and its involvement in regulating drought stress response in crops. Plants 2024, 13, 1400. [Google Scholar] [CrossRef]
- Wang, Y.; Huan, Q.; Li, K.; Qian, W. Single-cell transcriptome atlas of the leaf and root of rice seedlings. J. Genet. Genom. 2021, 48, 881–898. [Google Scholar] [CrossRef]
- Luo, W.; Tang, Y.; Li, S.; Zhang, L.; Liu, Y.; Zhang, R.; Diao, X.; Yu, J. The m6A reader SiYTH1 enhances drought tolerance by affecting the messenger RNA stability of genes related to stomatal closure and reactive oxygen species scavenging in Setaria italica. J. Integr. Plant Biol. 2023, 65, 2569–2586. [Google Scholar] [CrossRef]
- Wang, W.; Qin, Q.; Sun, F.; Wang, Y.; Xu, D.; Li, Z.; Fu, B. Genome-wide differences in DNA methylation changes in two contrasting rice genotypes in response to drought conditions. Front. Plant Sci. 2016, 7, 1675. [Google Scholar] [CrossRef]
- Hasan, M.; Nishat, Z.S.; Hasan, M.S.; Hossain, T.; Ghosh, A. Identification of m6A RNA methylation genes in Oryza sativa and expression profiling in response to different developmental and environmental stimuli. Biochem. Biophys. Rep. 2024, 38, 101677. [Google Scholar] [CrossRef] [PubMed]
- Lephatsi, M.M.; Meyer, V.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Plant responses to abiotic stresses and rhizobacterial biostimulants: Metabolomics and epigenetics perspectives. Metabolites 2021, 11, 457. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Y.; Sun, Y.; Jiang, S.; Li, J. N 6-methyladenosine RNA methylation: From regulatory mechanisms to potential clinical applications. Front. Cell Dev. Biol. 2022, 10, 1055808. [Google Scholar] [CrossRef] [PubMed]
- Vaschetto, L.M. DNA Methylation, Histone Modifications, and Non-coding RNA Pathways. In Epigenetics in Crop Improvement: Safeguarding Food Security in an Ever-Changing Climate; Springer: Berlin/Heidelberg, Germany, 2024; pp. 15–27. [Google Scholar]
- Zhang, W.; Wu, Y.; Schnable, J.C.; Zeng, Z.; Freeling, M.; Crawford, G.E.; Jiang, J. High-resolution mapping of open chromatin in the rice genome. Genome Res. 2012, 22, 151–162. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Cao, Z.; Ouyang, W.; Zhang, Q.; Xie, L.; Zheng, R.; Guo, M.; Ma, M.; Hu, Z. Chromatin loops associated with active genes and heterochromatin shape rice genome architecture for transcriptional regulation. Nat. Commun. 2019, 10, 3640. [Google Scholar] [CrossRef]
