Validation of Fiber-Dominant Expressing Gene Promoters in Populus trichocarpa
Abstract
1. Introduction
2. Results
2.1. Screen of Candidate Genes with Xylem Fiber-Dominant Expression Through scRNA-Seq Data
2.2. Expression of Candidate Genes Is Checked in the Fibers and/or Vessels Using LCM Technique and RT-qPCR Analysis
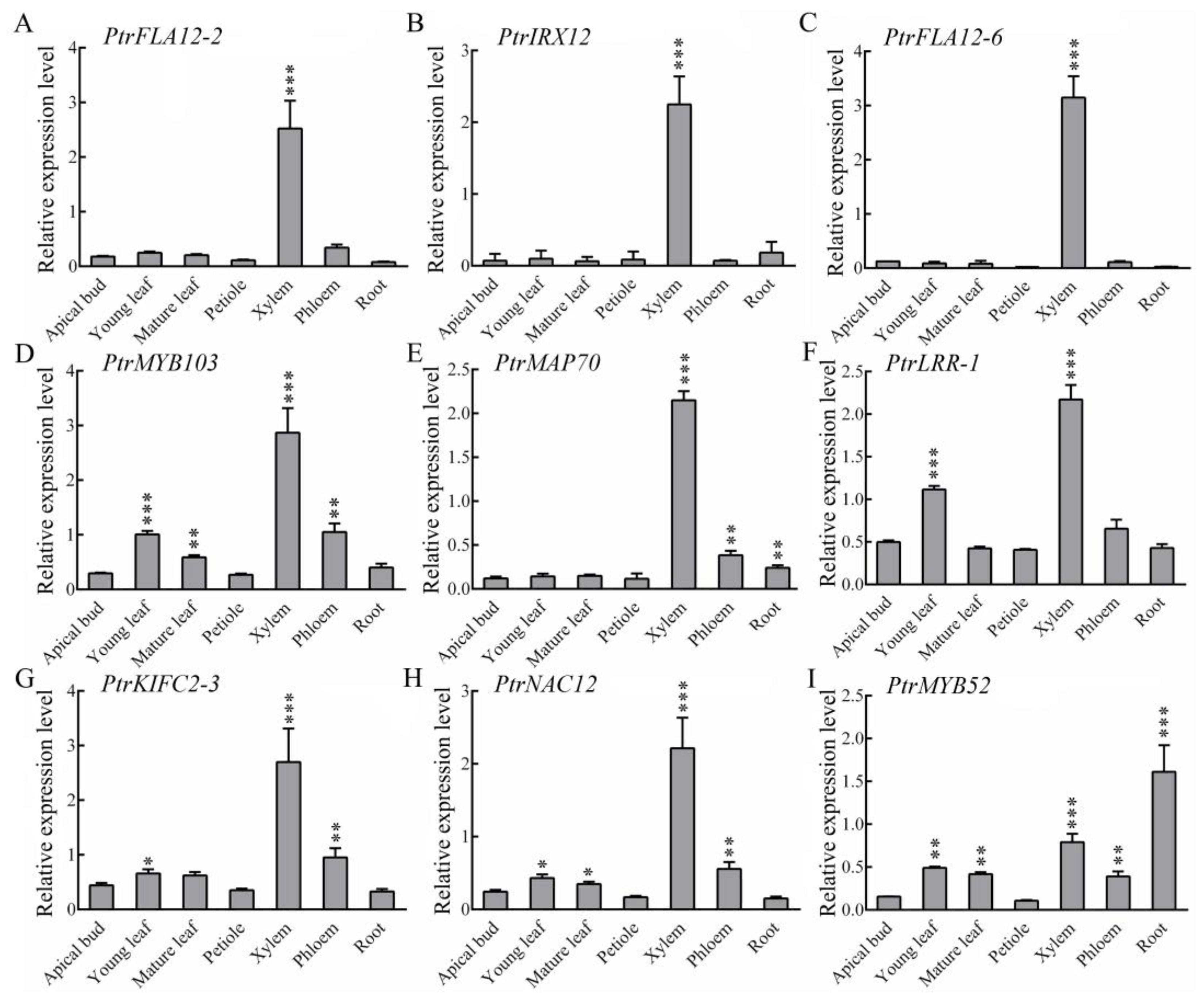
2.3. Transcript Levels of These Xylem Fiber-Dominant Expression Genes in Different Tissues
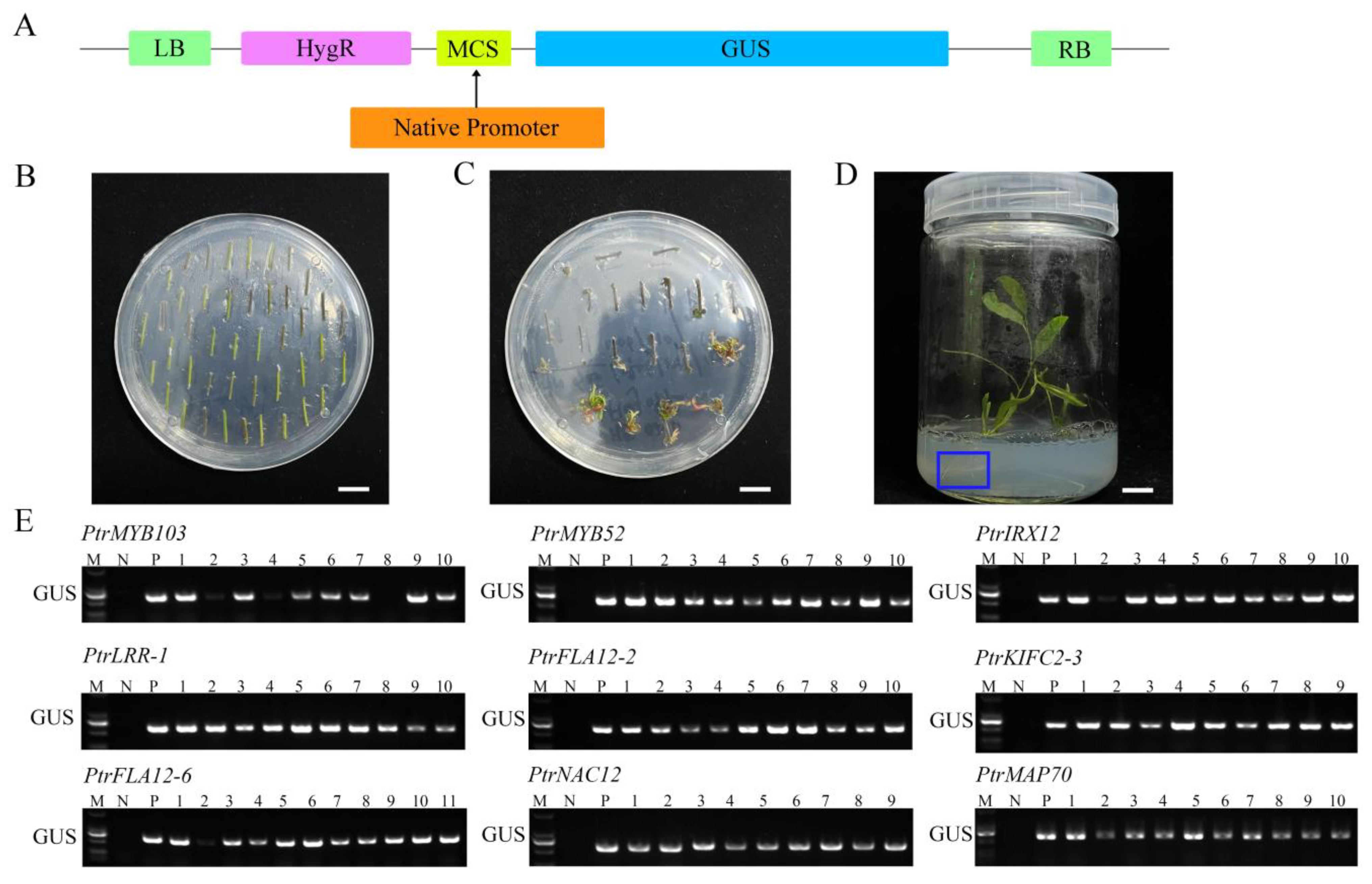
2.4. Cloning of Gene Promoter Fragments and Production of Promoter::GUS Transgenic Poplars
2.5. Tissue Expression Activities of Candidate Gene Promoters in Promoter::GUS Transgenic Poplars
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Tissue-Specific Expression Pattern Analysis
4.3. LCM
4.4. Extraction of RNA, and RT-qPCR
4.5. Vector Construction
4.6. Genetic Transformation
4.7. Identification of the Transgenic Plants
4.8. GUS Staining
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sarkanen, K.V. Renewable resources for the production of fuels and chemicals. Science 1976, 191, 773–776. [Google Scholar] [CrossRef]
- Richter, D.; Markewitz, D.; Trumbore, S.; Wells, C. Rapid accumulation and turnover of soil carbon in a re-establishing forest. Nature 1999, 400, 56–58. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J.; Hallett, J.P.; Leak, D.J.; Liotta, C.L.; et al. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef]
- Plomion, C.; Leprovost, G.; Stokes, A. Wood formation in trees. Plant Physiol. 2001, 127, 1513–1523. [Google Scholar] [CrossRef]
- Mellerowicz, E.J.; Sundberg, B. Wood cell walls: Biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 2008, 11, 293–300. [Google Scholar] [CrossRef]
- Yu, L.; Sun, J.; Li, L. PtrCel9A6, an endo-1,4-β-glucanase, is required for cell wall formation during xylem differentiation in populus. Mol. Plant 2013, 6, 1904–1917. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, J.; Xu, P.; Zhang, R.; Li, L. Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 2014, 164, 765–776. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Chiu, W.; Niwa, Y.; Zeng, W.; Hirano, T.; Kobayashi, H.; Sheen, J. Engineered GFP as a vital reporter in plants. Curr. Biol. 1996, 6, 325–330. [Google Scholar] [CrossRef]
- Odell, J.T.; Nagy, F.; Chua, N.H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef]
- McElroy, D.; Zhang, W.; Cao, J.; Wu, R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 1990, 2, 163–171. [Google Scholar]
