Molecular Mechanisms Underlying Defense Responses of Potato (Solanum tuberosum L.) to Environmental Stress and CRISPR/Cas-Mediated Engineering of Stress Tolerance
Abstract
1. Introduction
2. The Impact of Abiotic Stresses on Potato and the Application of CRISPR/Cas System to Enhance Tolerance
3. Impact of Biotic Stresses on Potato and the Application of CRISPR/Cas System to Enhance Tolerance
3.1. Late Blight Disease
3.2. Potato Virus Resistance
3.3. CRISPR/Cas System to Improve Nutritional Value of Potato
3.4. CRISPR/Cas System to Reduce of Postharvest Factors Affecting Potatoes
4. Conclusions and Future Perspectives
Funding
Data Availability Statement
Conflicts of Interest
References
- Křížkovská, B.; Viktorová, J.; Lipov, J. Approved Genetically Modified Potatoes (Solanum tuberosum) for Improved Stress Resistance and Food Safety. J. Agric. Food Chem. 2022, 70, 11833–11843. [Google Scholar] [CrossRef] [PubMed]
- Nasir, M.W.; Toth, Z. Effect of Drought Stress on Potato Production: A Review. Agronomy 2022, 12, 635. [Google Scholar] [CrossRef]
- Demirel, U. Chapter 4—Environmental requirements of potato and abiotic stress factors. In Potato Production Worldwide; Çalişkan, M.E., Bakhsh, A., Jabran, K., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 71–86. [Google Scholar]
- Gibson, S.; Kurilich, A.C. The nutritional value of potatoes and potato products in the UK diet. Nutr. Bull. 2013, 38, 389–399. [Google Scholar] [CrossRef]
- Haverkort, A.; Struik, P. Yield levels of potato crops: Recent achievements and future prospects. Field Crops Res. 2015, 182, 76–85. [Google Scholar] [CrossRef]
- Dahal, K.; Li, X.-Q.; Tai, H.; Creelman, A.; Bizimungu, B. Improving Potato Stress Tolerance and Tuber Yield Under a Climate Change Scenario—A Current Overview. Front. Plant Sci. 2019, 10, 563. [Google Scholar] [CrossRef]
- Abeytilakarathna, P. Factors affect to stolon formation and tuberization in potato: A review. Agric. Rev. 2022, 43, 91–97. [Google Scholar] [CrossRef]
- Chourasia, K.N.; Lal, M.K.; Tiwari, R.K.; Dev, D.; Kardile, H.B.; Patil, V.U.; Kumar, A.; Vanishree, G.; Kumar, D.; Bhardwaj, V. Salinity stress in potato: Understanding physiological, biochemical and molecular responses. Life 2021, 11, 545. [Google Scholar] [CrossRef]
- Chang, D.C.; Sohn, H.B.; Cho, J.H.; Im, J.S.; Jin, Y.I.; Do, G.R.; Kim, S.J.; Cho, H.M.; Lee, Y.B. Freezing and frost damage of potato plants: A case study on growth recovery, yield response, and quality changes. Potato Res. 2014, 57, 99–110. [Google Scholar] [CrossRef]
- Goutam, U.; Thakur, K.; Salaria, N.; Kukreja, S. Recent approaches for late blight disease management of potato caused by Phytophthora infestans. In Fungi and Their Role in Sustainable Development: Current Perspectives; Springer: Berlin/Heidelberg, Germany, 2018; pp. 311–325. [Google Scholar]
- Kumar, R.; Tiwari, R.K.; Sundaresha, S.; Kaundal, P.; Raigond, B. Potato viruses and their management. In Sustainable Management of Potato Pests and Diseases; Springer: Berlin/Heidelberg, Germany, 2022; pp. 309–335. [Google Scholar]
- Shah, M.A.; Subhash, S.; Naga, K.C.; Sharma, S. Biology and management of aphids infesting potato. In Sustainable Management of Potato Pests and Diseases; Springer: Berlin/Heidelberg, Germany, 2022; pp. 213–245. [Google Scholar]
- Saidi, A.; Hajibarat, Z. Phytohormones: Plant switchers in developmental and growth stages in potato. J. Genet. Eng. Biotechnol. 2021, 19, 89. [Google Scholar] [CrossRef]
- Kolachevskaya, O.O.; Lomin, S.N.; Arkhipov, D.V.; Romanov, G.A. Auxins in potato: Molecular aspects and emerging roles in tuber formation and stress resistance. Plant Cell Rep. 2019, 38, 681–698. [Google Scholar] [CrossRef]
- Shahzad, R.; Jamil, S.; Ahmad, S.; Nisar, A.; Amina, Z.; Saleem, S.; Zaffar Iqbal, M.; Muhammad Atif, R.; Wang, X. Harnessing the potential of plant transcription factors in developing climate resilient crops to improve global food security: Current and future perspectives. Saudi J. Biol. Sci. 2021, 28, 2323–2341. [Google Scholar] [CrossRef] [PubMed]
- Pieczynski, M.; Wyrzykowska, A.; Milanowska, K.; Boguszewska-Mankowska, D.; Zagdanska, B.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Genomewide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnol. J. 2018, 16, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, C.; Krannich, C.T.; Maletzki, L.; Köhl, K.; Kopka, J.; Sprenger, H.; Hincha, D.K.; Seddig, S.; Peters, R.; Hamera, S.; et al. Unravelling Differences in Candidate Genes for Drought Tolerance in Potato (Solanum tuberosum L.) by Use of New Functional Microsatellite Markers. Genes 2021, 12, 494. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide-Sequence of the Iap Gene, Responsible for Alkaline-Phosphatase Isozyme Conversion in Escherichia-Coli, and Identification of the Gene-Product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.-Y. CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 2021, 14, 127–150. [Google Scholar] [CrossRef]
- Kieu, N.P.; Lenman, M.; Wang, E.S.; Petersen, B.L.; Andreasson, E. Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 2021, 11, 4487. [Google Scholar] [CrossRef]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef]
