The CONSTANS-like 2 Gene Serves as a Pivotal Regulator of Flowering in Hemerocallis
Abstract
1. Introduction
2. Result
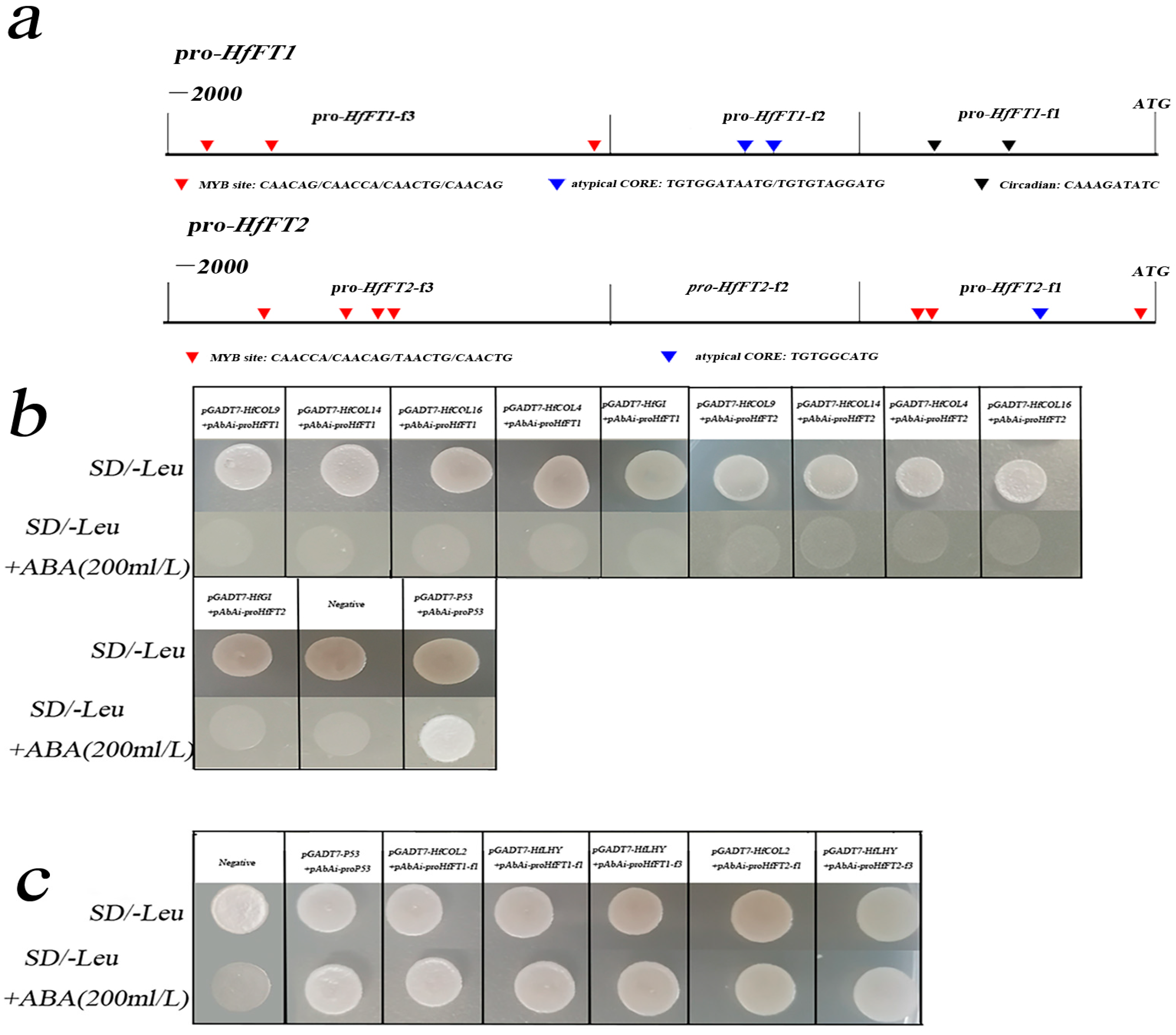
2.1. HfCOL2 and HfLHY Bind to the Promoter of HfFTs
2.2. The Rhythmicity of Gene Expression over a 24 h Period
2.3. Overexpression of HfCOL2 Accelerates the Flowering of Nicotiana tabacum
3. Discussion
4. Conclusions
5. Material and Method
5.1. Plant Material
5.2. RNA Isolation and Double-Strand cDNA Preparation
5.3. Yeast One-Hybrid Assay
5.4. Analysis of Gene Expression Rhythm
5.5. Isolation of HfCOL2, Vector Construction, and Plant Transformation
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gardner, M.J.; Hubbard, K.E.; Hotta, C.T.; Dodd, A.N.; Webb, A.A.R. How plants tell the time. Biochem. J. 2006, 397, 15–24. [Google Scholar] [CrossRef]
- Yakir, E.; Hilman, D.; Harir, Y.; Green, R.M. Regulation of output from the plant circadian clock. FEBS J. 2007, 274, 335–345. [Google Scholar] [CrossRef]
- McClung, C.R. Circadian rhythms in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 139–162. [Google Scholar] [CrossRef]
- Schultz, T.F.; Kay, S.A. Circadian clocks in daily and seasonal control of development. Science 2003, 301, 326–328. [Google Scholar] [CrossRef]
- Takagi, H.; Hempton, A.K.; Imaizumi, T. Photoperiodic flowering in Arabidopsis: Multilayered regulatory mechanisms of CONSTANS and the florigen FLOWERING LOCUS T. Plant Commun. 2023, 4, 100552. [Google Scholar] [CrossRef]
- Rez-Lopez, P.S.; Wheatley, K.; Robson, F.; Onouchi, H.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef]
- Romero, J.M.; Serrano-Bueno, G.; Camacho-Fernandez, C.; Vicente, M.H.; Ruiz, M.T.; Perez-Castineira, J.R.; Perez-Hormaeche, J.; Nogueira, F.T.S.; Valverde, F. CONSTANS, a HUB for all seasons: How photoperiod pervades plant physiology regulatory circuits. Plant Cell 2024, 36, 2086–2102. [Google Scholar] [CrossRef]
- Hayama, R.; Coupland, G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 2004, 135, 677–684. [Google Scholar] [CrossRef]
- Jarillo, J.A.; del Olmo, I.; Gomez-Zambrano, A.; Lazaro, A.; Lopez-Gonzalez, L.; Miguel, E.; Narro-Diego, L.; Saez, D.; Pineiro, M. Photoperiodic control of flowering time. Span. J. Agric. Res. 2008, 6, 221–244. [Google Scholar] [CrossRef]
- Wei, H.; Wang, X.; Xu, H.; Wang, L. Molecular basis of heading date control in rice. Abiotech 2020, 1, 219–232. [Google Scholar] [CrossRef]