- Christensen, A.H.; Sharrock, R.A.; Quail, P.H. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 1992, 18, 675–689. [Google Scholar] [CrossRef]
- Kasuga, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 2004, 45, 346–350. [Google Scholar] [CrossRef]
- Nguyen, V.P.; Cho, J.S.; Choi, Y.I.; Lee, S.W.; Han, K.H.; Ko, J.H. Evaluation of a novel promoter from Populus trichocarpa for mature xylem tissue specific gene delivery. Plant Physiol. Biochem. 2016, 104, 226–233. [Google Scholar] [CrossRef]
- Nguyen, V.P.; Cho, J.S.; Lee, J.H.; Kim, M.H.; Choi, Y.I.; Park, E.J.; Kim, W.C.; Hwang, S.; Han, K.H.; Ko, J.H. Identification and functional analysis of a promoter sequence for phloem tissue specific gene expression from Populus trichocarpa. J. Plant Biol. 2017, 60, 129–136. [Google Scholar] [CrossRef]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 2014, 111, 14097–14102. [Google Scholar] [CrossRef]
- Wang, Z.; Mao, Y.; Guo, Y.; Gao, J.; Liu, X.; Li, S.; Lin, Y.J.; Chen, H.; Wang, J.P.; Chiang, V.L.; et al. MYB Transcription Factor161 Mediates Feedback Regulation of Secondary wall-associated NAC-Domain1 Family Genes for Wood Formation. Plant Physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef]
- Zeng, J.; Yao, D.; Luo, M.; Ding, L.; Wang, Y.; Yan, X.; Ye, S.; Wang, C.; Wu, Y.; Zhang, J.; et al. Fiber-specific increase of carotenoid content promotes cotton fiber elongation by increasing abscisic acid and ethylene biosynthesis. Crop J. 2023, 11, 774–784. [Google Scholar] [CrossRef]
- Meng, Q.; Xie, P.; Xu, Z.; Tang, J.; Hui, L.; Gu, J.; Gu, X.; Jiang, S.; Rong, Y.; Zhang, J.; et al. Pangenome analysis reveals yield- and fiber-related diversity and interspecific gene flow in Gossypium barbadense L. Nat. Commun. 2025, 16, 4995. [Google Scholar] [CrossRef]
- Li, J.; Li, Z.; You, C.; Qi, Z.; You, J.; Grover, C.E.; Long, Y.; Huang, X.; Lu, S.; Wang, Y.; et al. Convergence and divergence of diploid and tetraploid cotton genomes. Nat. Genet. 2024, 56, 2562–2573. [Google Scholar] [CrossRef]
- Pires, R.C.; Ferro, A.; Capote, T.; Usié, A.; Correia, B.; Pinto, G.; Menéndez, E.; Marum, L. Laser Microdissection of Woody and Suberized Plant Tissues for RNA-Seq Analysis. Mol. Biotechnol. 2023, 65, 419–432. [Google Scholar] [CrossRef]
- Ludwig, Y.; Hochholdinger, F. Laser microdissection of plant cells. Methods Mol. Biol. 2014, 1080, 249–258. [Google Scholar] [PubMed]
