Abstract
The SEPALLATA3 (SEP3)-like MADS-box genes play crucial roles in determining petal identity and development in the petunia and tomato of Solanaceae. Solanum nigrum is a self-pollinating plant in the Solanaceae family, and produces white flowers. However, the mechanisms controlling the transition from green to white petals during flower development remain poorly understood. In this study, we isolated a flower-specific SEP3-like gene, SnMADS37, from S. nigrum, and investigated its potential role in chlorophyll metabolism during petal development. Our results show that quantitative RT-PCR analysis demonstrates that SnMADS37 is exclusively expressed in petals and stamens during early floral bud development. Overexpression of SnMADS37 clearly enhanced the number of petals, promoting the formation of additional petal-like tissues in stamens and extra organs in some fruits. Moreover, fully opened transformed petals exhibited notable chlorophyll accumulation at their tips and veins, whereas silencing of Snmads37 clearly inhibited petal expansion and reduced green pigmentation in early flower buds. Additionally, the transformed green petals exhibited distinct conical epidermal cells in the green regions, similar to wild type (WT) petals. Our results demonstrate that SnMADS37 plays a critical role in regulating petal identity, expansion, and chlorophyll metabolism during petal development. These findings provide new insights into the functional diversification of SEP3-like MADS-box genes in angiosperms.
1. Introduction
Flower color is an important trait in ornamental plants. Solanum nigrum is a member of the economically significant Solanaceae family with the eggplant, potato, and petunia [1]. It is globally important for agriculture, human health, and biotechnology [2,3,4]. The flower of S. nigrum exhibits a typical zygomorphic structure, consisting of five sepals, five fused white petals, five stamens, and a fused carpel. Notably, the petals initially contain chlorophyll, which diminishes as the flower matures, resulting in a transition to a white appearance.
MADS-box proteins are a class of transcription factors found in a wide range of organisms, from yeast to humans [5,6]. In plants, the MADS-box genes influence various aspects of development, including root [7], fiber [8], floral organ [9], and fruit development [10,11]. Furthermore, molecular genetic studies of the model plants such Arabidopsis, Antirrhinum, and petunia have demonstrated that MADS-box transcription factors play a crucial role in determining floral organ identity and are codified in the ABCE model, which postulates four gene functions, A, B, C, and E, that act in overlapping concentric domains of the meristem to specify the floral organs. For instance, A + E specifies sepal, A + B + E specifies petals, B + C + E specifies stamens, and C + E specifies carpel identity in the flowers [12,13,14,15].
SEPALLATA (SEP) is an E-class MADS-box gene that acts as an important mediator in various aspects of flower development, working in conjunction with A-, B-, and C-class MADS-box genes. The SEP-like genes have been isolated from numerous plant species, including Arabidopsis, petunia, and tomato [13,16,17,18]. In Arabidopsis, four SEP family genes, i.e., AtSEP1, AtSEP2, AtSEP3, and AtSEP4, exhibit slightly different expression patterns but are all essential for the specification of floral organs [13,19]. The Atsep3 single mutant displays only subtle phenotypic changes; however, the Atsep1/Atsep2/Atsep3 triple mutants exhibit an abnormal phenotype in which the inner three whorls of floral organs are converted into sepals. In the Atsep1/Atsep2/Atsep3/Atsep4 quadruple mutants, all the floral organs are converted into leaf-like structures [13,20]. In petunias, an AtSEP3 ortholog fbp2 mutant exhibit greenish petals and ectopic inflorescences originating from the third floral whorl [18]. In Gerbera of Asteraceae, the SEP3 gene GRCD5 (GERBERA REGULATOR OF CAPITULUM DEVELOPNT 5) shows unique functions in petal development. However, GRCD1/2 belongs to the SEP3 clade, and appears to have sub-functionalized to regulate stamen and carpel identity [21,22,23]. In contrast, overexpression of AtSEP3 induces early flowering and activates the AtAPETALA3 (AtAP3) and AtAGAMOUS (AtAG) genes [24]. Similarly, the ectopic expression of a LaSEP3-like gene in lavender results in early flowering in Arabidopsis [25]. Unlike, the ectopic expression of a wheat SEP3-like gene, TaMADS1 leads to early flowering, alters floral organ development including the conversion of sepals into leaf-like structures, and reduces the numbers of petals and stamens in transformed Arabidopsis [26]. Furthermore, the ectopic expression of a homolog of AtSEP3, BpMADS1, decreases the number of floral organs or whole whorls along with petaloid or carpelloid sepals in Arabidopsis [27]. Therefore, SEP genes play an important role in the activities of A-, B-, and C-function genes in the development of petals, stamens, and carpels in flowering plants.
In this study, we isolated the SnMADS37 gene and reported on its potential roles in S. nigrum. Our results indicate that the expression of SnMADS37 is restricted to the petals and stamens. Overexpression of this gene increased the number of petals and led to additional organ development in both the stamens and fruits. Furthermore, it influenced chlorophyll accumulation in the petals, suggesting that SnMADS37 plays an important role in the regulation of petal development, resulting in green petal-tip flowers in transformed S. nigrum.
2. Results
2.1. Isolation of SEPALLATA3-like Gene SnMADS37
Based on bioinformatics analysis of the transcriptome data of S. nigrum and BLAST searches (www.ncbi.nlm.nih.gov), two out of 88 MADS-box genes were found to be closely related to the AtSEP3 in Arabidopsis. Furthermore, a comparison of the amino acid sequences of AtSEP3 in Arabidopsis and LeSEP3 in tomato revealed a candidate gene, SnMADS37. The full amino acid sequence of SnMADS37 showed similarities of 63.7% with AtSEP3 and 98.3% with LeSEP3, respectively (Figure 1). This led to the identification of SnMADS37 as an SEP3-like gene in S. nigrum. To confirm the sequence of SnMADS37, the first strand cDNA from the inflorescence sample was amplified with the set of gene-specific primers SnMADS37-F2/SnMADS37-R2 (Table S1). The resulting 729 bp fragment encoded 242 amino acids, which corresponded to the primary transcriptome data (Figure 1).