- Li, Y.-F.; Zhao, Y.-Q.; Zhang, M.; Jia, G.-X.; Zaccai, M. Functional and Evolutionary Characterization of the CONSTANS-like Family in Lilium x formolongi. Plant Cell Physiol. 2018, 59, 1874–1888. [Google Scholar] [CrossRef]
- Xiao, G.; Li, B.; Chen, H.; Chen, W.; Wang, Z.; Mao, B.; Gui, R.; Guo, X. Overexpression of PvCO1, a bamboo CONSTANS-LIKE gene, delays flowering by reducing expression of the FT gene in transgenic Arabidopsis. BMC Plant Biol. 2018, 18, 232. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, Y.; Zhu, Y.; Su, W.; Long, T.; Huang, T.; Peng, J.; Yu, H.; Lin, S.; Gao, Y. Functional characterization of GI and CO homologs from Eriobotrya deflexa Nakai forma koshunensis. Plant Cell Rep. 2019, 38, 533–543. [Google Scholar] [CrossRef]
- Liu, L.; Ou, C.; Chen, S.; Shen, Q.; Liu, B.; Li, M.; Zhao, Z.; Kong, X.; Yan, X.; Zhuang, F. The Response of COL and FT Homologues to Photoperiodic Regulation in Carrot (Daucus carota L.). Sci. Rep. 2020, 10, 9984. [Google Scholar] [CrossRef]
- Yang, T.; He, Y.; Niu, S.; Yan, S.; Zhang, Y. Identification and characterization of the CONSTANS(CO)/CONSTANS like(COL) genes related to photoperiodic signaling and flowering in tomato. Plant Sci. 2020, 301, 110653. [Google Scholar] [CrossRef]
- Lopez, M.; Larrea, H.; Alvarenga, N.; Gonzalez, D.; Iehisa, J.C.M. CONSTANS-like genes are associated with flowering time in sesame. Theor. Exp. Plant Physiol. 2023, 35, 341–353. [Google Scholar] [CrossRef]
- Yin, X.; Liu, Y.; Zhao, H.; Su, Q.; Zong, J.; Zhu, X.; Bao, Y. GhCOL2 Positively Regulates Flowering by Activating the Transcription of GhHD3A in Upland Cotton (Gossypium hirsutum L.). Biochem. Genet. 2025, 63, 298–314. [Google Scholar] [CrossRef]
- Fenske, M.P.; Nguyen, L.P.; Horn, E.K.; Riffell, J.A.; Imaizumi, T. Circadian clocks of both plants and pollinators influence flower seeking behavior of the pollinator hawkmoth Manduca sexta. Sci. Rep. 2018, 8, 2842. [Google Scholar] [CrossRef]
- Yon, F.; Joo, Y.; Llorca, L.C.; Rothe, E.; Baldwin, I.T.; Kim, S.-G. Silencing Nicotiana attenuata LHY and ZTLalters circadian rhythms in flowers. New Phytol. 2016, 209, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Hassidim, M.; Dakhiya, Y.; Turjeman, A.; Hussien, D.; Shor, E.; Anidjar, A.; Goldberg, K.; Green, R.M. CIRCADIAN CLOCK ASSOCIATED1(CCA1) and the Circadian Control of Stomatal Aperture. Plant Physiol. 2017, 175, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Maguvu, T.E.; Higuchi, Y.; Shibata, M. Effect of Different Photoperiods on Flower Opening Time in Portulaca umbraticola. Hortic. J. 2018, 87, 124–131. [Google Scholar] [CrossRef]