- Li, H.; Dai, X.; Huang, X.; Xu, M.; Wang, Q.; Yan, X.; Sederoff, R.R.; Li, Q. Single-cell RNA sequencing reveals a high-resolution cell atlas of xylem in Populus. J. Integr. Plant Biol. 2021, 63, 1906–1921. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, J.; Zhou, J.; Cheng, X.; Hu, Y.; Wang, J.; Zou, J.; Zhao, Y.; Liu, C.; Hu, Z.; et al. Single-Cell RNA-Sequencing of Soybean Reveals Transcriptional Changes and Antiviral Functions of GmGSTU23 and GmGSTU24 in Response to Soybean Mosaic Virus. Plant Cell Environ. 2024. advance online publication. [Google Scholar] [CrossRef]
- Chen, Y.; Tong, S.; Jiang, Y.; Ai, F.; Feng, Y.; Zhang, J.; Gong, J.; Qin, J.; Zhang, Y.; Zhu, Y.; et al. Transcriptional landscape of highly lignified poplar stems at single-cell resolution. Genome Biol. 2021, 22, 319. [Google Scholar] [CrossRef]
- Conde, D.; Triozzi, P.M.; Pereira, W.J.; Schmidt, H.W.; Balmant, K.M.; Knaack, S.A.; Redondo-López, A.; Roy, S.; Dervinis, C.; Kirst, M. Single-nuclei transcriptome analysis of the shoot apex vascular system differentiation in Populus. Development 2022, 149, dev200632. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wang, Y.; Chen, W.; Xu, M.; Zhou, R.; Shou, H.; Chen, J. High-resolution anatomical and spatial transcriptome analyses reveal two types of meristematic cell pools within the secondary vascular tissue of poplar stem. Mol. Plant. 2023, 16, 809–828. [Google Scholar] [CrossRef]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-Spatial-Resolution Transcriptome Profiles Reveal Uncharacterized Modularity of Wood Formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef]
- Sreedasyam, A.; Plott, C.; Hossain, M.S.; Lovell, J.T.; Grimwood, J.; Jenkins, J.W.; Daum, C.; Barry, K.; Carlson, J.; Shu, S.; et al. JGI Plant Gene Atlas: An updateable transcriptome resource to improve functional gene descriptions across the plant kingdom. Nucleic Acids Res. 2023, 51, 8383–8401. [Google Scholar] [CrossRef]
- Dharmawardhana, P.; Brunner, A.M.; Strauss, S.H. Genome-wide analysis of Populus trichocarpa secondary stem development. Plant Physiol. 2010, 154, 1392–1404. [Google Scholar]
- Funk, V.; Kositsup, B.; Zhao, C.; Beers, E.P. The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 2002, 128, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170. [Google Scholar] [CrossRef]
- Harfouche, A.; Meilan, R.; Altman, A. Tree genetic engineering and applications to sustainable forestry and biomass production. Trends Biotechnol. 2011, 29, 9–17. [Google Scholar] [CrossRef]
