Meta-QTL Analysis and Genes Responsible for Plant and Ear Height in Maize (Zea mays L.)
Abstract
1. Introduction
2. Results
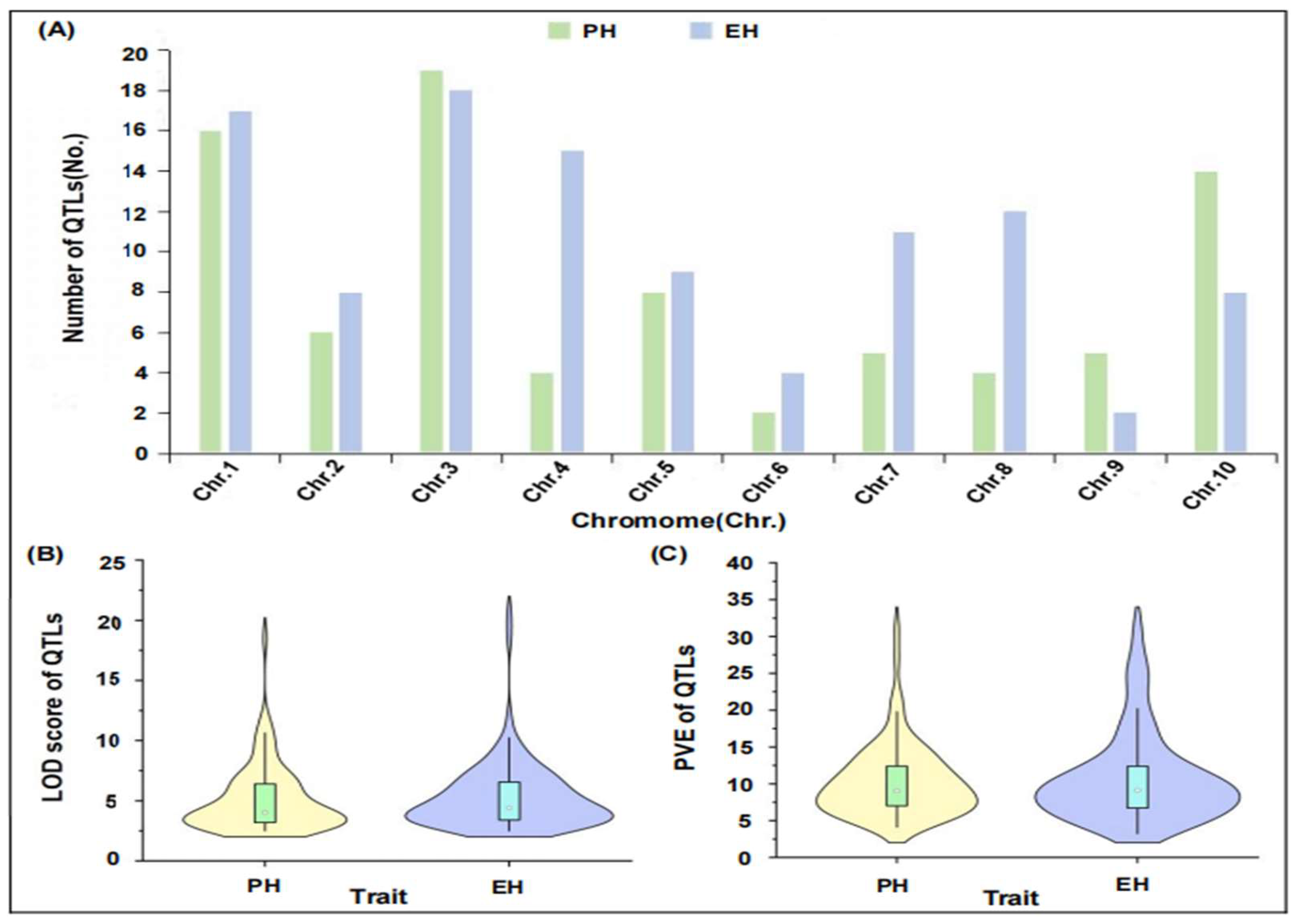
2.1. Integration of QTL-Related Information of PH and EH in Maize
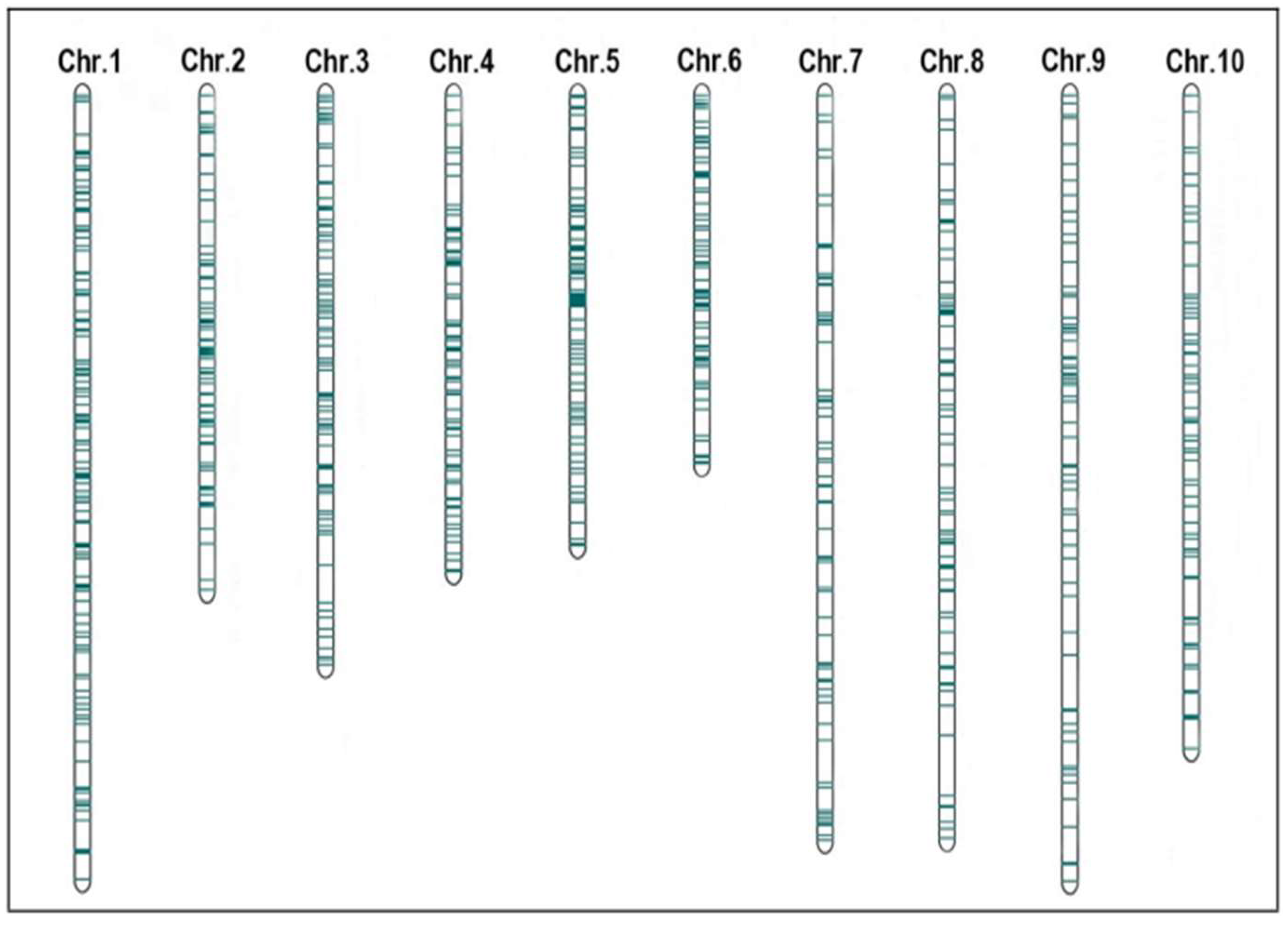
2.2. Construction and QTL Projection of Consensus Map of Maize PH and EH
2.3. Meta-QTL Analysis and Candidate Gene Identification in MQTL Region
3. Discussion
4. Materials and Methods
4.1. Literature Search and Information Collection for PH and EH QTL Localization of Maize
4.2. Consensus Map Construction and QTL Projection
4.3. Meta-QTL Analysis of Maize PH and EH
4.4. Identification and Functional Annotation of Candidate Genes in the MQTLs Interval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yin, X.F.; Bi, Y.Q.; Jiang, F.Y.; Guo, R.J.; Zhang, Y.D.; Fan, J.; Kang, M.S.; Fan, X.M. Fine mapping of candidate quantitative trait loci for plant and ear height in a maize nested-association mapping population. Front. Plant Sci. 2022, 13, 963985. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Wang, Y.L.; Wang, X.; Deng, D.X. Integrated Meta-QTL and genome-wide association study analyses reveal candidate genes for maize yield. J. Plant Growth Regul. 2020, 39, 229–238. [Google Scholar] [CrossRef]
- Shen, D.P.; Wang, K.R.; Zho, L.L.; Fang, L.; Wang, Z.; Fu, J.L.; Zhang, T.T.; Liang, Z.Y.; Xie, R.Z.; Ming, B.; et al. Increasing planting density and optimizing irrigation to improve maize yield and water-use efficiency in northeast China. Agronomy 2024, 14, 400. [Google Scholar] [CrossRef]
- Fei, J.B.; Lu, J.Y.; Jiang, Q.P.; Liu, Z.B.; Yao, D.; Qu, J.; Liu, S.Y.; Guan, S.Y.; Ma, Y.Y. Maize plant architecture trait QTL mapping and candidate gene identification based on multiple environments and double populations. BMC Plant Biol. 2022, 22, 110. [Google Scholar] [CrossRef]
- Zhang, G.X.; Cui, C.G.; Lv, Y.F.; Wang, X.Y.; Wang, X.F.; Zhao, D.H.; Hu, F.S.; Wen, X.X.; Han, J.; Liao, Y.C. Is it necessary to increase the maize planting density in China? Eur. J. Agron. 2024, 159, 127235. [Google Scholar] [CrossRef]
- Grassini, P.; Thorburn, J.; Burr, C.; Cassman, K.G. High-yield irrigated maize in the western U.S. Corn belt: I. On-farm yield, yield potential, and impact of agronomic practices. Field Crops Res. 2010, 120, 142–150. [Google Scholar] [CrossRef]
- Wang, X.Q.; Zhang, R.Y.; Shi, Z.; Zhang, Y.; Sun, X.; Ji, Y.L.; Zhao, Y.X.; Wang, J.D.; Zhang, Y.X.; Xing, J.F.; et al. Multi-omics analysis of the development and fracture resistance for maize internode. Sci. Rep. 2019, 9, 81–83. [Google Scholar] [CrossRef]
- Yang, J.S.; Geng, W.J.; Zhang, J.W.; Ren, B.Z.; Wang, L.C. Responses of the lodging resistance of summer maize with different gene types to plant density. Agronomy 2021, 12, 10. [Google Scholar] [CrossRef]