- Hayama, R.; Agashe, B.; Luley, E.; King, R.; Coupland, G. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 2007, 19, 2988–3000. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Tanaka, T.; Ogiwara, I.; Kanekatsu, M.; van Doorn, W.G.; Yamada, T. Expression of an AtNAP gene homolog in senescing morning glory (Ipomoea nil) petals of two cultivars with a different flower life span. J. Plant Physiol. 2014, 171, 633–638. [Google Scholar] [CrossRef]
- Xu, C.; Yu, Y.; Zhang, Y.; Li, Y.; Wei, S. Gibberellins are involved in effect of near-null magnetic field on Arabidopsis flowering. Bioelectromagnetics 2017, 38, 1–10. [Google Scholar] [CrossRef]
- Liu, R.; Gao, Y.; Fan, Z.; Guan, C.; Zhang, Q. Effects of different photoperiods on flower opening, flower closing and circadian expression of clock-related genes in Iris domestica and I. dichotoma. J. Plant Res. 2022, 135, 351–360. [Google Scholar] [CrossRef]
- Liu, R.; Gao, Y.; Guan, C.; Ding, L.; Fan, Z.; Zhang, Q. The Comparison of Temporal Transcriptome Changes Between Morning-Opening and Afternoon-Opening Iris Flowers Reveals the Candidate Genes Regulating Flower Opening and Closing. J. Plant Biol. 2023, 66, 455–473. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, Y.K.; Zhang, Q.X. Morning and evening alarm of the circadian clock for flower opening times in Hemerocallis. Plant Sci. 2021, 311, 110992. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, Y.; Gao, S.; Yuan, L.; Wang, X.; Zhang, Q. Genetic characteristics of circadian flowering rhythm in Hemerocallis. Sci. Hortic. 2019, 250, 19–26. [Google Scholar] [CrossRef]
- Ledger, S.; Strayer, C.; Ashton, F.; Kay, S.A.; Putterill, J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. Cell Mol. Biol. 2001, 26, 15–22. [Google Scholar] [CrossRef]
- Shin, B.S.; Lee, J.H.; Lee, J.H.; Jeong, H.J.; Yun, C.H.; Kim, J.K. Circadian regulation of rice (Oryza sativa L.) CONSTANS-like gene transcripts. Mol. Cells 2004, 17, 10–16. [Google Scholar] [CrossRef]
- Ito-Miwa, K.; Serikawa, M.; Kondo, T.; Oyama, T. Overexpression of a CO homologue disrupts the rhythmic expression of clock gene LgLHYH1in Lemna gibba. Plant Biotechnol. 2014, 31, 319–328. [Google Scholar] [CrossRef]