Figure 1.
Sequence comparison between SnMADS37 and related MADS-box proteins. The deduced amino acid sequence aligned with the sequences of AtSEP3 (Arabidopsis) and LeSEP3 (tomato). The MADS-box and K-box domains are indicated by lines above, and the three sub-domains in the K-box, namely K1, K2, and K3, are underlined.
2.2. Phylogenetic Tree Analysis
The alignment of the SnMADS37 protein sequence with the protein sequences of known MADS-box genes from various plant species revealed highly conserved MADS-box and K-box domains (Figure 1). The deduced amino acid sequence of SnMADS37 was 100% identical to both that of LeSEP3 from tomato and that of AtSEP3 from Arabidopsis in the MADS-box DNA-binding domain (Figure 1). The complete protein sequence of SnMADS37 exhibited 98.3% and 63.7% identity with LeSEP3 from tomato and AtSEP3 from Arabidopsis, respectively. Additionally, phylogenetic tree analysis demonstrated a homologous evolutionary relationship between SnMADS37 and SEP3 proteins from other plants. SnMADS37 was closely related to LeSEP3 from tomato and PhFBP2 from petunia within the same Solanaceae family, while SnMADS3 exhibited a highly conserved MADS-box domain and full sequence similarity with both SnMADS37 and AtSEP3 (Figure S1); phylogenetic analysis places it within the SEP1 subfamily (Figure 2A). Therefore, sequence analysis suggests that SnMADS37 is an SEP3-like MADS-box transcription factor in S. nigrum.
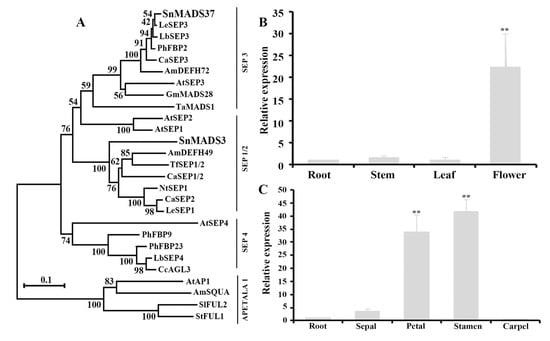
Figure 2.
Phylogenetic analysis and quantitative RT-PCR analysis of SnMADS37. (A) SnMADS37 identified in this study with selected SEP-like genes. The tree was constructed using the neighbor-joining method using MEGA 11 software and the bootstrap values for 1000 replicates. (B) Relative expression analysis of SnMADS37 in the roots, leaves, stem, and early small inflorescence (Inflo); (C) Relative expression analysis of SnMADS37 in the sepal, petal, stamen, and carpel of earlier stage 1 in supplemental Figure S2. Asterisks indicate statistically significant differences (n = 3; ** p < 0.01).
2.3. Expression Analysis of SnMADS37
To investigate the expression of SnMADS37 in WT S. nigrum plants, quantitative RT-PCR analysis was performed using total RNA isolated from roots, mature leaves, stems, and early small inflorescences (Figure 2B). A relatively strong expression of SnMADS37 was observed in the inflorescence; in contrast, its expression greatly reduced in the stems and leaves (Figure 2B). Additionally, strong expression of SnMADS37 was detected in the petals and stamens; however, its expression in the sepals and carpels of early stage 1 floral buds was remarkably decreased (Figure 2C and Figure S1). These results suggest that SnMADS37 is likely involved in the development of petals and stamens in S. nigrum.
Further, to investigate the relationship between green color and SnMADS37 expression during petal development, we performed qRT–PCR analysis at four stages of flower development (Figure 3A): stage 1, the earliest visible floral buds with small green petals; stage 2, elongated petals with an unopened flower; stage 3, initiation of flower opening; and stage 4, fully opened flower. The relative expressions of the B-function genes SnGlobosa (SnGLO) and SnDeficiens (SnDEF) exhibited similar patterns across all stages (Figure S2). However, strong expression of SnMADS37 was detected in stage 1, while its relative expression gradually decreased in stage 2. In stages 3 and 4, the expression of SnMADS37 was significantly suppressed (Figure 3B). We further investigated the chlorophyll content in the four developmental stages (Figure 3C) and identified about 0.23 mg∙g−1 chlorophyll in stage 1 petals. During the other three stages, the chlorophyll content gradually decreased, becoming 0.098, 0.045, and 0.051 mg∙g−1, respectively. In addition, the expression of SnMADS37 matched the chlorophyll decrease during four stages. These results suggest that SnMADS37 may be involved in the chlorophyll metabolism during petal development.
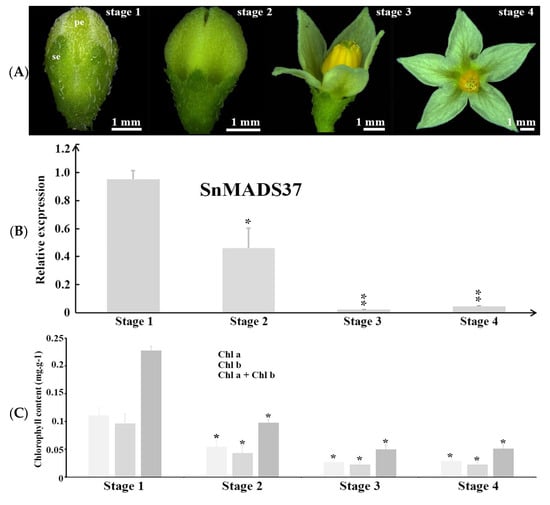
Figure 3.