- Zhan, X.; Tu, Z.; Song, W.; Nie, B.; Li, S.; Zhang, J.; Zhang, F. Cas13a-based multiplex RNA targeting for potato virus Y. Planta 2023, 258, 70. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Moon, K.B.; Park, S.J.; Park, J.S.; Lee, H.J.; Shin, S.Y.; Lee, S.M.; Choi, G.J.; Kim, S.G.; Cho, H.S.; Jeon, J.H.; et al. Editing of StSR4 by Cas9-RNPs confers resistance to Phytophthora infestans in potato. Front. Plant Sci. 2022, 13, 997888. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Jayarathna, S.; Turesson, H.; Fält, A.S.; Nestor, G.; González, M.N.; Olsson, N.; Beganovic, M.; Hofvander, P.; Andersson, R.; et al. Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci. Rep. 2021, 11, 4311. [Google Scholar] [CrossRef] [PubMed]
- Rather, G.A.; Ayzenshtat, D.; Teper-Bamnolker, P.; Kumar, M.; Forotan, Z.; Eshel, D.; Bocobza, S. Advances in protoplast transfection promote efficient CRISPR/Cas9-mediated genome editing in tetraploid potato. Planta 2022, 256, 14. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Yao, M.; Li, C.; Gong, M. Salt and osmotic stress can improve the editing efficiency of CRISPR/Cas9-mediated genome editing system in potato. PeerJ 2023, 11, e15771. [Google Scholar] [CrossRef]
- Chauhan, H.; Alok, A.; Aiana; Upadhyay, S.K.; Pandey, A.; Singh, K. CRISPR/Cas9 edited StbHLH47 lines exhibit altered expression profiling of iron regulating genes and increased iron content in Solanum tuberosum. Curr. Plant Biol. 2024, 38, 100354. [Google Scholar] [CrossRef]
- Wang, X.; Shi, M.; Zhang, R.; Wang, Y.; Zhang, W.; Qin, S.; Kang, Y. Dynamics of physiological and biochemical effects of heat, drought and combined stress on potato seedlings. Chem. Biol. Technol. Agric. 2024, 11, 109. [Google Scholar] [CrossRef]
- Razzaq, H.A.; Ijaz, S.; Haq, I.U.; Khan, I.A. Functional inhibition of the StERF3 gene by dual targeting through CRISPR/Cas9 enhances resistance to the late blight disease in Solanum tuberosum L. Mol. Biol. Rep. 2022, 49, 11675–11684. [Google Scholar] [CrossRef]
- Nourozi, M.; Nazarain-Firouzabadi, F.; Ismaili, A.; Ahmadvand, R.; Poormazaheri, H. CRISPR/Cas StNRL1 gene knockout increases resistance to late blight and susceptibility to early blight in potato. Front. Plant Sci. 2023, 14, 1278127. [Google Scholar] [CrossRef]
- Bi, W.; Liu, J.; Li, Y.; He, Z.; Chen, Y.; Zhao, T.; Liang, X.; Wang, X.; Meng, X.; Dou, D.; et al. CRISPR/Cas9-guided editing of a novel susceptibility gene in potato improves Phytophthora resistance without growth penalty. Plant Biotechnol. J. 2024, 22, 4–6. [Google Scholar] [CrossRef]
- Bi, W.; Chen, Y.; Song, Y.; Liu, J.; Zhao, T.; Sun, C.; Qin, J.; Tu, Z.; Li, Y.; Wang, X.; et al. Potato DMP2 positively regulates plant immunity by modulating endoplasmic reticulum homeostasis. J. Integr. Plant Biol. 2025, 67, 1568–1581. [Google Scholar] [CrossRef]
- Noureen, A.; Zuhaib Khan, M.; Amin, I.; Zainab, T.; Ahmad, N.; Haider, S.; Mansoor, S. Broad-spectrum resistance against multiple PVY-strains by CRSIPR/Cas13 system in Solanum tuberosum crop. GM Crops Food 2022, 13, 97–111. [Google Scholar] [CrossRef]
- Noureen, A.; Khan, M.Z.; Amin, I.; Zainab, T.; Mansoor, S. CRISPR/Cas9-Mediated Targeting of Susceptibility Factor eIF4E-Enhanced Resistance Against Potato Virus Y. Front. Genet. 2022, 13, 922019. [Google Scholar] [CrossRef]
- Karlsson, M.; Kieu, N.P.; Lenman, M.; Marttila, S.; Resjö, S.; Zahid, M.A.; Andreasson, E. CRISPR/Cas9 genome editing of potato StDMR6-1 results in plants less affected by different stress conditions. Hortic. Res. 2024, 11, uhae130. [Google Scholar] [CrossRef] [PubMed]
- Toinga-Villafuerte, S.; Vales, M.I.; Awika, J.M.; Rathore, K.S. CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers. Int. J. Mol. Sci. 2022, 23, 4640. [Google Scholar] [CrossRef] [PubMed]
- Abeuova, L.; Kali, B.; Tussipkan, D.; Akhmetollayeva, A.; Ramankulov, Y.; Manabayeva, S. CRISPR/Cas9-mediated multiple guide RNA-targeted mutagenesis in the potato. Transgenic Res. 2023, 32, 383–397. [Google Scholar] [CrossRef]
- Takeuchi, A.; Ohnuma, M.; Teramura, H.; Asano, K.; Noda, T.; Kusano, H.; Tamura, K.; Shimada, H. Creation of a potato mutant lacking the starch branching enzyme gene StSBE3 that was generated by genome editing using the CRISPR/dMac3-Cas9 system. Plant Biotechnol. 2021, 38, 345–353. [Google Scholar] [CrossRef]
- Yasmeen, A.; Shakoor, S.; Azam, S.; Bakhsh, A.; Shahid, N.; Latif, A.; Shahid, A.A.; Husnain, T.; Rao, A.Q. CRISPR/Cas-mediated knockdown of vacuolar invertase gene expression lowers the cold-induced sweetening in potatoes. Planta 2022, 256, 107. [Google Scholar] [CrossRef]
- Hu, S.; Song, L.; Yi, X.; Ni, X.; Cui, X.; Duan, S.; Jiang, R.; Lyu, D.; Wang, J.; Hu, B.; et al. Potato STARCH SYNTHEASE 5 is critical for simple starch granule initiation in amyloplasts and tuber development. Plant J. 2025, 122, e70206. [Google Scholar] [CrossRef]
- Du, H.; Zhai, Z.; Pu, J.; Liang, J.; Wang, R.; Zhang, Z.; Wang, P.; Zhu, Y.; Huang, L.; Li, D.; et al. Two tandem R2R3 MYB transcription factor genes cooperatively regulate anthocyanin accumulation in potato tuber flesh. Plant Biotechnol. J. 2025, 23, 1521–1534. [Google Scholar] [CrossRef]