- Zhao, X.H.; Wang, C.B.; Liu, J.; Han, B.; Huang, J.L. Molecular markers and molecular basis of plant type related traits in maize. Front. Genet. 2024, 15, 1487700. [Google Scholar] [CrossRef]
- Incognito, S.J.P.; Maddonni, G.A.; López, C.G. Genetic control of maize plant architecture traits under contrasting plant densities. Euphytica 2020, 216, 33–43. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Romay, M.C.; Gore, M.A.; Flint-Garcia, S.A.; Zhang, Z.W.; Millard, M.J.; Gardner, C.A.C.; McMullen, M.D.; Holland, J.B.; Bradbury, P.J.; et al. The genetic architecture of maize height. Genetics 2014, 196, 1337–1356. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.G.; Chu, Q.; Gu, R.L.; Yuan, L.X.; Liu, J.C.; Zhang, X.Z.; Chen, F.J.; Mi, G.H.; Zhang, F.S. Identification of QTLs for plant height, ear height and grain yield in maize (Zea mays L.) in response to nitrogen and phosphorus supply. Plant Breed. 2012, 131, 502–510. [Google Scholar] [CrossRef]
- Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Flint-Garcia, S.; Rocheford, T.R.; McMullen, M.D.; Holland, J.B.; Buckler, E.S. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011, 43, 159–162. [Google Scholar] [CrossRef]
- Yang, H.X.; Zhang, Z.R.; Zhang, N.; Li, T.; Wang, J.J.; Zhang, Q.; Xue, J.Q.; Zhu, W.C.; Xu, S.T. QTL mapping for plant height and ear height using bi-parental immortalized heterozygous populations in maize. Front. Plant Sci. 2024, 15, 1371394. [Google Scholar] [CrossRef]
- Li, X.P.; Zhou, Z.J.; Ding, J.Q.; Wu, Y.B.; Zhou, B.; Wang, R.X.; Ma, J.L.; Wang, S.W.; Zhang, X.C.; Xia, Z.L.; et al. Combined linkage and association mapping reveals QTL and candidate genes for plant and ear height in maize. Front. Plant Sci. 2016, 7, 833. [Google Scholar] [CrossRef]
- Choi, J.K.; Sa, K.J.; Park, D.H.; Lim, S.E.; Ryu, S.H.; Park, J.Y.; Park, K.J.; Rhee, H.I.; Lee, M.; Lee, J.K. Construction of genetic linkage map and identification of QTLs related to agronomic traits in DH population of maize (Zea mays L.) using SSR markers. Genes Genom. 2019, 41, 667–678. [Google Scholar] [CrossRef]
- Ku, L.X.; Zhang, L.K.; Tian, Z.Q.; Guo, S.L.; Su, H.H.; Ren, Z.Z.; Wang, Z.Y.; Li, G.H.; Wang, X.B.; Zhu, Y.G.; et al. Dissection of the genetic architecture underlying the plant density response by mapping plant height-related traits in maize (Zea mays L.). Mol. Genet. Genom. 2015, 290, 1223–1233. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, J.G.; Cao, J.S.; Cao, S.L.; Li, W.Y.; Yang, G.B. A meta-analysis of low temperature tolerance QTL in maize. Electron. J. Biotechnol. 2022, 58, 82–91. [Google Scholar] [CrossRef]