Quantitative RT-PCR analysis of SnMADS37 in four developmental stages of wild type (WT) flowers. (A) Comparison of the shapes and chlorophyll colorations of the petals among four flower developmental stages. se: sepal; pe: petal. (B) Analysis of the relative expression of SnMADS37 in four flower developmental stages. (C) Chlorophyll content comparison among four flower developmental stages. Asterisks indicate statistically significant differences (n = 3; * p < 0.05; ** p < 0.01).
2.4. Overexpression of SnMADS37 Leads to Morphological Changes in the Flower
To investigate the function of SnMADS37, it was ectopically expressed in S. nigrum, driven by the CaMV 35S promoter. Twelve overexpressing transgenic plants were selected following antibiotic selection (kanamycin) and PCR analysis. Among these, four independent transgenic lines with clear green petals were used for further examination. The transgenic plants exhibited complex phenotypic alterations in their flowers (Figure 4). The inflorescences and collations of early floral buds in most transgenic plants did not display visible differences compared to those of WT plants (Figure 4A–C). However, some flowers in the OE37-10 line developed into secondary inflorescences (Figure 4D). The sepals of the transgenic flowers closely resembled those of the WT flowers, with no obvious alterations in their number or shape observed (Figure 4A–D). Nevertheless, the fully opened petals exhibited greenish tips and vents (Figure 4B–D,F–H). Flowers with six to seven petals and stamens were present (Figure 4F,G,J,K), and additional petal-like tissues also developed on the stamens of the transformed flowers (Figure 4F–H,J–L). Furthermore, some fruits from the transgenic lines exhibited extra shoot like organ formation (Figure 4O,P), which was comparable to that of WT fruits (Figure 4N). Based on these results, SnMADS37 is implicated in the regulation of flower organ identity and development, particularly in petal development.

Figure 4.
Flower phenotype comparison between WT and transformed S. nigrum plants grown in a climate chamber (25 °C, 16 h light condition). (A) Inflorescence of WT; (B) inflorescence of OE37-5; (C) inflorescence of OE37-8; (D) inflorescence of OE37-10; (E) flower of WT, showing five petals and stamens; (F) flower, showing seven petals and stamens; (G) flower, showing six petals and stamens; (H) flower, showing notable greenish petals; (I) five stamens of WT plant from panel (E); (J) seven stamens of the case in panel (F); (K) six stamens of the case in panel (G); (L) five stamens of the case in panel (H); (M) magnified petaloid anther of the case in panel (F); (N) WT fruit; (O) Extra organ developing transformed fruits of OE37-8; (P) Extra organ developing transformed fruits of OE37-10. The increased petal-like tissue in panels (F–H,J,K), and (L) formed an additional organ on the fruit in panels (O,P), which is indicated by the white arrow (se: sepal; pe: petal; st: stamen; fil: filament; an: anther; fr: fruit; plt: petal-like tissue, and es: extra shoot like organ).
2.5. SEM and Sliced Section Analysis
Similar to the WT petal, most of the region of the transformed petal appeared whitish; however, the tip and vein regions exhibited a greenish hue (Figure 4A–D and Figure 5A,B). To determine how SnMADS37 influences petal-tip coloration and structure, we chose the remarkable OE37-10 line and a WT plant was investigated using SEM and sliced sections (Figure 5A,B). We observed distinct conical epidermal cells and trichomes on the adaxial surface of the petal-tip in the OE37-10 line, which resembled those of the WT petal-tip (Figure 5C,D). Furthermore, the cross-section of the petal-tip section in the OE37-10 line was thicker, with a noticeable accumulation of chlorophyll observed on the inner side of the petal (Figure 5E,F), in contrast to the thinner, whitish WT petal that lacked chlorophyll accumulation (Figure 5C,E).

Figure 5.
Petal-tip comparison between the WT and OE37-10 flower. (A) WT flower; (B) OE37-10 flower; (C) SEM image of adaxial surface of petal-tip in panel (A); (D) SEM image of adaxial surface of petal-tip in panel (B); (E) transection of petal-tip in panel (A); (F) transection of petal-tip in panel (B) (Ad: adaxial of petal; Ab: abaxial of petal; Tri: trichome; Cc: conical cell).
2.6. Expression of SnMADS37 Affects Chlorophyll Content in Flower
To determine whether the expression of SnMADS37 affects chlorophyll content in flowers, we investigated the total chlorophyll content in the petals of the WT and four transformed plants using a spectrophotometer. Chlorophyll was detected in the petals of all transformed plants, with its content being approximately 1.5- to 4.4-fold higher than that in the WT petals. This chlorophyll content and coloration corresponded with the expression of SnMADS37 in WT and transgenic petals (Figure 6A,B). However, no significant differences in the expression of B-function genes SnGLO and SnDEF were detected in the petal of WT and transgenic lines (Figure 6C,D). These results indicate that the expression of SnMADS37 likely affects chlorophyll metabolism, resulting in chlorophyll accumulation in the transformed petals.
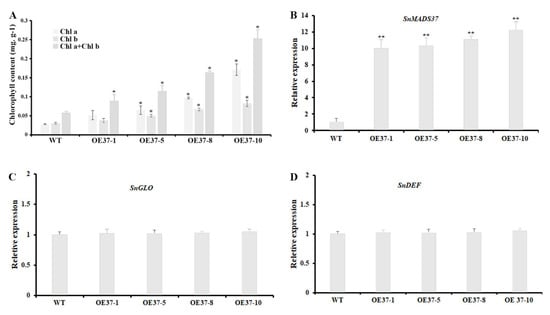
Figure 6.