- Ohnuma, M.; Ito, K.; Hamada, K.; Takeuchi, A.; Asano, K.; Noda, T.; Watanabe, A.; Hokura, A.; Teramura, H.; Takahashi, F.; et al. Peculiar properties of tuber starch in a potato mutant lacking the α-glucan water dikinase 1 gene GWD1 created by targeted mutagenesis using the CRISPR/dMac3-Cas9 system. Plant Biotechnol. 2023, 40, 219–227. [Google Scholar] [CrossRef]
- Wan, M.; Xie, H.; Guo, H.; Jing, S.; Zeng, D.; Li, B.; Zhu, B.; Zeng, Z. Developing a pipeline for identification, characterization and molecular editing of cis-regulatory elements: A case study in potato. Abiotech 2025, 6, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Ye, M.; Li, C.; Gong, M. H2O2 participates in the induction and formation of potato tubers by activating tuberization-related signal transduction pathways. Agronomy 2023, 13, 1398. [Google Scholar] [CrossRef]
- Wulff-Vester, A.; Andersson, M.; Brurberg, M.B.; Hofvander, P.; Alsheikh, M.; Harwood, W.; Hvoslef-Eide, T. Colour change in potato (Solanum tuberosum L.) tubers by disruption of the anthocyanin pathway via ribonucleoprotein complex delivery of the CRISPR/Cas9 system. Plant Cell Tissue Organ Cult. 2024, 157, 25. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Li, L.; Yan, L.; Wang, X.; Wang, Q.; Lai, X. StSN2 enhances tuber formation in potato via upregulating of the ABA signaling pathway. Front. Plant Sci. 2025, 16, 1566237. [Google Scholar] [CrossRef]
- Zheng, Z.; Ye, G.; Zhou, Y.; Pu, X.; Su, W.; Wang, J. Editing sterol side chain reductase 2 gene (StSSR2) via CRISPR/Cas9 reduces the total steroidal glycoalkaloids in potato. All Life 2021, 14, 401–413. [Google Scholar] [CrossRef]
- Ly, D.N.P.; Iqbal, S.; Fosu-Nyarko, J.; Milroy, S.; Jones, M.G.K. Multiplex CRISPR-Cas9 Gene-Editing Can Deliver Potato Cultivars with Reduced Browning and Acrylamide. Plants 2023, 12, 379. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, A.; Butler, N.M.; Zeng, Z.; Xin, H.; Wang, L.; Lv, Z.; Eshel, D.; Douches, D.S.; Jiang, J. Molecular dissection of an intronic enhancer governing cold-induced expression of the vacuolar invertase gene in potato. Plant Cell 2024, 36, 1985–1999. [Google Scholar] [CrossRef]
- Jayakody, T.B.; Zarka, D.; Cho, K.H.; Jensen, J.; Sikora, S.; Buell, C.R.; Douches, D.S.; Nadakuduti, S.S. Genome-wide evaluation of gene editing outcomes using CRISPR/Cas9 in seed propagated Camelina sativa and vegetatively propagated Solanum tuberosum. Front. Plant Sci. 2024, 15, 1496861. [Google Scholar] [CrossRef]
- Massa, G.A.; Décima Oneto, C.A.; González, M.N.; Poulsen Hornum, A.; Arizmendi, A.; Sucar, S.; Divito, S.B.; Feingold, S.E. CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance. Biology 2025, 14, 445. [Google Scholar] [CrossRef]
- Pfotenhauer, A.C.; Occhialini, A.; Harbison, S.A.; Li, L.; Piatek, A.A.; Luckett, C.R.; Yang, Y.; Stewart, C.N., Jr.; Lenaghan, S.C. Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers. Plants 2023, 12, 1878. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kang, B.; Venkatesh, J.; Lee, J.H.; Lee, S.; Kim, J.M.; Back, S.; Kwon, J.K.; Kang, B.C. Development of virus-induced genome editing methods in Solanaceous crops. Hortic. Res. 2024, 11, uhad233. [Google Scholar] [CrossRef] [PubMed]
- Carlsen, F.M.; Johansen, I.E.; Yang, Z.; Liu, Y.; Westberg, I.N.; Kieu, N.P.; Jørgensen, B.; Lenman, M.; Andreasson, E.; Nielsen, K.L.; et al. Strategies for Efficient Gene Editing in Protoplasts of Solanum tuberosum Theme: Determining gRNA Efficiency Design by Utilizing Protoplast (Research). Front. Genome Ed. 2021, 3, 795644. [Google Scholar] [CrossRef]
- Lukan, T.; Veillet, F.; Križnik, M.; Coll, A.; Mahkovec Povalej, T.; Pogačar, K.; Stare, K.; Chauvin, L.; Chauvin, J.E.; Gruden, K. CRISPR/Cas9-mediated fine-tuning of miRNA expression in tetraploid potato. Hortic. Res. 2022, 9, uhac147. [Google Scholar] [CrossRef] [PubMed]
- Khmeleva, S.A.; Kurbatov, L.K.; Ptitsyn, K.G.; Timoshenko, O.S.; Morozova, D.D.; Suprun, E.V.; Radko, S.P.; Lisitsa, A.V. Detection of Potato Pathogen Clavibacter sepedonicus by CRISPR/Cas13a Analysis of NASBA Amplicons. Int. J. Mol. Sci. 2024, 25, 12218. [Google Scholar] [CrossRef]
- Demirel, U.; Morris, W.L.; Ducreux, L.J.M.; Yavuz, C.; Asim, A.; Tindas, I.; Campbell, R.; Morris, J.A.; Verrall, S.R.; Hedley, P.E.; et al. Physiological, Biochemical, and Transcriptional Responses to Single and Combined Abiotic Stress in Stress-Tolerant and Stress-Sensitive Potato Genotypes. Front. Plant Sci. 2020, 11, 169. [Google Scholar] [CrossRef]
- Gervais, T.; Creelman, A.; Li, X.Q.; Bizimungu, B.; De Koeyer, D.; Dahal, K. Potato Response to Drought Stress: Physiological and Growth Basis. Front. Plant Sci. 2021, 12, 698060. [Google Scholar] [CrossRef]
- Lal, M.K.; Tiwari, R.K.; Kumar, A.; Dey, A.; Kumar, R.; Kumar, D.; Jaiswal, A.; Changan, S.S.; Raigond, P.; Dutt, S.; et al. Mechanistic Concept of Physiological, Biochemical, and Molecular Responses of the Potato Crop to Heat and Drought Stress. Plants 2022, 11, 2857. [Google Scholar] [CrossRef]
- Zinta, R.; Tiwari, J.K.; Buckseth, T.; Thakur, K.; Goutam, U.; Kumar, D.; Challam, C.; Bhatia, N.; Poonia, A.K.; Naik, S.; et al. Root system architecture for abiotic stress tolerance in potato: Lessons from plants. Front. Plant Sci. 2022, 13, 926214. [Google Scholar] [CrossRef]