- Fanata, W.I.D.; Lee, S.Y.; Lee, K.O. The unfolded protein response in plants: A fundamental adaptive cellular response to internal and external stresses. J. Proteom. 2013, 93, 356–368. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Jain, M. Gene regulatory networks in abiotic stress responses via single-cell sequencing and spatial technologies: Advances and opportunities. Curr. Opin. Plant Biol. 2024, 82, 102662. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, G.; Tang, R.; Wang, W.; Wang, Y.; Tian, S. m(6) A-mediated regulation of crop development and stress responses. Plant Biotechnol. J. 2022, 20, 1447–1455. [Google Scholar] [CrossRef]
- Zhu, T.; Xia, C.; Yu, R.; Zhou, X.; Xu, X.; Wang, L.; Zong, Z.; Yang, J.; Liu, Y.; Ming, L. Comprehensive mapping and modelling of the rice regulome landscape unveils the regulatory architecture underlying complex traits. Nat. Commun. 2024, 15, 6562. [Google Scholar] [CrossRef]
- Singh, A.; Jain, D.; Pandey, J.; Yadav, M.; Bansal, K.C.; Singh, I.K. Deciphering the role of miRNA in reprogramming plant responses to drought stress. Crit. Rev. Biotechnol. 2023, 43, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.M.; Mir, R.A.; Ebinezer, L.B.; Masi, A.; Hami, A.; Manzoor, M.; Salgotra, R.K.; Sofi, N.R.; Mushtaq, R.; Rohila, J.S. Physiological and multi-omics approaches for explaining drought stress tolerance and supporting sustainable production of rice. Front. Plant Sci. 2022, 12, 803603. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Kapoor, U.; Jantsch, M.F. Understanding RNA modifications: The promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017, 7, 170077. [Google Scholar] [CrossRef]

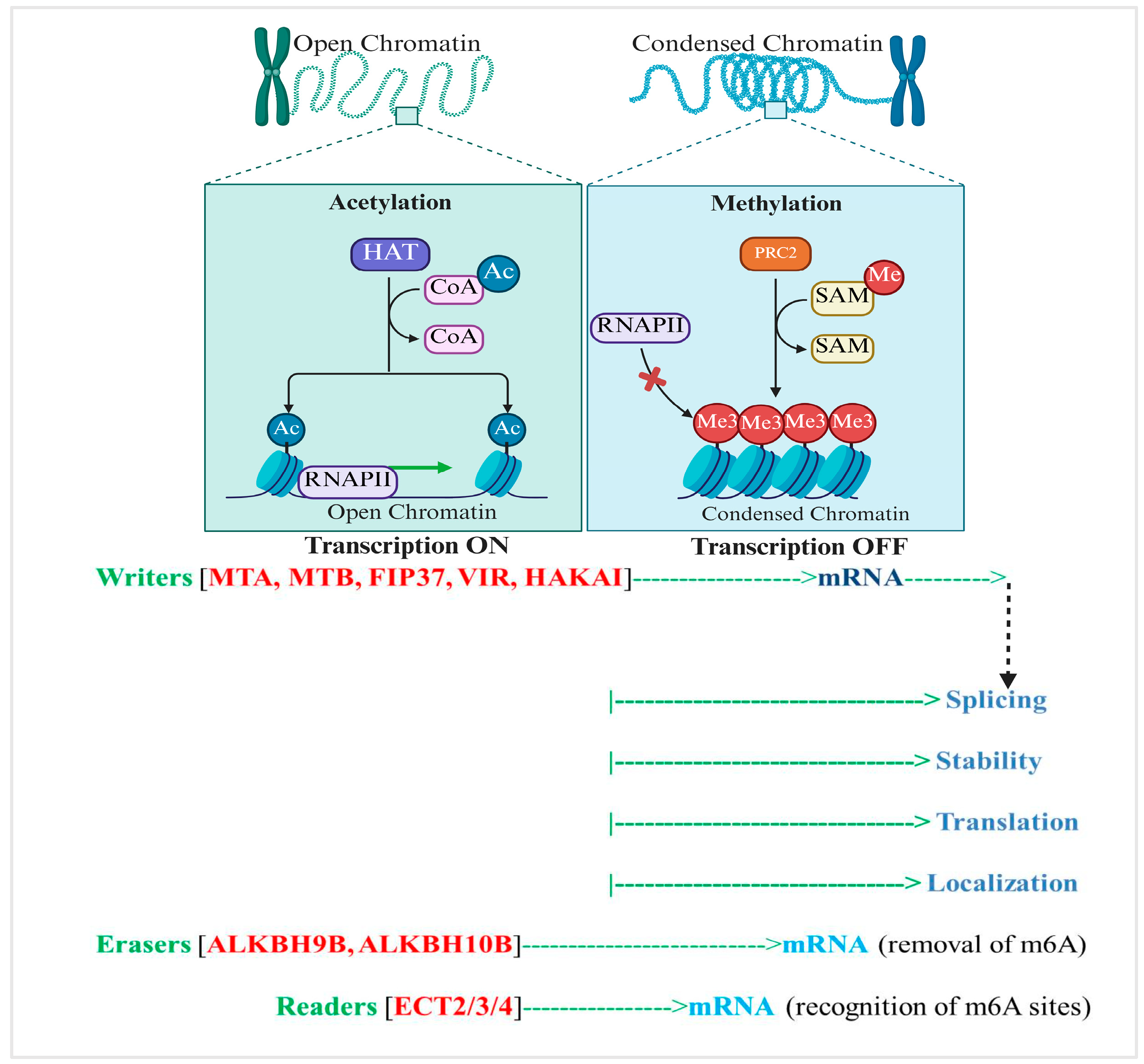
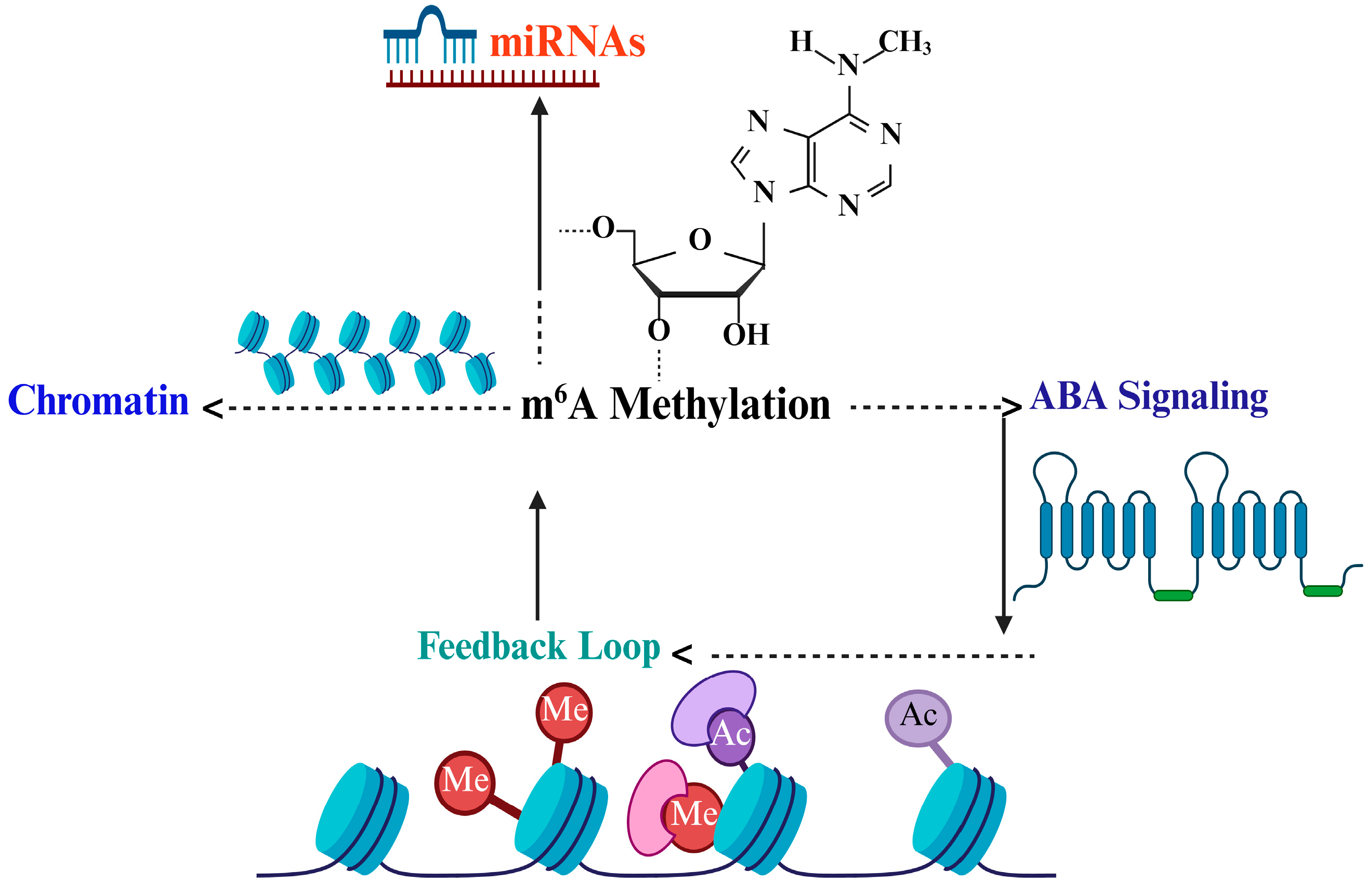
| Regulatory Level | Example Targets or Pathways | Impact of m6A Modification | Evidence (Species) | Identified Gap in Rice | Reference |
|---|---|---|---|---|---|
| Transcription factor regulation | DREB2A, NAC, bZIP TFs | m6A increases mRNA stability or enhances translation | Arabidopsis thaliana, tomato | Functional validation of TFs under m6A control is lacking | [56] |
| ABA signaling pathway | PYR/PYL/RCAR, PP2Cs, SnRK2s | Fine-tunes ABA response by modulating key mRNA levels | A. thaliana, maize | ABA components under m6A control in rice not fully mapped | [57] |