Comparison of chlorophyll content and relative expression analysis of B-function genes in fully opened stage 4 petals among a WT and four transgenic S. nigrum plants. (A) Analysis of the total chlorophyll content from petals of a WT and four transgenic plants; (B) SnMADS37 expression analysis from petals of a WT and four transgenic plants; (C) B-function gene SnGLO expression analysis from petals of a WT and four transgenic plants; (D) B-function gene SnDEF expression analysis from petals of a WT and four transgenic plants. OE37-1, 5, 8, and 10: four transformed plants with pBI-35S::SnMADS37. Asterisks indicate statistically significant differences between the WT and transgenic lines (n = 3; * p < 0.05; ** p < 0.01). Chl a: chlorophyll a; Chl b: chlorophyll b.
To confirm our hypothesis, we employed the virus-induced gene silencing (VIGS) method to investigate the potential role of SnMADS37 in flower development. A 260-bp fragment from the 3′ end of SnMADS37 was used to construct the VIGS vector pTRV2::SnMADS37, with the empty pTRV2 vector as a control. These constructs were infiltrated into the apical tips of 5-week-old S. nigrum plants. Subsequently, early altered floral buds were observed in 4 out of 12 treated plants after 2 weeks of culture in a growth chamber at 25 °C under 16/8 h of light/dark conditions (Figure 7). We found that the control flower bud treated with GV3101/TRV2 showed green sepals and closed greenish petals, which were similar to wild type flower buds (Figure 7A and Figure S1). Compared to the GV3101/TRV2 control flower bud, some flower buds treated with GV3101/TRV2::mads37 displayed clear morphological changes, such as shrunk and reduced green color petals, and wide and pale green sepals and some floral buds displayed distinct stigma; we also observed that the relative expression of SnMADS37 was greatly downregulated in the Snmads37-silenced floral buds compared to the control floral buds (Figure 7B,C). Furthermore, we also found likely reduced relative expression of a chlorophyll-related biosynthetic gene SnCHLH (Mg-chelatase subunit H gene) and two chlorophyll-related degradation genes SnCLH (hydroxymethyl chlorophyll a reductase) and SnPPH (pheophytinase) by qRT-PCR analysis (Figure S3). These results suggest that SnMADS37 is not only involved in the regulation of petal expansion but also negatively affects chlorophyll degradation during the flower development; the green petal-tip flower may be caused by SnMADS37 expression.
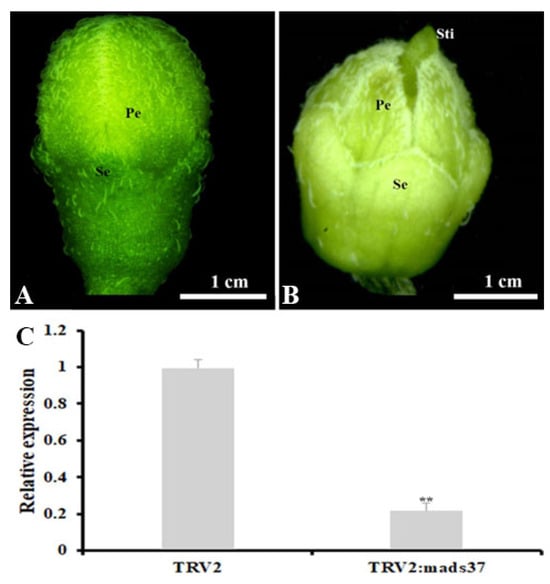
Figure 7.
Comparison of phenotype between a control and an SnMADS37-silenced stage 1 floral bud of S. nigrum by VIGS treatment. (A) a control stage 1 floral bud treated with GV3101/TRV2; (B) an Snmads37-silenced stage 1 floral bud treated with GV3101/TRV2::Snmads37; (C) total RNA was extracted from stage 1 floral bud in (A) and (B), respectively, and relative expression of SnMADS37 was detected by qRT-PCR analysis. Asterisks indicate statistically significant differences (n = 3; ** p < 0.01). Se, sepal; Pe. petal; Sti, stigma.
3. Discussion
Flower morphogenesis is a highly robust and standardized developmental process. In the past three decades, significant transcriptional regulators that govern floral development have been identified in model organisms such as Antirrhinum and Arabidopsis [9,12]. The “floral quartet model” posits that floral homeotic proteins form organ-specific tetrameric complexes, with their interactions being facilitated by functionally redundant SEP proteins (SEP1–SEP4) in Arabidopsis. Within this model, a quartet complex consisting of APETALA1, APETALA3, PISTILLATA, and SEP is responsible for specifying petal identity. Additionally, previous research has also demonstrated that SEP3 family genes are involved in petal development regulation in the tomato and petunia of Solanaceae [18,28]. In this study, we isolated and characterized the MADS-box gene SnMADS37 from S. nigrum. Phylogenetic analysis of SnMADS37 within a clade with LeSEP3 from tomato [28], PhFBP2 from petunia [18], and AtSEP3 from Arabidopsis [13] (Figure 2A) was conducted. Moreover, the deduced amino acid sequence of SnMADS37 contains the highly conserved MADS-box and K-box domains, similar to LeSEP3 of the tomato (Figure 1). These structural similarities in the amino acid sequence and phylogenetic analysis suggest that SnMADS37 may function as an SEP3-like MADS-box gene in S. nigrum.