- Mańkowska, D.; Zarzyńska, K.; Wasilewska-Nascimento, B. Potato (Solanum tuberosum L.) Plant Shoot and Root Changes under Abiotic Stresses—Yield Response. Plants 2022, 11, 3568. [Google Scholar] [CrossRef]
- Hill, D.; Nelson, D.; Hammond, J.; Bell, L. Morphophysiology of potato (Solanum tuberosum) in response to drought stress: Paving the way forward. Front. Plant Sci. 2021, 11, 597554. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Gao, M.; Han, M.; Liu, H. Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L. BMC Genom. 2022, 23, 548. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, X.; Li, N.; Nie, H.; Ma, Y.; Wu, J.; Zhang, Z.; Ma, Y. The StbHLH47 transcription factor negatively regulates drought tolerance in potato (Solanum tuberosum L.). BMC Plant Biol. 2025, 25, 14. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2009, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional Dependence, Cliques, and Predictive Motifs in the bHLH Protein Domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Kanwar, P.; Baby, D.; Bauer, P. Interconnection of iron and osmotic stress signalling in plants: Is FIT a regulatory hub to cross-connect abscisic acid responses? Plant Biol. 2021, 23, 31–38. [Google Scholar] [CrossRef]
- Zhang, J.-C.; Wang, X.-F.; Wang, X.-N.; Wang, F.-P.; Ji, X.-L.; An, J.-P.; Yang, K.; Zhao, Q.; You, C.-X.; Hao, Y.-J. Abscisic acid alleviates iron deficiency by regulating iron distribution in roots and shoots of apple. Sci. Hortic. 2020, 262, 109018. [Google Scholar] [CrossRef]
- Alam, M.S.; Kong, J.; Tao, R.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.-H. CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef]
- Liang, Y.; Yang, C.; Ming, F.; Yu, B.; Cheng, Z.; Wang, Y.; Qiu, Z.; Zhang, X.; Cao, B.; Yan, S. A bHLH transcription factor, CsSPT, regulates high-temperature resistance in cucumber. Hortic. Plant J. 2024, 10, 503–514. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Ding, Y.; Chen, C.; Chen, X.; Su, N.; Zhang, X.; Pan, Y.; Li, J. Novel flavin-containing monooxygenase protein FMO1 interacts with CAT2 to negatively regulate drought tolerance through ROS homeostasis and ABA signaling pathway in tomato. Hortic. Res. 2023, 10, uhad037. [Google Scholar] [CrossRef]
- Zhao, H.; Li, D.; Liu, Y.; Zhang, T.; Zhao, X.; Su, H.; Li, J. Flavin-containing monooxygenases FMOGS-OXs integrate flowering transition and salt tolerance in Arabidopsis thaliana. Physiol. Plant. 2024, 176, e14287. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Li, J.; Yu, Q.; Cang, W.; Xu, R.; Wang, Y.; Ji, W. Two Novel Flavin-Containing Monooxygenases Involved in Biosynthesis of Aliphatic Glucosinolates. Front. Plant Sci. 2016, 7, 1292. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yao, B.; Peng, Z.; Yang, X.; Li, K.; Zhang, X.; Zhu, H.; Zhou, X.; Wang, M.; Jiang, L.; et al. Splicing defect of StDRO2 intron 1 promotes potato root growth by disturbing auxin transport to adapt to drought stress. Hortic. Plant J. 2025, 11, 706–720. [Google Scholar] [CrossRef]
- Uga, Y.; Okuno, K.; Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 2011, 62, 2485–2494. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, L.; Bai, H.; Dai, Y.; Li, K.; Li, S.; Wang, X.; Wu, L.; Fu, Q.; Yang, Y. Characteristics of members of IGT family genes in controlling rice root system architecture and tiller development. Front. Plant Sci. 2022, 13, 961658. [Google Scholar] [CrossRef]
- Tiwari, J.K. Genome sequence analysis provides insights on genomic variation and late blight resistance genes in potato somatic hybrid (parents and progeny). Mol. Biol. Rep. 2021, 48, 623–635. [Google Scholar] [CrossRef]
- Turnbull, D.; Yang, L.; Naqvi, S.; Breen, S.; Welsh, L.; Stephens, J.; Morris, J.; Boevink, P.C.; Hedley, P.E.; Zhan, J.; et al. RXLR Effector AVR2 Up-Regulates a Brassinosteroid-Responsive bHLH Transcription Factor to Suppress Immunity. Plant Physiol. 2017, 174, 356–369. [Google Scholar] [CrossRef]
- Sun, K.; Schipper, D.; Jacobsen, E.; Visser, R.G.F.; Govers, F.; Bouwmeester, K.; Bai, Y. Silencing susceptibility genes in potato hinders primary infection of Phytophthora infestans at different stages. Hortic. Res. 2022, 9, uhab058. [Google Scholar] [CrossRef]
- Sun, K.; Wolters, A.-M.A.; Vossen, J.H.; Rouwet, M.E.; Loonen, A.E.H.M.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 2016, 25, 731–742. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.-J.; Zhang, K. S5H/DMR6 Encodes a Salicylic Acid 5-Hydroxylase That Fine-Tunes Salicylic Acid Homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef]
- Tian, Z.; He, Q.; Wang, H.; Liu, Y.; Zhang, Y.; Shao, F.; Xie, C. The Potato ERF Transcription Factor StERF3 Negatively Regulates Resistance to Phytophthora infestans and Salt Tolerance in Potato. Plant Cell Physiol. 2015, 56, 992–1005. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M. Repression Domains of Class II ERF Transcriptional Repressors Share an Essential Motif for Active Repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Zhao, C.; Wu, G.; Wu, Y.; Chen, Y.; Tang, D. SR1, a Calmodulin-Binding Transcription Factor, Modulates Plant Defense and Ethylene-Induced Senescence by Directly Regulating NDR1 and EIN3. Plant Physiol. 2012, 158, 1847–1859. [Google Scholar] [CrossRef]