- Sheoran, S.; Gupta, M.; Kumari, S.; Kumar, S.; Rakshit, S. Meta-QTL analysis and candidate genes identification for various abiotic stresses in maize (Zea mays L.) and their implications in breeding programs. Mol. Breed. 2022, 42, 26. [Google Scholar] [CrossRef]
- Tang, R.; Zhuang, Z.L.; Bian, J.W.; Ren, Z.P.; Ta, W.; Peng, Y.L. GWAS and meta-QTL analysis of kernel quality-related traits in maize. Plants 2024, 13, 2730. [Google Scholar] [CrossRef]
- Li, R.L.; Wang, Y.L.; Li, D.D.; Guo, Y.H.; Zhou, Z.P.; Zhang, M.; Zhang, Y.F.; Wuerschum, T.; Liu, W.X. Meta-quantitative trait loci analysis and candidate gene mining for drought tolerance-associated traits in maize (Zea mays L.). Int. J. Mol. Sci. 2024, 25, 4295. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.G.; Qi, H.D.; Chen, Q.S.; Zhang, Z.G.; Jiang, H.W.; Zhu, R.S.; Hu, Z.B.; Wu, X.X.; Li, C.D.; Zhang, Y.; et al. Soybean plant height QTL mapping and meta-analysis for mining candidate genes. Plant Breed. 2017, 136, 688–698. [Google Scholar] [CrossRef]
- Wang, Z.H. QTL Mapping and Analysis for Plant Type and Ear Traits in Maize. Master’s Thesis, Southwest University, Chongqing, China, 2011. [Google Scholar]
- Zhang, X. The QTL Analysis of Important Agronomic Traits for a RIL Population in Maize. Master’s Thesis, Sichuan Agricultural University, Ya’an, China, 2012. [Google Scholar]
- Tang, Z.X.; Yang, Z.F.; Hu, Z.Q.; Zhang, D.; Lu, X.; Jia, B.; Deng, D.; Xu, C.W. Cytonuclear epistatic quantitative trait locus mapping for plant height and ear height in maize. Mol. Breed. 2013, 31, 1–14. [Google Scholar] [CrossRef]
- Sun, T.T. Construction of Genetic Linkage Map and Indentification of QTLs Important Agronomic Trait in Maize (Zea mays L.). Master’s Thesis, Yanbian University, Yanji, China, 2013. [Google Scholar]
- Shi, L.G. QTL Mapping of Four Plant Type Traits in Maize. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2013. [Google Scholar]
- Fang, H.Y. QTL Analysis of Lodging Resietance-Related Traits in Maize (Zea mays L.) Under High Plant Density. Master’s Thesis, Yanbian University, Yanji, China, 2017. [Google Scholar]
- Niu, Q.K.; Yang, C.; Shi, Z.W.; Cao, M.J. QTL mapping of dwarf-associated traits in the maize male sterile mutant obtained by space flight. J. Sichuan Agric. Univer. 2018, 36, 429–435. [Google Scholar]
- Shen, T.; Tan, K.; Li, C.H.; Yang, M.; Hu, X.L.; Jiang, T.; Zhang, Z.; Qiu, H.B. QTL mapping for plant type related traits in maize. Mol. Plant Breed. 2022, 20, 155–162. [Google Scholar]
- Zhu, L.Y.; Chen, J.T.; Li, D.; Zhang, J.H.; Huang, Y.Q.; Zhao, Y.F.; Song, Z.Q.; Liu, Z.Z. QTL mapping for stalk related traits in maize (Zea mays L.) under different densities. J. Integr. Agric. 2013, 12, 218–228. [Google Scholar] [CrossRef]