Members of the MADS-box transcription factor play an essential role in regulating floral organ identity, which is specified by the combined protein interactions of ABCE-class MADS-box domain transcription factors. The E-class SEP transcription factors are particularly significant in the regulation of floral organ identity and development across various plants [9,12,17,29]. In this study, the overexpression of the SEP3-like gene SnMADS37 resulted in various floral developmental alterations in transgenic plants, such as increased numbers of flowers (Figure 4C,D), the development of flowers with 6 to 7 petals and stamens (Figure 4F–H), and the occurrence of ectopic petal formation on staminal anthers. Furthermore, we observed the emergence of additional organs on early green fruits, while certain flowers appeared to revert to secondary inflorescences in transgenic S. nigrum (see Figure 4D). These findings imply that SnMADS37 is critical for floral organ specification and petal development in S. nigrum. Similar functions of SEP3-like genes have been identified and characterized in transformed tobacco. For example, overexpression of the soybean SEP3-like gene GmMADS28 in tobacco induces early flowering and alters the floral organs’ morphology, including stamens, sepals, and petals [30]. Similarly, PlacSEP3 overexpression in transformed tobacco promotes early flowering and produces lateral branches [31]. In Arabidopsis, overexpression of the wheat SEP3-like gene TaMADS1 leads to early flowering, converts the sepal into a leaf-like structure, and reduces petal and stamen numbers [26]. Moreover, loss of function of FBP2 in petunias led to secondary inflorescence development in the third whorl [32,33], TM5 downregulation in the tomato resulted in a loss of determinacy in the center of the flower [34], and overexpression of JcSEP3 in Jatropha curcas induced male floral organ formation despite broad expression across floral tissues [35]. These results suggest that SnMADS37, probably an SEP-like gene, plays multifaceted roles in floral development, with phenotypic outcomes likely modulated by expression levels. However, since S. nigrum may harbor additional undetected SEP-like genes with redundant functions, which specific contributions it makes to floral organ identity and development requires further investigation.
In S. nigrum, the petals of closed flower buds are green (Figure 3A). Once the flower opens, the upper part of the petal expands, and chlorophyll degrades, resulting in whitish petals (Figure 3A). In this study, the petals of transformed S. nigrum with the 35S::SnMADS37 showed clear chlorophyll accumulation, displaying greenish tips and veins that were comparable to those of WT petals (Figure 5). The morphological changes are similar to the loss of function seen in the SEP3 ortholog FBP2 mutant in petunias [18]. However, while the petals of the fbp2 mutant exhibit leaf or sepal-like characteristics, such as stomata in the greenish segments of petals, the petals of transformed SnMADS37 showed conical epidermal cells on the adaxial side and lacked stomatal structures. This is indicative of petal identity in angiosperms [36], and the epidermal cell structure closely resembles that of WT petals (Figure 5A,B). Therefore, the chlorophyll accumulation observed in the petals of transformed S. nigrum is distinct from that in leaf-like flowers, resulting from loss-of-function mutations in SEP-like genes, such as fbp2 in petunias [18] and the sep1/sep2/sep3 triple mutant in Arabidopsis [19].
In addition, we observed that floral buds with reduced SnMADS37 expression exhibited wider and pale green sepals, along with shriveled petals showing decreased green pigmentation, and the floral buds displayed a more prominent stigma compared to both the control pTRV2-treated and overexpression flower buds (Figure 5B–D and Figure 7), implying that SnMADS37 is directly or indirectly involved in chlorophyll degradation during petal development. On the other hand, we focused solely on SnMADS37; however, other undetected SEP-like genes, such as SnMADS3 (Figure 2A), may have redundant functions in regulating floral organ identity in S. nigrum. Thus, the role of SnMADS37 in floral organ identity and petal maturation warrants further investigation through transcriptomic and metabolomic analyses, genome editing, and in situ techniques.
Previous studies have reported that the MADS-box gene SOC1 is involved in regulating morphological development and contributes to chloroplast biogenesis and biosynthesis, resulting in green petal formation in transformed tobacco [37]. Furthermore, the overexpression of BpMADS from Betula platyphylla significantly enhances chloroplast division, and the rate of photosynthesis in transformed tobacco [38]. In this study, the B-function genes SnGLO and SnDEF were likely not involved in the accumulation of chlorophyll during petal development (Figure 3 and Figure 6C,D). Conversely, chlorophyll accumulation correlates with the expression of SnMADS37 during the petal development stages of both WT and transformed petals (Figure 3 and Figure 7A,B). We also found reduced expression of certain SnCLH and SnPPH genes, which are involved in chlorophyll degradation in the petals of SnMADS37 overexpressed lines (Figure S3). Additionally, SnMADS37 alone could not stimulate chlorophyll accumulation in the roots of transformed lines (Figure S4). Moreover, compared to WT and control floral buds, the silenced Snmads37 floral petals reduced green coloration (Figure 7 and Figure S1). These findings suggest that SnMADS37 negatively influences chlorophyll degradation during the petal development process. Consequently, the reduced rate of chlorophyll degradation associated with the expression of SnMADS37 likely contributes to the accumulation of chlorophyll in the green petal-tips of transformed S. nigrum.
In this study, we observed that the overexpression of SnMADS37 affected floral organ development and influenced chlorophyll accumulation in the petals. This finding indicates that SnMADS37 plays a crucial role in floral organ identity and the petal development process in S. nigrum. Clearly, elucidating the role of the SnMADS37 gene will not only enhance our understanding of the biological functions of SEF family genes but also provide novel insights into the significance of SnMADS37 in regulating petals and other floral organ developments.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
Solanum nigrum plants were cultivated on solid Murashige and Skoog (MS) medium [39] at 25 °C under a 16/8 h of light/dark conditions in a tissue culture room at Zhejiang A&F University. Fully expanded young leaves were used for transformation and other experiments.
4.2. SnMADS37 Gene Cloning
Total RNA was isolated from 100 mg of floral bud powder of S. nigrum using the Eastep® Super Total RNA Extraction Kit (Promega, Madison, WI, USA). First-strand complementary DNA (cDNA) was synthesized from 5 µg of total RNA using the TransScript® II First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Cloning of full-length SnMADS37 was performed using the TransStart® FastPfu DNA Polymerase (TransGen Biotech) and a set of gene-specific primers SnMADS37-F2/SnMADS37-R2 (Table S1), which were designed based on MADS-box-like transcription factor-derived transcriptome data of S. nigrum. Polymerase chain reaction (PCR) was performed under the following conditions: preliminary denaturation at 97 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 50 s, annealing at 60 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The amplified fragment was cloned using the pEASY-Blunt simple vector (TransGen Biotech), dubbed pEASY-MADS37, and confirmed via sequencing. The amplified 729-bp full-length cDNA was dubbed SnMADS37 (DDBJ: LC782345).