- Yuan, P.; Tanaka, K.; Poovaiah, B.W. Calmodulin-binding transcription activator AtSR1/CAMTA3 fine-tunes plant immune response by transcriptional regulation of the salicylate receptor NPR1. Plant Cell Environ. 2021, 44, 3140–3154. [Google Scholar] [CrossRef]
- Yang, L.; McLellan, H.; Naqvi, S.; He, Q.; Boevink, P.C.; Armstrong, M.; Giuliani, L.M.; Zhang, W.; Tian, Z.; Zhan, J.; et al. Potato NPH3/RPT2-Like Protein StNRL1, Targeted by a Phytophthora infestans RXLR Effector, Is a Susceptibility Factor. Plant Physiol. 2016, 171, 645–657. [Google Scholar] [CrossRef]
- He, Q.; Naqvi, S.; McLellan, H.; Boevink, P.C.; Champouret, N.; Hein, I.; Birch, P.R. Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl. Acad. Sci. USA 2018, 115, E7834–E7843. [Google Scholar] [CrossRef]
- Li, X.; Zhong, J.; Li, B.; Luo, Y.; Wang, K.; Wang, Y.; Ye, Z.; Sun, L.; Zhang, J.; Yang, L.; et al. Two putative calcium-dependent protein kinases are involved in the regulation of sugarcane defense genes. Phytopathol. Res. 2024, 6, 22. [Google Scholar] [CrossRef]
- Quenouille, J.; Vassilakos, N.; Moury, B. Potato virus Y: A major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol. Plant Pathol. 2013, 14, 439–452. [Google Scholar] [CrossRef]
- Singh, R.P.; Valkonen, J.P.; Gray, S.M.; Boonham, N.; Jones, R.A.; Kerlan, C.; Schubert, J. Discussion paper: The naming of Potato virus Y strains infecting potato. Arch. Virol. 2008, 153, 1–13. [Google Scholar] [CrossRef]
- Zlobin, N.; Taranov, V. Plant eIF4E isoforms as factors of susceptibility and resistance to potyviruses. Front. Plant Sci. 2023, 14, 1041868. [Google Scholar] [CrossRef] [PubMed]
- Lucioli, A.; Tavazza, R.; Baima, S.; Fatyol, K.; Burgyan, J.; Tavazza, M. CRISPR-Cas9 Targeting of the eIF4E1 Gene Extends the Potato Virus Y Resistance Spectrum of the Solanum tuberosum L. cv. Desirée. Front. Microbiol. 2022, 13, 873930. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Zhang, M.; Yang, W.; Li, C.; Liu, Y.; Li, C.; He, J.; Wu, W. Starch phosphorylation and the in vivo regulation of starch metabolism and characteristics. Int. J. Biol. Macromol. 2020, 159, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Nazarian-Firouzabadi, F.; Visser, R.G.F. Potato starch synthases: Functions and relationships. Biochem. Biophys. Rep. 2017, 10, 7–16. [Google Scholar] [CrossRef]
- Abeuova, L.S.; Kali, B.R.; Rakhimzhanova, A.O.; Bekkuzhina, S.S.; Manabayeva, S.A. High frequency direct shoot regeneration from Kazakh commercial potato cultivars. PeerJ 2020, 8, e9447. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 2014, 66, 546–558. [Google Scholar] [CrossRef]
- Yu, J.; Wang, K.; Beckles, D.M. Starch branching enzymes as putative determinants of postharvest quality in horticultural crops. BMC Plant Biol. 2021, 21, 479. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Cappellini, F.; Marinelli, A.; Toccaceli, M.; Tonelli, C.; Petroni, K. Anthocyanins: From Mechanisms of Regulation in Plants to Health Benefits in Foods. Front. Plant Sci. 2021, 12, 748049. [Google Scholar] [CrossRef]
- Bhaskar, P.B.; Wu, L.; Busse, J.S.; Whitty, B.R.; Hamernik, A.J.; Jansky, S.H.; Buell, C.R.; Bethke, P.C.; Jiang, J. Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiol. 2010, 154, 939–948. [Google Scholar] [CrossRef]
- Zhu, X.; Richael, C.; Chamberlain, P.; Busse, J.S.; Bussan, A.J.; Jiang, J.; Bethke, P.C. Vacuolar invertase gene silencing in potato (Solanum tuberosum L.) improves processing quality by decreasing the frequency of sugar-end defects. PLoS ONE 2014, 9, e93381. [Google Scholar] [CrossRef] [PubMed]
- Clasen, B.M.; Stoddard, T.J.; Luo, S.; Demorest, Z.L.; Li, J.; Cedrone, F.; Tibebu, R.; Davison, S.; Ray, E.E.; Daulhac, A.; et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 2016, 14, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, D.W.K.; Yevtushenko, D.P.; Konschuh, M.; Bizimungu, B.; Lu, Z.-X. Processing strategies to decrease acrylamide formation, reducing sugars and free asparagine content in potato chips from three commercial cultivars. Food Control 2021, 119, 107452. [Google Scholar] [CrossRef]
- Lomelino, C.L.; Andring, J.T.; McKenna, R.; Kilberg, M.S. Asparagine synthetase: Function, structure, and role in disease. J. Biol. Chem. 2017, 292, 19952–19958. [Google Scholar] [CrossRef]
- Fronk, P.; Hartmann, H.; Bauer, M.; Solem, E.; Jaenicke, E.; Tenzer, S.; Decker, H. Polyphenoloxidase from Riesling and Dornfelder wine grapes (Vitis vinifera) is a tyrosinase. Food Chem. 2015, 183, 49–57. [Google Scholar] [CrossRef]
- Lourenco, E.J.; Neves, V.A.; Da Silva, M.A. Polyphenol oxidase from sweet potato: Purification and properties. J. Agric. Food Chem. 1992, 40, 2369–2373. [Google Scholar] [CrossRef]
- Pérez-Gilabert, M.; García Carmona, F. Characterization of catecholase and cresolase activities of eggplant polyphenol oxidase. J. Agric. Food Chem. 2000, 48, 695–700. [Google Scholar] [CrossRef]