- Bian, Y.L.; Sun, D.L.; Gu, X.; Wang, Y.J.; Yin, Z.T.; Deng, D.X.; Wang, Y.Q.; Wu, F.F.; Li, G.S. Identification of QTL for stalk sugar-related traits in a population of recombinant inbred lines of maize. Euphytica 2014, 198, 79–89. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Peng, Y.L.; Zhang, J.W.; Fang, P.; Wu, B.Y. Identification of QTLs and Meta-QTLs for seven agronomic traits in multiple maize populations under well-watered and water-stressed conditions. Crop Sci. 2018, 58, 507–520. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, Q.; Zhang, Y.N.; Li, X.; Huang, X.Q. Dissection of the genetic architecture of plant height and ear height in maize (Zea mays L.). Fudan Univ. J. Med. Sci. (Nat. Sci.) 2016, 55, 605–613. [Google Scholar]
- Zhao, B.W.; Tong, L.L.; Liu, H.L.; Hao, M.Y.; Zhang, R.H. Optimizing root morphology is a key to improving maize yield under nitrogen reduction and densification cultivation. Field Crops Res. 2024, 329, 109958. [Google Scholar] [CrossRef]
- Jia, Q.M.; Sun, L.F.; Mou, H.Y.; Ali, S.; Liu, D.H.; Zhang, Y.; Zhang, P.; Ren, X.L.; Jia, Z.K. Effects of planting patterns and sowing densities on grain-filling, radiation use efficiency and yield of maize (Zea mays L.) in semi-arid regions. Agric. Water Manag. 2018, 201, 287–298. [Google Scholar] [CrossRef]
- Yang, X.J.; Lu, M.; Zhang, S.H.; Zhou, F.; Qu, Y.Y.; Xie, C.X. QTL mapping of plant height and ear position in maize (Zea mays L.). Hereditas 2008, 30, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Yang, Q.S.; Fan, N.N.; Zhang, M.; Zhai, H.J.; Ni, Z.F.; Zhang, Y.R. Quantitative trait locus analysis of heterosis for plant height and ear height in an elite maize hybrid zhengdan 958 by design III. BMC Genet. 2017, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.S.; Shiri, M.; Mostafavi, K.; Mohammadi, A.; Miri, S.M. Genetic analysis and association detection of agronomic traits in maize genotypes. Sci. Rep. 2025, 15, 399. [Google Scholar] [CrossRef]
- Wang, B.B.; Lin, Z.C.; Li, X.; Zhao, Y.P.; Zhao, B.B.; Wu, G.X.; Ma, X.J.; Wang, H.; Xie, Y.R.; Li, Q.Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef]
- Li, C.H.; Guan, H.H.; Jing, X.; Li, Y.Y.; Wang, B.B.; Li, Y.X.; Liu, X.Y.; Zhang, D.F.; Liu, C.; Xie, X.Q.; et al. Genomic insights into historical improvement of heterotic groups during modern hybrid maize breeding. Nat. Plants 2022, 8, 750–763. [Google Scholar] [CrossRef]
- Phillips, K.A.; Skirpan, A.L.; Liu, X.; Christensen, A.; Slewinski, T.L.; Hudson, C.; Barazesh, S.; Cohen, J.D.; Malcomber, S.; McSteen, P. Vanishing tassel encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 2011, 23, 550–566. [Google Scholar] [CrossRef]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef]