| Gene networks | LEA proteins, HSPs, RD29A | Stabilizes stress-inducible transcripts | A. thaliana | Transcriptome-wide validation in rice not yet available | [58] |
| Epigenetic–epitranscriptomic interface | Histone-modifying enzymes, chromatin remodelers | Co-regulation of gene expression with histone marks and RNA methylation | A. thaliana | Epigenomic integration with m6A data is absent in rice | [54] |
| Oxidative stress response | SOD, CAT, APX (antioxidant enzymes) | Modulates ROS-scavenging enzyme transcripts | Wheat, A. thaliana | Role of m6A in rice oxidative stress management remains unexplored | [59] |
| Protein translation efficiency | eIFs (eukaryotic initiation factors), ribosomal proteins | Regulates translation under stress | Human cells, A. thaliana | No direct evidence in rice under drought | [60] |
| Circadian clock regulation | CCA1, TOC1 (circadian clock genes) | Alters mRNA turnover of clock-related genes | A. thaliana | Potential link between m6A and drought-responsive circadian shifts in rice is unknown | [61] |
| Long non-coding RNAs (lncRNAs) | Drought-responsive lncRNAs | m6A marks influence lncRNA stability and function | Maize, A. thaliana | No studies on m6A-modified lncRNAs in rice drought response | [62] |
| Alternative polyadenylation | Poly(A) site selection regulators | Affects mRNA stability and processing efficiency | Mammalian cells, A. thaliana | Whether m6A influences polyadenylation in rice under drought is unclear | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, X.; Zhang, Y.; Gu, P.; Naz, M. Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation. Plants 2025, 14, 2002. https://doi.org/10.3390/plants14132002
Fan X, Zhang Y, Gu P, Naz M. Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation. Plants. 2025; 14(13):2002. https://doi.org/10.3390/plants14132002
Chicago/Turabian StyleFan, Xiaoru, Yong Zhang, Pengyuan Gu, and Misbah Naz. 2025. "Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation" Plants 14, no. 13: 2002. https://doi.org/10.3390/plants14132002
APA StyleFan, X., Zhang, Y., Gu, P., & Naz, M. (2025). Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation. Plants, 14(13), 2002. https://doi.org/10.3390/plants14132002