- Tussipkan, D.; Manabayeva, S.A. Employing CRISPR/Cas Technology for the Improvement of Potato and Other Tuber Crops. Front. Plant Sci. 2021, 12, 747476. [Google Scholar] [CrossRef]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 2020, 15, e0243376. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Kang, Z.; Mao, H. Improvement of wheat drought tolerance through editing of TaATX4 by CRISPR/Cas9. J. Genet. Genom. 2023, 50, 913–916. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, T.; Cao, L.; Jiao, Z.; Ku, L.; Dou, D.; Liu, Z.; Fu, J.; Xie, X.; Zhu, Y.; et al. Molecular mechanism analysis of ZmRL6 positively regulating drought stress tolerance in maize. Stress Biol. 2023, 3, 47. [Google Scholar] [CrossRef] [PubMed]



| Cultivar | Method of Delivery | Technology | Promoters | Transgenic or DNA-Free Genome Editing | Target Gene (s) | Trait Associated with the Genes | Type of Mutation | Results | Reference |
|---|---|---|---|---|---|---|---|---|---|
| CRISPR/Cas system for the enhancement of tolerance abiotic stress | |||||||||
| Cv.CIP 149 | Agrobacterium-mediated transformation (A. rhizogenes); pKESE401 vector and pCBC-DT1T2 intermediate vector | CRISPR/Cas9 system | U6 promoter | transgenic | StFMO GS-OX-Like3 (StLike3) | tolerance to salt stress | chimeras, deletion, insertion and replacement | Significantly increased mutation efficiency under appropriate NaCl and mannitol concentrations; no off-target effects were found, but root regeneration was inhibited. | [28] |
| Cv. Kufri jyoti | Agrobacterium-mediated transformation (A. rhizogenes); pHSE401 vector | CRISPR/Cas9 system | AtU6 promoter | transgenic | StbHLH47 | iron regulation | deletion | Showed reduced ferric chelate reductase (FCR) activity; but increased expression of iron uptake-related genes, resulting in significantly higher Fe(II) accumulation in tuber tissues; changes in phenotype with short and thin trichomes on stem. | [29] |
| Cv. Li Shu 6 | Agrobacterium-mediated transformation; pBWA(V)KS vector | CRISPR/Cas9 system | 35S promoter | transgenic | Deeper Rooting 1 (StDRO2) | regulation of the root growth | deletion and insertion | Mutant lines exhibited higher plant height, longer root length, smaller root growth angle, and increased tuber weight than the wild-type. | [30] |
| CRISPR/Cas system for the enhancement of tolerance biotic stress | |||||||||
| Cv. Desiree and King Edward | Agrobacterium-mediated transformation; Csy4 multi-gRNA vector | CRISPR/Cas9 system | - | transgenic | StDND1 and StCHL1, StDMR6-1, StDMR6-2 | tolerance to late blight pathogen (P. infestans) | deletion (indel) | StDND1, StCHL1, and StDMR6-1 mutants showed increased resistance to late blight. | [21] |
| Cv. Lady Rosetta | Agrobacterium-mediated transformation; pYLCRISPR/Cas9Pubi-B binary vector | CRISPR/Cas9 system | ubiquitin promoter derived from Oryza sativa (OsU6a) | transgenic | ERF transcription factor (StERF3) | tolerance to late blight pathogen (P. infestans) | deletion (indel) | Improved resistance to P. infestans and relatively high expression of StPR1 and StNPR1. | [31] |
| Cv. Desiree | PEG-mediated protoplast transfection of ribonucleoprotein (RNPs) | CRISPR/Cas9 system | StSR4 binds to the promoters of EDS1 and NDR1 | DNA-free genome editing | Signal Responsive 4 (StSR4) | tolerance to late blight pathogen (P. infestans) | insertion and deletion (indel) | Improved resistance to P. infestans and the expression of StEDS1, StPAD4, and StPR1; resulted in stunted growth and a dwarf phenotype. | [25] |
| Cv. Agria | Agrobacterium-mediated transformation | CRISPR/Cas9 system | U6-26 promotor | transgenic | NPH3/ RPT2-LIKE1 protein (StNRL1) | tolerance to late blight pathogen (P. infestans) and early blight (A. alternata) | deletion (indel) | Improved resistance to P. infestans and sensitivity to A. alternate. | [32] |
| Phureja S15-65 clone | Agrobacterium mediated transformation; pTC212, pTC241, and pCGS752 vectors | CRISPR/Cas9 system | 35S promoter | transgenic | Plasma membrane protein 1 (StPM1) | tolerance to late blight pathogen (P. infestans) | deletions and the consequent frameshift mutations | Milder disease symptoms and smaller lesions than wild types; overexpressing StPM1 and more susceptible to P. infestans and P. capsici. | [33] |
| Phureja S15-65 clone | Agrobacterium mediated transformation; pCAMBIA-1300-35S vectors | CRISPR/Cas9 system | 35S promoter | transgenic | DOMAIN OF UNKNOWN FUNCTION 679 membrane protein (DMP) (StDMP2) | tolerance to late blight pathogen (P. infestans) | - | First, subsequently challenged with P. infestans. Second, overexpression of StDMP2 was significantly reduced the P. infestans infection symptoms and enhanced resistance to infection. Third, the mutants exhibited more susceptibility to P. infestans than wild-type plants. | [34] |
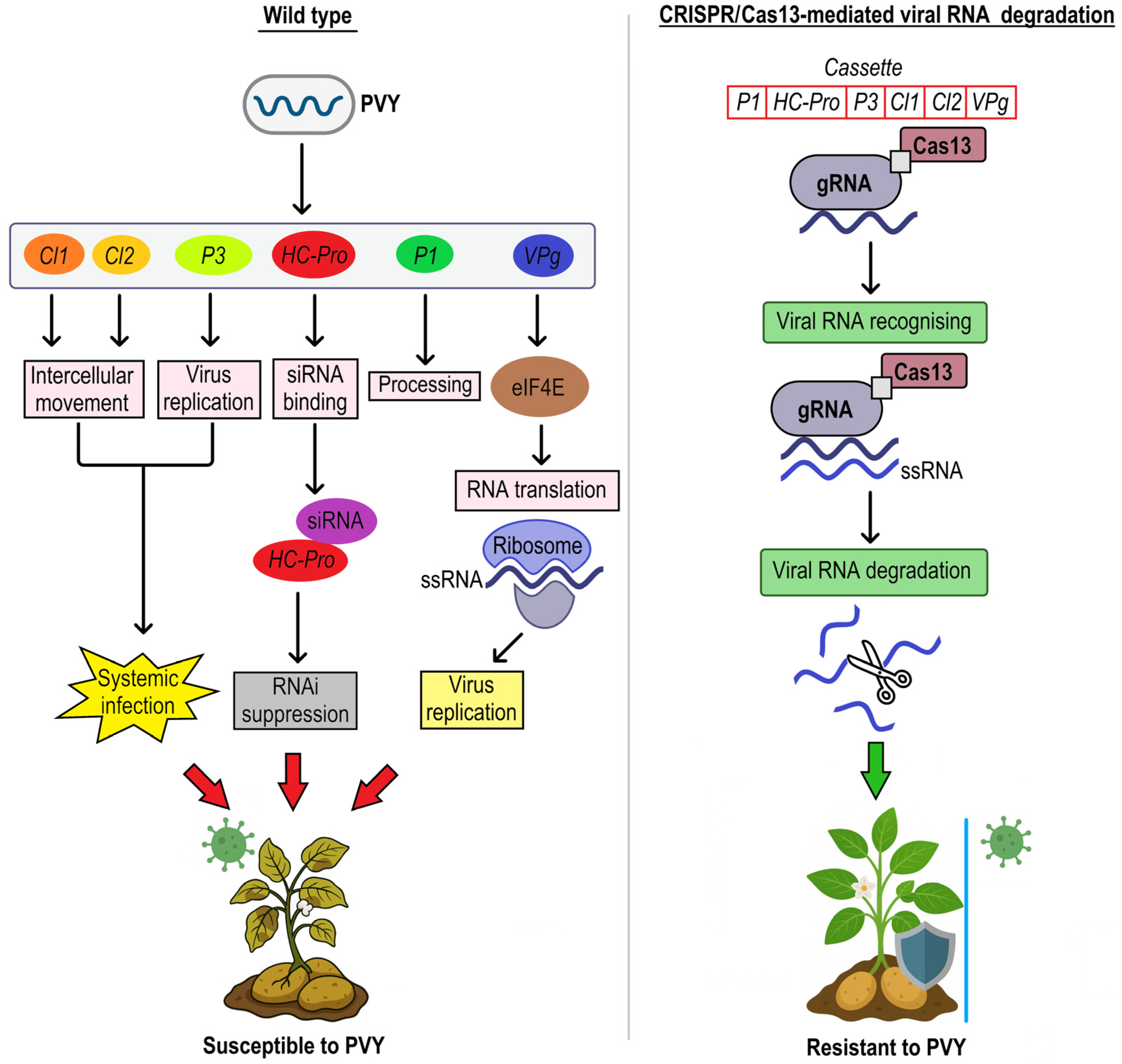
| Cv. Kruda | Agrobacterium mediated transformation; pK2GW7-pCas13a vector and PTZ57Rkana vector | CRISPR/Cas13a system | Arabidopsis U6 promoter, 35S promoter | transgenic | six genes (PI, HC-Pro, P3, CI1, CI2, and VPg) of potato virus Y (PVY) | tolerance to broad-spectrum resistance | - | High expression of Cas13a/sgRNA resulting in resistance to PVY strains. | [35,36] |
| Cv. Kruda | Agrobacterium-mediated transformation; binary expression vector pK2GW7 | CRISPR/Cas9 system | 35S promoter | transgenic | eukaryotic translation initiation factor (eIF4E) | tolerance to potato virus Y resistance | insertion, deletion, and point mutation, and conversion events | Exhibited a gradual decrease in virus titer and resistance to PVY. | [36] |