- Xu, F.; Rong, X.; Huang, X.; Cheng, S. Recent Advances of Flowering Locus T Gene in Higher Plants. Int. J. Mol. Sci. 2012, 13, 3773–3781. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, T.; Zeng, X. Genetic and Epigenetic Understanding of the Seasonal Timing of Flowering. Plant Commun. 2020, 1, 100008. [Google Scholar] [CrossRef]
- Sun, L.; Nie, T.; Chen, Y.; Yin, Z. From Floral Induction to Blooming: The Molecular Mysteries of Flowering in Woody Plants. Int. J. Mol. Sci. 2022, 23, 10959. [Google Scholar] [CrossRef]
- Li, C.; Liu, X.-J.; Yan, Y.; Alam, M.S.; Liu, Z.; Yang, Z.-K.; Tao, R.-F.; Yue, E.-k.; Duan, M.-H.; Xu, J.-H. OsLHY is involved in regulating flowering through the Hd1-and Ehd1-mediated pathways in rice (Oryza sativa L.). Plant Sci. 2022, 315, 111145. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Shoko, K.; Yuji, T.; Yasushi, K.; Lisa, M.; Takuji, S.; Takashi, A.; Masahiro, Y. Hd3a, a Rice Ortholog of the Arabidopsis FT Gene, Promotes Transition to Flowering Downstream of Hd1 under Short-Day Conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar]
- Liang, R.-Z.; Luo, C.; Liu, Y.; Hu, W.-L.; Guo, Y.-H.; Yu, H.-X.; Lu, T.-T.; Chen, S.-Q.; Zhang, X.-J.; He, X.-H. Overexpression of two CONSTANS-like 2 (MiCOL2) genes from mango delays flowering and enhances tolerance to abiotic stress in transgenic Arabidopsis. Plant Sci. 2023, 327, 111541. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Q.; Zhu, H.; Cai, C.; Li, S. Characterization of Mungbean CONSTANS-LIKE Genes and Functional Analysis of CONSTANS-LIKE 2 in the Regulation of Flowering Time in Arabidopsis. Front. Plant Sci. 2021, 12, 608603. [Google Scholar] [CrossRef]
- Kim, S.J. Molecular cloning and expression analysis of a CONSTANS homologue, PnCOL1, from Pharbitis nil. J. Exp. Bot. 2003, 54, 1879–1887. [Google Scholar] [CrossRef]
- Serrano-Bueno, G.; de los Reyes, P.; Chini, A.; Ferreras-Garrucho, G.; Sánchez de Medina-Hernández, V.; Boter, M.; Solano, R.; Valverde, F. Regulation of floral senescence in Arabidopsis by coordinated action of CONSTANS and jasmonate signaling. Mol. Plant 2022, 15, 1710–1724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jiang, Y.; Dong, H.; Shan, X.; Tian, J.; Sun, M.; Ma, F.; Ren, C.; Yuan, Y. Transcriptomic response for revealing the molecular mechanism of oat flowering under different photoperiods. Front. Plant Sci. 2023, 14, 1279107. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Primer Name | Primer Sequence (5′ → 3′) | Accession Number |