- Avila, L.M.; Cerrudo, D.; Swanton, C.; Lukens, L. Brevis plant1, a putative inositol polyphosphate 5-phosphatase, is required for internode elongation in maize. J. Exp. Bot. 2016, 67, 1577–1588. [Google Scholar] [CrossRef]
- Yue, K.; Li, L.L.; Xie, J.H.; Coulter, J.A.; Luo, Z.Z. Synthesis and regulation of auxin and abscisic acid in maize. Plant Signal. Behav. 2021, 16, 1891756. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Park, S.; Han, K.H. Transcriptional regulation of secondary growth in Arabidopsis thaliana. J. Expt. Bot. 2003, 54, 2709–2722. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Tanveer, M.; Anjum, S.A.; Iqbal, J.; Ahmad, R. Lodging stress in cereal-effects and management: An overview. Environ. Sci. Pollut. Res. 2017, 24, 5222–5237. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.; Liu, D.C.; Guo, X.L.; Yang, W.L.; Sun, J.Z.; Li, X.; Zhan, K.H.; Cui, D.Q.; Lin, J.X.; Zhang, A.M. Anatomical and chemical characteristics associated with lodging resistance in wheat. Crop J. 2013, 1, 43–49. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, X.; Lin, Z.; Wang, J.; Liu, H.; Zhou, L.; Zhong, S.; Li, Y.; Zhu, C.; Lai, J.; et al. A large transposon insertion in the stiff 1 promoter increases stalk strength in maize. Plant Cell 2020, 32, 152–165. [Google Scholar] [CrossRef]
- Guo, W.B.; Zhao, J.; Li, X.X.; Qin, L.; Yan, X.L.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Cheng, M.Z.; Meng, F.Y.; Mo, F.L.; Qi, H.A.; Wang, P.W.; Chen, X.L.; Liu, J.Y.; Ghanizadeh, H.; Zhang, H.; Wang, A.X. Slym1 control the color etiolation of leaves by facilitating the decomposition of chlorophyll in tomato. Plant Sci. 2022, 324, 111457. [Google Scholar] [CrossRef]
- Chen, S.Z.; Fan, X.C.; Song, M.F.; Yao, S.T.; Liu, T.; Ding, W.S.; Liu, L.; Zhang, M.L.; Zhan, W.M.; Yan, L.; et al. Cryptochrome 1b represses gibberellin signaling to enhance lodging resistance in maize. Plant Physiol. 2024, 194, 902–917. [Google Scholar] [CrossRef]
- Tang, H.J.; Jing, D.; Liu, C.; Xie, X.Q.; Zhang, L.; Chen, X.J.; Li, C.Y.; Bhadauria, V. Genome-wide identification and expression analyses of the FAR1/FHY3 gene family provide insight into inflorescence development in maize. Curr. Issues Mol. Biol. 2024, 46, 430–449. [Google Scholar] [CrossRef]
- Inada, N. Plant actin depolymerizing factor: Actin microfilament disassembly and more. J. Plant Res. 2018, 131, 567. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Li, H.Y.; Zhang, D.F.; Liu, Y.H.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Characterization and expression analysis of six MADS-box genes in maize (Zea mays L.). J. Plant Physiol. 