| Cv. Desiree | Agrobacterium mediated transformation | CRISPR/Cas13a system | Arabidopsis U6 promoter | transgenic | four genes (P3, CI, Nib, and CP) of PVY | tolerance to potato virus Y resistance | - | Similar viral accumulation and reduced symptoms, consistent resistance to various PVY strains. | [23] |
| Cv. KingEdward | Agrobacterium mediated transformation; Csy4 multi-gRNA vector | CRISPR/Cas9 system | - | transgenic | Downy mildew Resistance 6 (StDMR6-1) | tolerance to bacterial scab, salt stress and drought stress | - | Exhibited fewer scab lesions, greater fresh weight and survival rates; improved tuber quality and faster adaptation in the field. | [37] |
| CRISPR/Cas system for the enhancement of nutrient contents in potato | |||||||||
| Cv.Yukon Gold | Agrobacterium-mediated transformation; pCGS752 (base binary vector), | CRISPR/Cas9 system | - | transgenic | Granule-bound starch synthase (GBSSI) | amylose-free starch | indels (insertions and deletions) | A reduction or complete elimination of amylose. | [38] |
| Cv. Astanalyk, Tokhtar, and Aksor | Agrobacterium-mediated transformation; pEn-Chimera, pMR203, pMR204, and pMR205 vectors | CRISPR/Cas9 system | AtU6 promoter | transgenic | Granule-bound starch synthase (StGBSS) | amylose-free starch | substitutions and indels | Resulting in an amylose-free phenotype. | [39] |
| Cv. Desiree | PEG-mediated protoplast transfection of ribonucleoprotein (RNPs) | CRISPR/Cas9 system | - | DNA-free genome editing | Starch branching enzyme (SBE1 and SBE2) | amylopectin-free starch | deletion, insertion | Resulting in no amylopectin with no detectable branching and a high mutation frequency. | [26] |
| Cv. Sayaka | Agrobacterium-mediated transformation; pMR203, pMR204, and pMR205 | CRISPR/dMac3-Cas9 system | AtU6-26 promoter | transgenic | Starch-branching enzyme (SBE) (StSBE3) | amylopectin- free starch | deletion, frameshift mutations | Resulting in 8% target efficiency and loss of function of SBE3, mutants grew normally, and yielded sufficient amounts of tubers. | [40] |
| Cv. AGB Purple | Agrobacterium-mediated transformation; pDES vector | CRISPR/Cas9 system | TA cloning kit dual Promoter (PCR® II) | transgenic | Vacuolar invertase (VInv) | tolerance to cold-induced sweetening | indels, frame shift mutations | Resulting reducing sugars and editing efficiencies; no morphological variations were observed in the edited lines; however, there were notable differences in physical characteristics. | [41] |
| Cv. AC142 | Agrobacterium-mediated transformation; pH7LIC-N-eGFP vector | CRISPR/Cas9 system | potato U6 promoter and CaMV 35S promoter | transgenic | Starch Synthase 5 (StSS5) | number and morphology of starch granules | deletion, insertion | In tubers, the ss5 mutation increased starch granule initiation sites, producing compound and more small granules. | [42] |
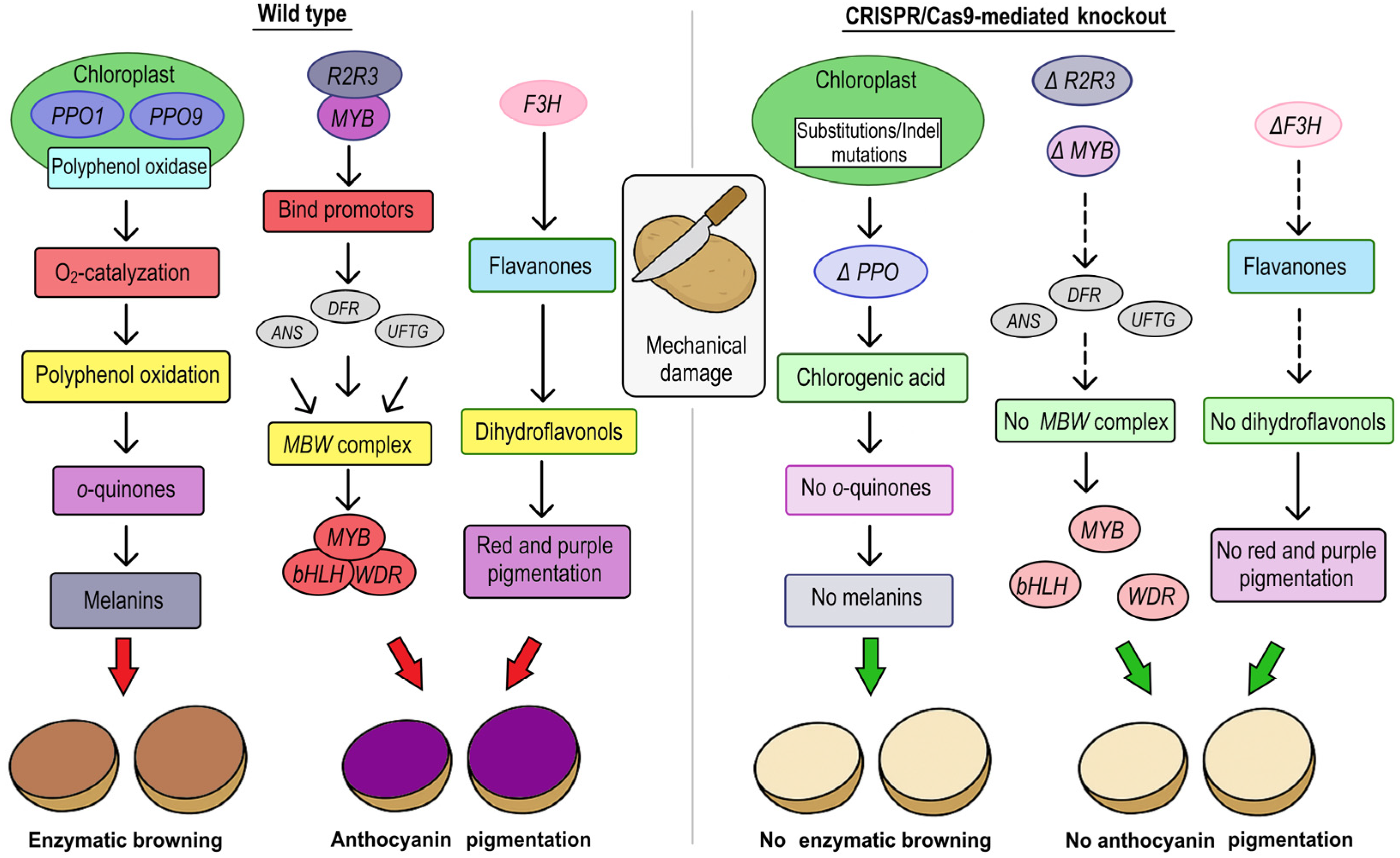
| Clone 01–58 | Agrobacterium-mediated transformation; pCAMBIA2300MGFPuv-sgRNACas vector | CRISPR/Cas9 system | StMYB210 promoter | Tandem R2R3 MYB genes (StMYB200 and StMYB210) | regulation anthocyanin accumulation in tuber flesh | deletion, insertion | Both StMYB200 and StMYB210 activate the expression of the bHLH TF gene StbHLH1 and interact with it to regulate anthocyanin biosynthesis. Analysis of the StMYB210 promoter in various diploid potato accessions revealed that insertion events were associated with flesh color. | [43] | |
| Cv. Sayaka | Agrobacterium-mediated transformation | CRISPR/Cas9 system | AtU6-26 promoter | transgenic | α-glucan water dikinase 1 gene (GWD1) | retaining moisture and stabilization of the starch structure | deletion, insertion, and substitution | The gwd1 mutant tubers showed decreased phosphorus content, significantly less water loss, and a higher amylose content. | [44] |
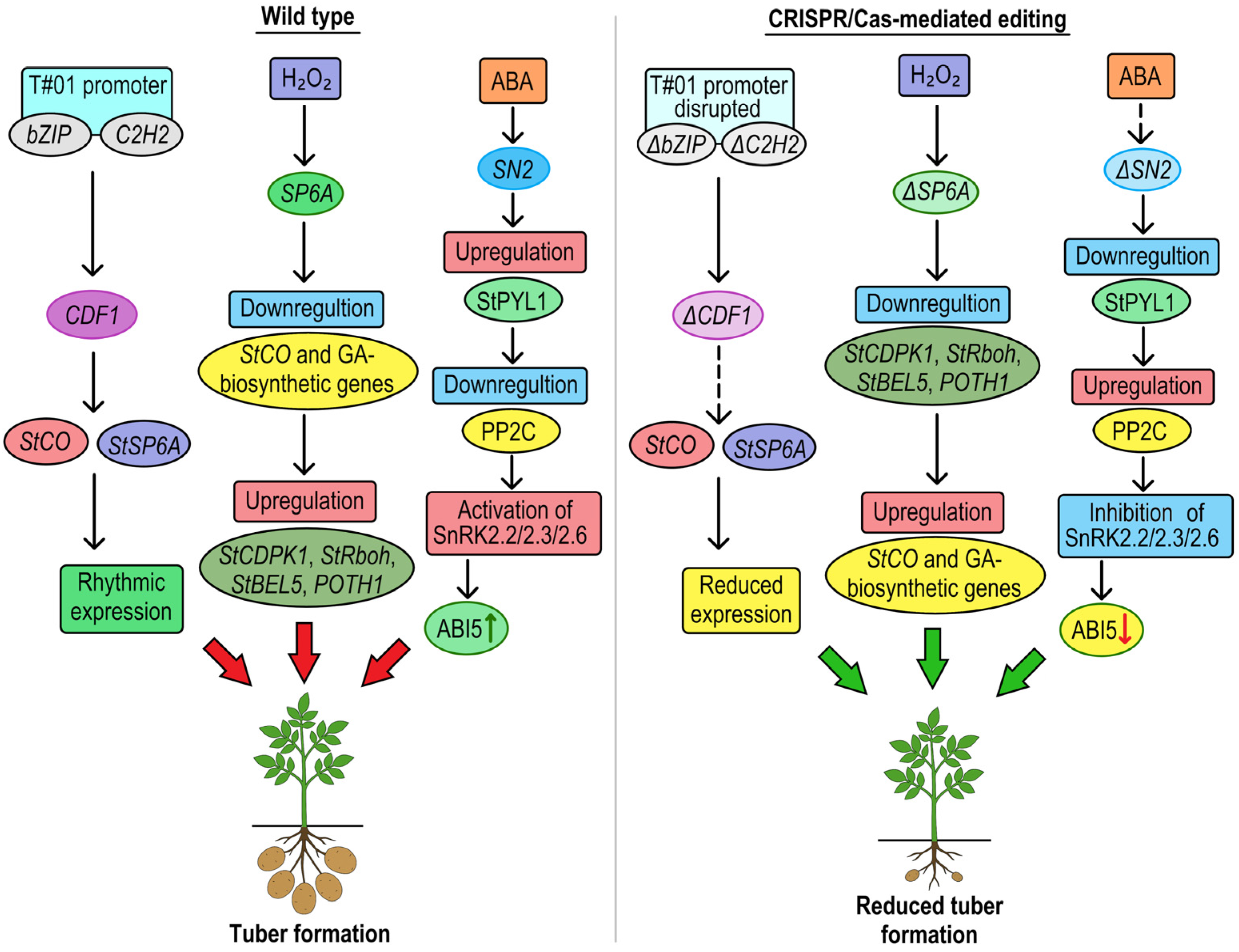
| Cv. DM 1–3 516 R44 | Agrobacterium-mediated transformation; pKGWFS 7.0 vector | CRISPR/Cas9 system | 35S promoter | transgenic | CYCLING DOF FACTOR 1 (StCDF1) | late tuberization | deletions | Displayed a reduced expression level, resulting in late tuberization under both long-day and short-day conditions. | [45] |
| Cvs. CIP-149 and CIP-178 | Agrobacterium-mediated transformation | CRISPR/Cas9 system | GA20ox1 promoter | transgenic | Flowering locus T (FT)-like self-pruning 6A (StSP6A) | induction and for formation of potato tubers | deletions and insertions | The mutants showed a marked drop in tuber formation. | [46] |
| Cvs. Desirée and Nansen | PEG-mediated protoplast transfection of ribonucleoprotein (RNPs) | CRISPR/Cas9 system | - | DNA-free genome editing | flavanone 3-hydroxylase (F3H) | formation of the anthocyanidins | deletion | Observed changes in skin pigmentation, temperature-dependent tuber phenotypes, and instances of somaclonal variation. | [47] |
| Cv. Desirée | Agrobacterium-mediated transformation | CRISPR/Cas9 system | StSN2promoter | transgenic | Snf1-related protein kinase 2.2 (snrk2.2) or StSN2 | regulation of tuber formation | deletions, Insertions, and SNPs | Mutation lines promote tuber formation by enhancing ABA signaling, specifically through upregulation of StPYL1, StSnRK2.2/2.3/2.6, and StABI5 genes. | [48] |