|---|---|---|---|
| HfCOL2 | HfCOL2-F1 | GAGGCACTTTTCGGTGAGGA | PQ149922 |
| HfCOL2-R1 | GCACCCTCGCTTCTCTATCC | ||
| HfCOL4 | HfCOL14-F1 | TCCAAACTCAAACTGAACCATATCC | PQ149925 |
| HfCOL14-R1 | TTACTACAAATTATCCGAACACCGG | ||
| HfCOL9 | HfCOL9-F1 | CTTCTTCAATGTGGTCGCAGAGTTA | PQ149923 |
| HfCOL9-R1 | ACCTGACTGGGCAAAAGTTAGACCT | ||
| HfCOL14 | HfCOL4-F1 | TCAAACAGCGTCAAAAGCGACTGAC | PQ149924 |
| HfCOL4-R1 | TGATGGTTGGTACATGGGCCACTGG | ||
| HfCOL16 | HfCOL16-F1 | GCTCCCAGATTGCCCTTTAT | PQ149926 |
| HfCOL16-R1 | ACCGGGCTTGCTAGAGT | ||
| HfGI | HfGI-F1 | ATGTCTACTTGTGCTTCTTGTCATGAG | PQ149920 |
| HfGI-R1 | CTATCATCATCAGATCTGAGACACCG | ||
| HfLHY | HfLHY-F1 | ATGGATACGAAAACGTTGGGGGATGA | PQ149921 |
| HfLHY-R1 | CTGCAAAGACGGTTGTTTGTGAAAATG | ||
| HfCOL2 | 35S:HfCOL2-F | agaacacgggggactcttgaccatgg ATGATGAGGCATTGCGATTCGTG | |
| 35S:HfCOL2-R | tgaaaagttcttctcctttactagt TTAGAATGAGGGCACAATCC |
| Gene Name | Primer Name | Primer Sequence (5′ → 3′) | Fragment Length |
|---|---|---|---|
| HfCOL2 | HfCOL2-F | GAGGCACTTTTCGGTGAGGA | 240 |
| HfCOL2-R | GCACCCTCGCTTCTCTATCC | ||
| HfGI | HfGI-F | CTTGCGGCCTCTATCTTCGT | 233 |
| HfGI-R | CTACAACTTGTCGGGGGCTT | ||
| HfLHY | HfLHY-F | TGTGAGTTTTGTGGGGAGCA | 252 |
| HfLHY-R | AAAGAGTTCGAGAGTGGCGG | ||
| HfTCTP | HfTCTP-F | GGTTGCTCCTGAAGCCTGAA | 211 |
| HfTCTP-R | TCAGCGGAAGGAGGAGAAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, C.; Gao, Y.; Wang, Z.; Zhang, Q. The CONSTANS-like 2 Gene Serves as a Pivotal Regulator of Flowering in Hemerocallis. Plants 2025, 14, 1996. https://doi.org/10.3390/plants14131996
Guan C, Gao Y, Wang Z, Zhang Q. The CONSTANS-like 2 Gene Serves as a Pivotal Regulator of Flowering in Hemerocallis. Plants. 2025; 14(13):1996. https://doi.org/10.3390/plants14131996
Chicago/Turabian StyleGuan, Chunjing, Yike Gao, Ziyi Wang, and Qixiang Zhang. 2025. "The CONSTANS-like 2 Gene Serves as a Pivotal Regulator of Flowering in Hemerocallis" Plants 14, no. 13: 1996. https://doi.org/10.3390/plants14131996
APA StyleGuan, C., Gao, Y., Wang, Z., & Zhang, Q. (2025). The CONSTANS-like 2 Gene Serves as a Pivotal Regulator of Flowering in Hemerocallis. Plants, 14(13), 1996. https://doi.org/10.3390/plants14131996