2012, 169, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A.; Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 1997, 27, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Swamy, B.P.M.; Vikram, P.; Dixit, S.; Ahmed, H.U.; Kumar, A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genom. 2011, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Jasinski, S.; Durandet, M.; Le’cureuil, A.; Soulay, F.; Bedu, M.; Guerche, P.; Masclaux-Daubresse, C. QTL meta-analysis in Arabidopsis reveals an interaction between leaf senescence and resource allocation to seeds. J. Exp. Bot. 2014, 65, 3949–3962. [Google Scholar] [CrossRef]
- Woodhouse, M.R.; Sen, S.; Schott, D.; Portwood, J.L.; Freeling, M.; Walley, J.W.; Andorf, C.M.; Schnable, J.C. qTeller: A tool for comparative multi-genomic gene expression analysis. Bioinformatics 2021, 38, 236–242. [Google Scholar] [CrossRef]



| Population | Population Phenotypes | QTL Number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cross Group | Type | Size | Env. | Marker | Length (cM) | Range of PH (cm) | Range of EH (cm) | Mean PH (cm) | Mean EH (cm) | PH | EH | Reference |
| MO17 × SDM | F2 | – | 1 | 180/SSR | 985.71 | 104.00–285.00 | 34.00–116.00 | 230.70 | 79.97 | 3 | 2 | [23] |
| MU6 × SDM | F2 | – | 1 | 150/SSR | 1085.81 | 21.00–290.00 | 34.00–116.00 | 224.10 | 74.85 | 3 | 3 | |
| 178 × 9782 | RIL | 271 | 1 | 259/SSR | 1440.00 | 128.65–224.76 | 45.90–99.49 | 172.88 | 70.27 | 4 | 6 | [24] |
| JB × Y53 | F2 | – | 1 | 154/SSR | 1735.00 | 96.00–211.00 | 18.00–75.00 | 173.53 | 50.31 | 3 | 10 | [25] |
| F2:3 | 211 | 1 | 130.40–210.38 | 27.00–95.78 | 168.41 | 62.62 | 6 | 11 | ||||
| Chuan-287 × Chuan-144 | F2 | 187 | 1 | 152/SSR | 1268.10 | 172.25–306.00 | 54.50–144.56 | 241.02 | 90.53 | 3 | 3 | [26] |
| JS037 × JS133 | F2 | 192 | 1 | 136/SSR | 1148.40 | 143.10–245.90 | 38.20–107.70 | 196.27 | 70.71 | 4 | 1 | [27] |
| Chuan-287 × Chuan-144 | F3 | 187 | 2 | 152/SSR | 1268.10 | 142.25–256.23 | 44.50–124.56 | 172.20 | 65.88 | 3 | 3 | [28] |
| 156.81–285.23 | 35.40–125.82 | 206.74 | 65.38 | 1 | 1 | |||||||
| F4 | 187 | 1 | 151.42–274.81 | 39.56–121.33 | 202.99 | 68.57 | 3 | 3 | ||||
| ms39ms39 × B73 | F2 | 120 | 1 | 71/Indel,109/SSR | 1173.40 | 134.80–273.00 | 41.10–101.40 | – | – | 4 | 2 | [29] |
| T32 × HuangC | F2:3 | 184 | 3 | 193/SSR | 1103.36 | 84.00–206.33 | 24.67–87.00 | 63.16 | 59.58 | 2 | 2 | [30] |
| 103.23–201.72 | 33.67–84.28 | 161.70 | 57.93 | 3 | 3 | |||||||
| 112.38–207.41 | 28.58–90.73 | 160.98 | 57.10 | 2 | 3 | |||||||
| NX110 × NX531 | DH | 162 | 4 | 178/SSR | 1721.19 | 121.00–224.00 | 32.00–99.67 | 170.82 | 64.39 | 1 | 2 | [31] |
| 128.60–247.40 | 33.40–114.80 | 183.41 | 73.01 | 1 | 3 | |||||||
| 122.00–231.67 | 37.33–109.67 | 170.24 | 65.95 | 1 | 3 | |||||||
| 124.00–274.40 | 49.40–131.20 | 196.67 | 82.57 | 1 | 2 | |||||||