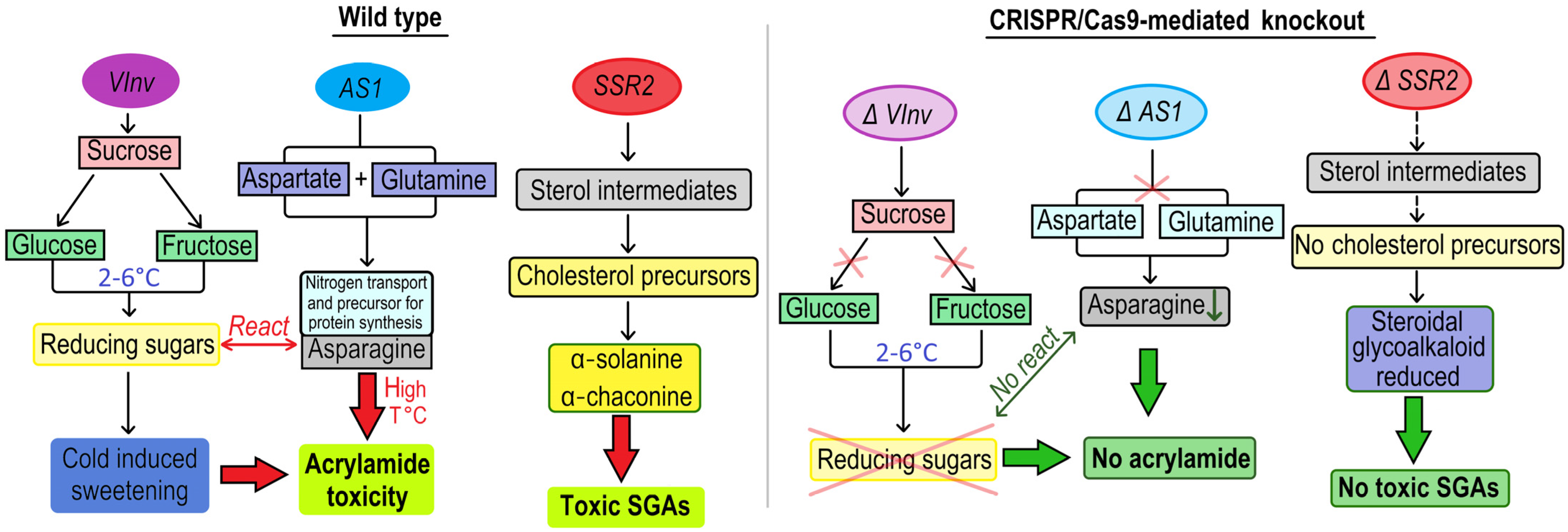
| Cv. Atlantic | Agrobacterium-mediated transformation; VK005-StSSR2 vector | CRISPR/Cas9 system | AtU6 promoter | sterol side-chain reductase 2 enzyme (StSSR2) | reduction in steroidal glycoalkaloid content | deletions and insertions | Leads to a significant reduction in steroidal glycoalkaloid content with a 46% mutation efficiency. | [49] | |
| CRISPR/Cas system for the reduced of postharvest factors affecting potato | |||||||||
| Cv. Atlantic and Desiree | Agrobacterium-mediated transformation; pFGC-Cas9-ASVI vector | CRISPR/Cas9 system | 35S promoter | transgenic | Vacuolar invertase (VInv) and asparagine synthetase 1 (AS1) | storing potato tubers at cold temperatures | deletions, insertions and substitutions | Reduced fructose and glucose concentrations after cold storage; less acrylamide. | [50] |
| Cv. Katahdin | Agrobacterium-mediated transformation; | CRISPR/Cas9 system | 35S promoter | transgenic | Vacuolar invertase (VInv) | storing potato tubers at cold temperatures | deletion | Led to a 54% reduction in VInv expression in the mutated lines. | [51] |
| Clone DRH195 | Agrobacterium-mediated transformation | CRISPR/Cas9 system | - | transgenic | Polyphenol oxidases (StPPO) | resistance to tuber bruising | indels (insertions and deletions) and SNP | No significant evidence of off-target effects. | [52] |
| Cvs. Atlantic and Spunta | Protoplast transfection; pTRANS_100 vector | CRISPR/Cas9 system | AtU6 promoter | DNA-free genome editing | Vacuolar invertase (VInv) and Polyphenol oxidases (StPPO) | resistance to tuber bruising | deletions, insertions and in-frame mutations | Improved chip quality, reduced browning, and largely preserved tuber traits in both gene edited lines. | [53] |
| Cv. Desiree | Protoplast transfection; pTRANS_100 vector | CRISPR/Cas9 system | - | transgenic | Tubulin-like GTPase (FtsZ1) | morphology of starch granules | deletions and insertions | Mutated lines showed reduced FtsZ1 gene expression with increased starch granule size without nutritional quality change. | [54] |
| CRISPR/Cas9 system applied for fundamental research | |||||||||
| Cv. Desiree | protoplasts’ transgene expression and protoplasts’ regeneration | CRISPR/Cas9 system | UBIQUITIN10 promoters | DNA-free gene-editing | Neomycin phosphotransferase2 (NPT2) | improving genome editing efficiencies | Not given | Editing efficiency reached to 95%. | [27] |
| Cv. Desiree | Agrobacterium-mediated transformation; a potato virus X (PVX) vector (SlPDS, St&mPDS1, StPDS2, StPDS3, SmPDS2) | CRISPR/Cas9 system | - | transgenic | Phytoene desaturase gene (PDS): StPDS | improving genome editing efficiencies | Not given | Editing efficiency increased from 22.1% to 30.5%. | [55] |
| Cv. Saturna and Wotan | Protoplasts’ transgene expression | CRISPR/Ca system | DNA-free gene-editing | α-glucan water dikinase 1 gene (GWD1) and downy mildew resistant 6 (DMR6-1) genes | improving genome editing efficiencies | indels and single nucleotide polymorphisms (SNPs) | Resulted significantly better editing in the GWD1 mutant lines with regions targeted comprising the 5′ end, while editing efficiency was more balanced between the 5′ and 3′ ends the GWD1 mutant lines. | [56] | |
| Cvs. Desiree and Rywal | Agrobacterium-mediated transformation | CRISPR/Cas9 system | - | transgenic | MicroRNAs (MIR160a, MIR160b, and MIR390a) | To establish fast and efficient protocol for CRISPR/Cas9- mediated modulation of miRNA expression | deletions and insertions | Suggesting high editing efficiency. | [57] |
| many cultivars | Agrobacterium-mediated transformation; p2CT-His-MBPLbu_C2c2_WT plasmid vector | CRISPR/Cas13a system | T7 promoter | transgenic | Clavibacter sepedonicus detection | to determine viable bacteria of Clavibacter sepedonicus in potato tubers | Not given | Effective, easy, time consuming. | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutula, M.; Tussipkan, D.; Kali, B.; Manabayeva, S. Molecular Mechanisms Underlying Defense Responses of Potato (Solanum tuberosum L.) to Environmental Stress and CRISPR/Cas-Mediated Engineering of Stress Tolerance. Plants 2025, 14, 1983. https://doi.org/10.3390/plants14131983
Sutula M, Tussipkan D, Kali B, Manabayeva S. Molecular Mechanisms Underlying Defense Responses of Potato (Solanum tuberosum L.) to Environmental Stress and CRISPR/Cas-Mediated Engineering of Stress Tolerance. Plants. 2025; 14(13):1983. https://doi.org/10.3390/plants14131983
Chicago/Turabian StyleSutula, Maxim, Dilnur Tussipkan, Balnur Kali, and Shuga Manabayeva. 2025. "Molecular Mechanisms Underlying Defense Responses of Potato (Solanum tuberosum L.) to Environmental Stress and CRISPR/Cas-Mediated Engineering of Stress Tolerance" Plants 14, no. 13: 1983. https://doi.org/10.3390/plants14131983
APA StyleSutula, M., Tussipkan, D., Kali, B., & Manabayeva, S. (2025). Molecular Mechanisms Underlying Defense Responses of Potato (Solanum tuberosum L.) to Environmental Stress and CRISPR/Cas-Mediated Engineering of Stress Tolerance. Plants, 14(13), 1983. https://doi.org/10.3390/plants14131983