4.3. Phylogenetic Tree Construction
The amino acid sequence of SnMADS37 was aligned with certain SEPALLATA-like MADS-box gene sequences. Neighbor-joining phylogenetic trees with 1000 bootstrap replicates were constructed using Clustal W version 2.0 [40] and MEGA version 11 [41]. The bars indicate an evolutionary distance of 0.1%. The GenBank accession numbers of these proteins are as follows: Arabidopsis thaliana AtAP1 (NP_177074), AtSEP1 AK118608), AtSEP2 (AY727601), AtSEP3 (AF015552), and AtSEP4 (NM126418); Antirrhinum majus AmSQUA (CAA45228), AmDEFH49 (ACAA64741), and AmDEFH72 (CAA64742); Solanum lycopersicum SlFUL2 (ART88618), LeSEP1 (NP_001233911), and LeSEP3 (NP_001234384); Petunia hybrida PhFBP2 (M91666), PhFBP9 (AAK21249), and PhFBP23 (AF335241); Solanum tuberosum StFUL1 (NP_001275142); Coffea arabica CaSEP1/2 (AHW58036) and CaSEP3 (AHW58034); Lycium barbarum LbSEPL3 (ADP09004); Torenia fournieri TfSEP1/2 (BAO74162); Nicotiana tabacum NtSEPL 1 (XP_016466966); Capsicum annuum CaSEP2 (KAF3682829); Capsicum chinense CcAGL3 (PHU23937); Lycium barbarum LbSEP4 (AJW87597); Glycin max GmMADS28 (AJ878424); and Triticum aestivum TaMADS1 (AF543316).
4.4. Transcriptional Analysis of SnMADS37
Total RNA was isolated from the roots, leaves, stems, and inflorescences as well as the sepals, petals, stamens, and carpels of wild type S. nigrum (Figure S1) using an Eastep® Super Total RNA Extraction Kit (Promega, Madison, WI, USA). Reverse transcription from 2 µg of total RNA to cDNA was performed using EasyScript® First-Strand cDNA Synthesis Super Mix (TransGen Biotech) according to the manufacturer’s instructions. Each reaction volume was 10 μL, and PCRs were conducted as follows: 95 °C for 30 s, followed by 30 cycles of 5 s at 95 °C and 30 s at 60 °C. The relative expression levels of the target genes were calculated using the 2−ΔΔCt method [42]. SnActin (Table S1) was used as an internal control to normalize the transcription levels of the target genes.
4.5. Construction of the Overexpression Vector pBI-35S::SnMADS37
The full-length SnMADS37 cDNA coding region was amplified from the pEASY-MADS37 plasmid using a BamH I-SnMADS37F and Sac I-SnMADS37R gene-specific primer set (Table S1) in combination with TransStart® FastPfu DNA Polymerase (TransGen Biotech). The resultant was sub-cloned into a pEASY-Blunt vector for confirmation via sequencing. Using BamH I and Sac I restriction sites, SnMADS37 substituted the GUS gene in the pBI121 vector to produce pBI-35S::SnMADS37, containing the full-length SnMADS37 coding region between the 35S promoter and CaMV terminator. The pBI-35S::SnMADS37 expression vector was electroporated into Agrobacterium GV3101 cells [43].
4.6. Transformation of S. nigrum
Young leaves of S. nigrum were used for transformation. Transformation was performed according to the method of Piao et al. [44], albeit slightly modified. The Agrobacterium strain GV3101/pBI-35S::SnMADS37 was grown in 5 mL of liquid LB medium containing 50 mg∙L−1 kanamycin at 28 °C, shaken at 200 rpm for 24 h, and subsequently 30-fold diluted with liquid MS medium for inoculation. After four weeks of infection, the obtained adventitious shoots were transferred to a fresh solid MS medium containing 250 mg∙L−1 cefotaxime and 50 mg∙L−1 kanamycin, and transformed positive shoots were selected for PCR analysis. The selected transformed plants were maintained at 25 °C under a 16/8 h light/dark photoperiod condition in an artificial climate chamber and used in the subsequent experiments.
4.7. SnMADS37 Expression Analysis in the Transgenic S. nigrum
Total RNA was extracted from 100 mg of fresh young wild type (WT) leaves and five independent T1 transgenic S. nigrum plants, and first-strand cDNA synthesis was performed as described above. Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis was performed using 20 ng first cDNA as a template and the set of gene-specific primers, namely SnMADS37-F2/SnMADS37-R2, and Actin gene of S. nigrum as a positive control (Table S1).
4.8. VIGS-Mediated Silencing of SnMADS37 in Solanum nigrum Flowers
To construct the SnMADS37 VIGS vector, we designed a primer set targeting a 260-bp fragment at the 3′ end of the gene, a region exhibiting low sequence identity with the SnMADS3 gene. Using pTRV2 [45] as the backbone, we generated the VIGS vector pTRV2::SnMADS37. Both this construct and the helper vector pTRV1 were introduced into A. tumefaciens strain GV3101 via electroporation.