| YXD053 × Y6-1 | RIL | 202 | 2 | 200/SSR,12/AFLP | 1648.60 | 109.83–163.67 | – | 136.84 | – | 3 | [32] | |
| 91.02–188.98 | – | 143.14 | – | 3 | ||||||||
| Langhuang × TS141 | F2:3 | 202 | 4 | 213/SSR | 1542.50 | 135.00–261.73 | 49.32–138.52 | 204.61 | 94.27 | 3 | 3 | [33] |
| 121.82–251.62 | 35.20–130.33 | 182.62 | 78.53 | 3 | 3 | |||||||
| 156.00–264.00 | 62.39–148.26 | 214.57 | 103.12 | 2 | 5 | |||||||
| 154.00–261.00 | 46.00–123.33 | 202.84 | 83.37 | 3 | 3 | |||||||
| Chang7-2 × TS141 | F2:3 | 218 | 4 | 217/SSR | 1648.80 | 100.28–51.21 | 50.00–165.30 | 171.75 | 94.24 | 4 | 5 | [33] |
| 84.80–241.27 | 34.20–146.60 | 157.36 | 89.33 | 2 | 4 | |||||||
| 97.20–261.06 | 33.20–131.53 | 161.36 | 76.19 | 3 | 5 | |||||||
| 89.70–244.56 | 30.10–126.10 | 152.22 | 62.66 | 2 | 4 | |||||||
| B73 × Zheng58 | RIL | 165 | 1 | 189/SSR | 2058.80 | 106.80–297.30 | 35.00–120.00 | 207.10 | 71.80 | 5 | 6 | [34] |
| S112 × H132 | F2:3 | 217 | 5 | 171/SSR | 4734.51 | 135.00–300.00 | 43.00–179.00 | 227.67 | 100.26 | 1 | 1 | [4] |
| 143.00–297.00 | 55.00–145.00 | 225.81 | 96.00 | 1 | 1 | |||||||
| 155.00–345.00 | 53.00–136.00 | 232.35 | 92.59 | 1 | 1 | |||||||
| 157.00–300.00 | 55.00–136.00 | 225.32 | 93.70 | – | 1 | |||||||
| 157.00–292.00 | 59.00–165.00 | 229.96 | 96.91 | 1 | 3 | |||||||
| Trait | MQTL | Chr. | Position (cM) | QTLs Number | Bin | Marker Interval | CI | Physical Interval (Mb) | Contig |
|---|---|---|---|---|---|---|---|---|---|
| PH, EH | MQTL1–1 | 1 | 30.90 | 6 | 1.01 | umc2546–umc1292 | 5.40–59.20 | 2.09–5.41 | ctg2–ctg3 |
| PH, EH | MQTL1–2 | 1 | 130.30 | 2 | 1.02 | bnlg1178–bnlg1429 | 125.00–143.50 | 14.07–16.56 | ctg6–ctg7 |
| PH, EH | MQTL1–3 | 1 | 232.50 | 5 | 1.03 | umc1073–bnlg1203 | 208.60–259.30 | 32.87–43.71 | ctg11 |
| EH | MQTL1–4 | 1 | 402.70 | 2 | 1.04 | bnlg2295–umc1243 | 398.20–405.00 | 80.17–83.54 | ctg20 |
| PH | MQTL1–5 | 1 | 488.10 | 3 | 1.05–1.06 | umc1603–umc1972 | 475.90–503.30 | 165.42–178.04 | ctg33–ctg38 |
| PH, EH | MQTL1–6 | 1 | 779.40 | 4 | 1.08 | bnlg1643–umc1991 | 768.50–800.70 | 232.80–245.37 | ctg50 |
| PH, EH | MQTL2–1 | 2 | 54.90 | 4 | 2.01–2.02 | umc1542–umc1227 | 54.5–55.30 | 4.67–4.71 | ctg69 |
| PH, EH | MQTL2–2 | 2 | 89.10 | 5 | 2.02 | umc1961–mmc0111 | 88.90–90.20 | 8.02–8.30 | ctg70 |
| PH, EH | MQTL2–3 | 2 | 404.00 | 3 | 2.07 | bnlg1329–umc2129 | 383.5–403.40 | 184.75–188.81 | ctg98 |
| PH, EH | MQTL3–1 | 3 | 108.40 | 6 | 3.02–3.04 | bnlg1647–bnlg1904 | 102.50–126.50 | 8.21–9.81 | ctg112 |
| PH, EH | MQTL3–2 | 3 | 193.90 | 10 | 3.04 | umc1655–umc1504 | 189.90–227.40 | 26.31–58.49 | ctg116–ctg119 |
| PH, EH | MQTL3–3 | 3 | 350.10 | 8 | 3.04–3.05 | umc1839–umc1087 | 343.50–364.60 | 155.29–161.90 | ctg117–ctg131 |