- An, Y.; Jiao, X.; Yang, S.; Wang, S.; Chen, N.; Huang, L.; Jiang, C.; Lu, M.; Zhang, J. Evaluation of novel promoters for vascular tissue-specific gene expression in Populus. Plant Sci. 2024, 344, 112083. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Lebovka, I.; López-Salmerón, V.; Sanchez, P.; Greb, T. Bifacial cambium stem cells generate xylem and phloem during radial plant growth. Development 2019, 146, dev171355. [Google Scholar] [CrossRef] [PubMed]
- Twayana, M.; Girija, A.M.; Mohan, V.; Shah, J. Phloem: At the center of action in plant defense against aphids. J. Plant Physiol. 2022, 273, 153695. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; McCarthy, R.L.; Reeves, C.K.; Jones, E.G.; Ye, Z.H. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011, 52, 1856–1871. [Google Scholar] [CrossRef]
- Hussey, S.G.; Mizrachi, E.; Spokevicius, A.V.; Bossinger, G.; Berger, D.K.; Myburg, A.A. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011, 11, 173. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, M.; Tuskan, G.A.; Muchero, W.; Chen, J.G. Recent Advances in the Transcriptional Regulation of Secondary Cell Wall Biosynthesis in the Woody Plants. Front. Plant Sci. 2018, 9, 1535. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.R.; Helariutta, Y.; He, X.Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Alonso Serra, J.; Koskinen, P.; Zhang, J.; Elo, A.; Mähönen, A.P.; Street, N.; et al. Cytokinin and Auxin Display Distinct but Interconnected Distribution and Signaling Profiles to Stimulate Cambial Activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Help, H.; El-Showk, S.; Weijers, D.; Scheres, B.; Friml, J.; Benková, E.; Mähönen, A.P.; Helariutta, Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 2011, 21, 917–926. [Google Scholar] [CrossRef]
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Sánchez Rodríguez, F.; Wu, M.F.; Solé-Gil, A.; Leal Gavarrón, M.; Siligato, R.; Miyashima, S.; et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef]
- Miyashima, S.; Roszak, P.; Sevilem, I.; Toyokura, K.; Blob, B.; Heo, J.O.; Mellor, N.; Help-Rinta-Rahko, H.; Otero, S.; Smet, W.; et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490–494. [Google Scholar] [CrossRef]
- Lu, S.; Li, Q.; Wei, H.; Chang, M.J.; Tunlaya-Anukit, S.; Kim, H.; Liu, J.; Song, J.; Sun, Y.H.; Yuan, L.; et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2013, 110, 10848–10853. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef]