The silencing of SnMADS37 in the flower of S. nigrum was performed with a slightly modified procedure described by Hartl et al. [46]. The A. strains GV3101/TRV1, GV3101/TRV2, and GV3101/TRV2::SnMADS37 were cultured in 100 mL LB medium supplemented with 50 mg·L−1 kanamycin and 25 mg·L−1 Rifampicin at 28 °C with shaking 200 rpm for 24 h. Cells were then harvested by centrifugation at 5000 rpm for 10 min, and the pellet was resuspended in 100 mL infiltration medium (1% sucrose, 10 mM MgCl2, 10 mM MES, 200 µM acetosyringone). After 3 h of incubation at 28 °C and 200 rpm, the GV3101/TRV1 suspension was mixed in equal volumes with either GV3101/TRV2::SnMADS37 or GV3101/TRV2 (control). The mixtures were diluted with infiltration medium to an OD600 of 0.3 for infiltration. For each treatment, six 5-week-old plants were used. The apical meristems were vacuum-infiltrated for a pressure of 60–70 mbar and 1 min slowly. The vacuum infiltration experiment was performed twice, and phenotypic observations were performed 10 days post-infiltration.
4.9. Scanning Electron Microscopy (SEM) Observations
Petal tissue samples of both wild type and T1 transgenic SnMADS37-10 were prepared for SEM as previously described by Krizek [47]. SEM analyses were performed using a TM4000 instrument (Hitachi, Chiyoda City, Japan).
4.10. Slice Analysis of Petals
Fresh WT and T1 transgenic SnMADS37-10 petal-tips were sliced into 40 μm thick sections using a Leica VT1000 slicer. The sections were covered with a cover glass for conducting microscopic observations. Images were captured using a Leica light microscope DM2500. Digital image processing was performed using Adobe Photoshop 7.0.
4.11. Measurement of Chlorophyll Pigment Content
The chlorophyll a and b contents were determined using the method of Zhou et al. [48], albeit slightly modified. Briefly, 50 mg of excised leaf disks or fully opened petals of wild type or T1 transgenic S. nigrum were fully submerged in 10 mL acetone:ethanol (2:1, v/v) buffered at 4 °C, and were left in the dark overnight to extract the chlorophyll pigments. The absorbances at 663 and 645 nm were measured for the chlorophyll extract, and the chlorophyll a and b levels were calculated following Zhou et al. [47]. Each sample was replicated three times, and statistical analyses were performed using SPSS 25.0.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants14131891/s1, Table S1: the primers used for SnMADS37 gene cloning, detection of transformed plants, and RT-PCR analysis in this study; Figure S1: the green color of early wild type stage 1 floral bud (A), and with stamens (B) that removed the sepals and petals in Figure 4A, Figure S2: expression analysis of B-function gene in petal development stage of WT, Figure S3: relative expression analysis of chlorophyll-related genes in fully opened stage 4 petals among a WT and four transgenic S. nigrum plants, Figure S4: comparison of root coloration between a WT and five overexpressed SnMASD37 plants on MS medium + 200 mg/L cefotaxime after three weeks culture.
Author Contributions
S.Y.: Data curation, Formal analysis, Investigation, Writing—original draft. C.-L.P.: Methodology, Investigation, Resources. X.Z.: Investigation. M.-L.C.: Conceptualization, Writing—original draft, and Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Scientific Research Foundation of Zhejiang A&F University (2018FR004) to MLC.
Data Availability Statement
All data generated or analyzed during this study are included in the article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Edmonds, J.; Chweya, J.A. Black nightshades. Solanum nigrum L. and related species. In Promoting the Conservation and Use of Underutilized and Neglected Crops, 15th ed.; Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute: Rome, Italy, 1997; ISBN 92-9043-321-3. [Google Scholar]
- Lehmann, C.; Biela, C.; Töpfl, S.; Jansen, G.; Vögel, R. Solanum scabrum a potential source of a coloring plant extract. Euphytica 2007, 158, 189–199. [Google Scholar] [CrossRef]
- Poczai, P.; Hyvönen, J. On the origin of Solanum nigrum, can networks help? Mol. Biol. Rep. 2011, 38, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Jabamalairaj, A.; Priatama, R.A.; Heo, J.; Park, S.J. Medicinal metabolites with common biosynthetic pathways in Solanum nigrum. Plant Biotech. Rep. 2019, 13, 315–327. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Lönnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Defciens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus, the protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Malone, K.M.; Timmis, J.N.; Orford, S.J. Evidence for alternative splicing of MADS-box transcripts in developing cotton fibre cells. Mol. Gent. Genom. 2008, 279, 75–85. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Garceau, D.C.; Batson, M.K.; Pan, I.L. Variations on theme in fruit development the PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta 2017, 246, 313–321. [Google Scholar] [CrossRef]
- Martin-Pizarro, C.; Trivino, J.C.; Pose, D. Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 2019, 70, 885–895. [Google Scholar] [CrossRef]
- Theißen, G.; Saedler, H. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Krizek, B.A.; Fletcher, J.C. Molecular mechanisms of flower development, an armchair guide. Nat. Rev. Genet. 2005, 6, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, M.; Zethof, J.; Souer, E.; Koes, R.; Tornielli, G.B.; Pezzotti, M.; Ferrario, S.; Angenent, G.C.; Gerats, T. Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 2003, 15, 2680–2693. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef]