| PH, EH | MQTL3–4 | 3 | 474.70 | 3 | 3.06 | umc1644–umc2269 | 472.30–478.00 | 183.94–184.73 | ctg132–ctg138 |
| PH, EH | MQTL3–5 | 3 | 567.20 | 7 | 3.07 | umc1489–umc1286 | 566.80–569.60 | 202.17–202.87 | ctg142–ctg143 |
| PH, EH | MQTL3–6 | 3 | 803.50 | 3 | 3.09–3.10 | umc1052–umc2048 | 789.00–817.20 | 226.86–230.27 | ctg151–ctg151 |
| EH | MQTL4–1 | 4 | 312.70 | 8 | 4.06 | bnlg1741–bnlg1784 | 296.60–336.30 | 154.65–170.00 | ctg182–ctg431 |
| EH | MQTL4–2 | 4 | 535.10 | 4 | 4.09 | umc1940–umc1650 | 524.30–544.60 | 221.18–230.89 | ctg196–ctg200 |
| PH, EH | MQTL5–1 | 5 | 586.90 | 9 | 5.06–5.07 | umc2305–bnlg1346 | 459.20–534.40 | 193.36–208.04 | ctg247–ctg251 |
| EH | MQTL6–1 | 6 | 307.00 | 3 | 6.05 | npi252–bnlg1702 | 304.00–312.70 | 143.66–145.85 | ctg281–ctg285 |
| PH, EH | MQTL7–1 | 7 | 187.20 | 6 | 7.02 | umc1666–umc1932 | 181.10–202.50 | 47.95–78.03 | ctg301–ctg308 |
| PH, EH | MQTL7–2 | 7 | 401.80 | 5 | 7.03–7.04 | umc2329–bnlg1666 | 382.10–428.20 | 151.28–158.98 | ctg322–ctg323 |
| EH | MQTL8–1 | 8 | 264.90 | 3 | 8.03–8.04 | umc1457–umc1858 | 257.80–285.60 | 101.65–112.06 | ctg345–ctg349 |
| PH, EH | MQTL8–2 | 8 | 348.40 | 3 | 8.05 | bnlg666–umc2210 | 340.20–366.30 | 133.37–152.00 | ctg354–ctg358 |
| PH | MQTL8–3 | 8 | 505.00 | 3 | 8.07–8.08 | umc1384–bnlg1056 | 482.40–523.60 | 169.21–171.33 | ctg363–ctg365 |
| PH, EH | MQTL9–1 | 9 | 254.10 | 3 | 9.03–9.04 | bnlg1626–bnlg1209 | 230.20–268.20 | 88.14–109.64 | ctg376 |
| PH, EH | MQTL10–1 | 10 | 68.70 | 7 | 10.01–10.02 | umc1319–umc1432 | 47.40–91.40 | 4.61–5.77 | ctg392 |
| PH | MQTL10–2 | 10 | 144.90 | 2 | 10.03 | umc1863–bnlg210 | 139.40–166.80 | 13.34–26.78 | ctg394–ctg398 |
| PH, EH | MQTL10–3 | 10 | 200.40 | 3 | 10.03–10.04 | bnlg1079–umc1938 | 196.70–204.50 | 63.83–77.35 | ctg400–ctg402 |
| EH | MQTL10–4 | 10 | 232.10 | 2 | 10.03–10.04 | umc2348–umc1077 | 229.40–236.90 | 93.28–102.71 | ctg409–ctg411 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhao, X.; Sun, S.; Tao, K.; Niu, Y. Meta-QTL Analysis and Genes Responsible for Plant and Ear Height in Maize (Zea mays L.). Plants 2025, 14, 1943. https://doi.org/10.3390/plants14131943
Li X, Zhao X, Sun S, Tao K, Niu Y. Meta-QTL Analysis and Genes Responsible for Plant and Ear Height in Maize (Zea mays L.). Plants. 2025; 14(13):1943. https://doi.org/10.3390/plants14131943
Chicago/Turabian StyleLi, Xin, Xiaoqiang Zhao, Siqi Sun, Kejin Tao, and Yining Niu. 2025. "Meta-QTL Analysis and Genes Responsible for Plant and Ear Height in Maize (Zea mays L.)" Plants 14, no. 13: 1943. https://doi.org/10.3390/plants14131943
APA StyleLi, X., Zhao, X., Sun, S., Tao, K., & Niu, Y. (2025). Meta-QTL Analysis and Genes Responsible for Plant and Ear Height in Maize (Zea mays L.). Plants, 14(13), 1943. https://doi.org/10.3390/plants14131943