- Hinchee, M.; Rottmann, W.; Mullinax, L.; Zhang, C.; Chang, S.; Cunningham, M.; Pearson, L.; Nehra, N. Short-rotation woody crops for bioenergy and biofuels applications. Vitr. Cell. Dev. Biol. Plant 2009, 45, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Plomion, C.; Bastien, C.; Bogeat-Triboulot, M.B.; Bouffier, L.; Déjardin, A.; Duplessis, S.; Fady, B.; Heuertz, M.; Le Gac, A.L.; Le Provost, G.; et al. Forest tree genomics: 10 achievements from the past 10 years and future prospects. Ann. For. Sci. 2016, 73, 77–103. [Google Scholar] [CrossRef]
- Chen, H.C.; Song, J.; Wang, J.P.; Lin, Y.C.; Ducoste, J.; Shuford, C.M.; Liu, J.; Li, Q.; Shi, R.; Nepomuceno, A.; et al. Systems biology of lignin biosynthesis in Populus trichocarpa: Heteromeric 4-coumaric acid:coenzyme A ligase protein complex formation, regulation, and numerical modeling. Plant Cell 2014, 26, 876–893. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guo, M.; Ma, X.; Xu, S.; Cheng, J.; Xu, W.; Elsheery, N.I.; Cheng, Y. Genome-Wide Identification of TLP Gene Family in Populus trichocarpa and Functional Characterization of PtTLP6, Preferentially Expressed in Phloem. Int. J. Mol. Sci. 2024, 25, 5990. [Google Scholar] [CrossRef]
- Li, S.; Zhen, C.; Xu, W.; Wang, C.; Cheng, Y. Simple, rapid and efficient transformation of genotype Nisqually-1: A basic tool for the first sequenced model tree. Sci. Rep. 2017, 7, 2638. [Google Scholar] [CrossRef]





| Gene ID | Define |
|---|---|
| Potri.016G006900 | PtrMAP70 |
| Potri.003G132000 | PtrMYB103 |
| Potri.009G012200 | PtrFLA12-2 |
| Potri.005G186400 | PtrMYB52 |
| Potri.010G193100 | PtrIRX12 |
| Potri.013G011500 | PtrKIFC2-3 |
| Potri.013G151500 | PtrFLA12-6 |
| Potri.011G153300 | PtrNAC12 |
| Potri.005G188700 | PtrLRR-1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Wang, R.; Wang, B.; Xu, W.; Hou, H.; Cheng, H.; Zhang, Y.; Wang, C.; Cheng, Y. Validation of Fiber-Dominant Expressing Gene Promoters in Populus trichocarpa. Plants 2025, 14, 1948. https://doi.org/10.3390/plants14131948
Guo M, Wang R, Wang B, Xu W, Hou H, Cheng H, Zhang Y, Wang C, Cheng Y. Validation of Fiber-Dominant Expressing Gene Promoters in Populus trichocarpa. Plants. 2025; 14(13):1948. https://doi.org/10.3390/plants14131948
Chicago/Turabian StyleGuo, Mengjie, Ruxia Wang, Bo Wang, Wenjing Xu, Hui Hou, Hao Cheng, Yun Zhang, Chong Wang, and Yuxiang Cheng. 2025. "Validation of Fiber-Dominant Expressing Gene Promoters in Populus trichocarpa" Plants 14, no. 13: 1948. https://doi.org/10.3390/plants14131948
APA StyleGuo, M., Wang, R., Wang, B., Xu, W., Hou, H., Cheng, H., Zhang, Y., Wang, C., & Cheng, Y. (2025). Validation of Fiber-Dominant Expressing Gene Promoters in Populus trichocarpa. Plants, 14(13), 1948. https://doi.org/10.3390/plants14131948