- Matsubara, K.; Shimamura, K.; Kodama, H.; Kokubun, H.; Watanabe, H.; Basualdo, I.L.; Ando, T. Green corolla segments in a wild Petunia species caused by a mutation in FBP2, a SEPALLATA-like MADS box gene. Planta 2008, 228, 401–409. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Kotilainen, M.; Elomaa, P.; Uimari, A.; Albert, V.A.; Yu, D.; Teeri, T.H. GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 2000, 12, 1893–1902. [Google Scholar] [CrossRef]
- Uimari, A.; Kotilainen, M.; Elomaa, P.; Yu, D.; Albert, V.A.; Teeri, T.H. Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Pro. Natl. Acad. Sci. USA 2004, 101, 15817–15822. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Juntheikki, I.; Mouhu, K.; Broholm, S.K.; Rijpkema, A.S.; Kins, L.; Lan, T.; Albert, V.A.; Teeri, T.H.; et al. Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera Hybrida. New Phytol. 2017, 216, 939–954. [Google Scholar] [CrossRef]
- Castillejo, C.; Romera-Branchat, M.; Pelaz, S. A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 2005, 43, 586–596. [Google Scholar] [CrossRef]
- Adal, A.M.; Binson, E.; Remedios, L.; Mahmoud, S.S. Expression of lavender AGAMOUS-like and SEPALLATA3-like genes promote early flowering and alter leaf morphology in Arabidopsis thaliana. Planta 2021, 254, 54. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Cheng, Z.J.; Zhang, X.S. Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 2006, 223, 698–707. [Google Scholar] [CrossRef]
- Lemmetyinen, J.; Hassinen, M.; Elo, A.; Porali, I.; Sopanen, T. Functional characterization of SEPALLATA3 and AGAMOUS orthologues in silver birch. Physiol. Plant. 2004, 121, 149–162. [Google Scholar] [CrossRef]
- Slugina, M.A.; Dyachenko, E.A.; Kochieva, E.Z.; Shchennikova, A.V. Structural and functional diversification of SEPALLATA genes TM5 and RIN in tomato species (Section Lycopersicon). Dokl. Biochem.Biophys. 2020, 492, 152–158. [Google Scholar] [CrossRef]
- Ruelens, P.; Zhang, Z.; van Mourik, H.; Maere, S.; Kaufmann, K.; Geuten, K. The origin of floral organ identity quartets. Plant Cell 2017, 29, 229–242. [Google Scholar] [CrossRef]
- Huang, F.; Xu, G.; Chi, Y.; Liu, H.; Xue, Q.; Zhao, T.; Gai, J.; Yu, D. A soybean MADS-box protein modulates floral organ numbers, petal identity and sterility. BMC Plant Biol. 2014, 14, 89. [Google Scholar] [CrossRef]
- Zhang, S.; Lu, S.; Yi, S.; Han, H.; Liu, L.; Zhang, J.; Bao, M.; Liu, G. Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia. Planta 2017, 245, 439–457. [Google Scholar]
- Angenent, G.C.; Franken, J.; Busscher, M.; Weiss, D.; Van Tunen, A.J. Co-suppression of the Petunia homeotic gene Fbp2 affects the identity of the generative meristem. Plant J. 1994, 5, 33–44. [Google Scholar] [CrossRef]
- Ferrario, S.; Immink, R.G.H.; Shchennikova, A.; Busscher-Lange, J.; Angenent, G.C. The MADS-box gene FBP2 is required for SEPALLATA function in Petunia. Plant Cell 2003, 15, 914–925. [Google Scholar] [CrossRef]
- Pnueli, L.; Hareven, D.; Broday, L.; Hurwitz, C.; Lifschitz, E. The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 1994, 6, 175–186. [Google Scholar] [CrossRef]
- Zhao, M.L.; Zhou, Z.F.; Chen, M.S.; Xu, C.J.; Xu, Z.F. An ortholog of the MADS-box gene SEPALLATA3 regulates stamen development in the woody plant Jatropha curcas. Planta 2022, 255, 111. [Google Scholar] [CrossRef]
- Whitney, H.M.; Bennett, K.M.; Dorling, M.; Sandbach, L.; Prince, D.; Chittka, L.; Glover, B.J. Why do so many petals have conical epidermal cells? Annal. Bot. 2011, 108, 609–616. [Google Scholar] [CrossRef]
- Wang, Z.; Shen, Y.; Yang, X.; Pan, Q.; Ma, G.; Bao, M.; Zheng, B.; Duanmu, D.; Lin, R.; Larkin, R.M.; et al. Overexpression of particular MADS-box transcription factors in heat-stressed plants induces chloroplast biogenesis in petals. Plant Cell Environ. 2019, 42, 1545–1560. [Google Scholar] [CrossRef]
- Qu, G.Z.; Zheng, T.; Liu, G.; Wang, W.; Zang, L.; Liu, H.; Yang, C. Overexpression of a MADS-Box Gene from Birch (Betula platyphylla) Promotes Flowering and Enhances Chloroplast Development in Transgenic Tobacco. PLoS ONE 2013, 8, e63398. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962, 15, 81–84. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetic analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.L.; Liu, C.; Piao, C.; Liu, C.L. A stable Agrobacterium rhizogenes-mediated transformation of cotton (Gossypium hirsutum L.) and plant regeneration from transformed hairy root via embryogenesis. Front. Plant Sci. 2020, 11, 604255. [Google Scholar] [CrossRef] [PubMed]
- Piao, C.; Gao, Z.; Yuan, S.; Cui, M.L. The R2R3-MYB gene CgMYB4 is involved in the regulation of cell differentiation and fiber development in the stamens of Chelone glabra L. Protoplasma 2022, 259, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Zhang, C.; Jiang, X.; Kang, M.; Yin, X.; Lü, P.; Zhang, X.; Zheng, Y.; Gao, J. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 2012, 160, 2064–2082. [Google Scholar] [CrossRef]
- Hartl, M.; Merker, H.; Schmidt, D.D.; Baldwin, I.T. Optimized virus-induced gene silencing in Solanum nigrum reveals the defensive function of leucine aminopeptidase against herbivores and the shortcomings of empty vector controls. New Phytol. 2008, 179, 356–365. [Google Scholar] [CrossRef]
- Krizek, B.A. Overexpression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 1999, 25, 224–236. [Google Scholar] [CrossRef]
- Zhou, J.; Han, P.; Pan, Y.; Wu, M.; Zhao, Y.; Jia, Y.; Jiang, B.; Zhang, L.; Xu, Q.; Liu, S.; et al. Effects of cadmium stress on photosynthetic physiology and chlorophyll fluorescence in Solanum nigrum and Solanum americanum. J. Agro-Environ. Sci. 2021, 40, 26–34. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).