Artificial Sweet Cherry miRNA 396 Promotes Early Flowering in Vernalization-Dependent Arabidopsis Edi-0 Ecotype
Abstract
1. Introduction
2. Results
2.1. Determination of Dormancy Stages in ‘Regina’
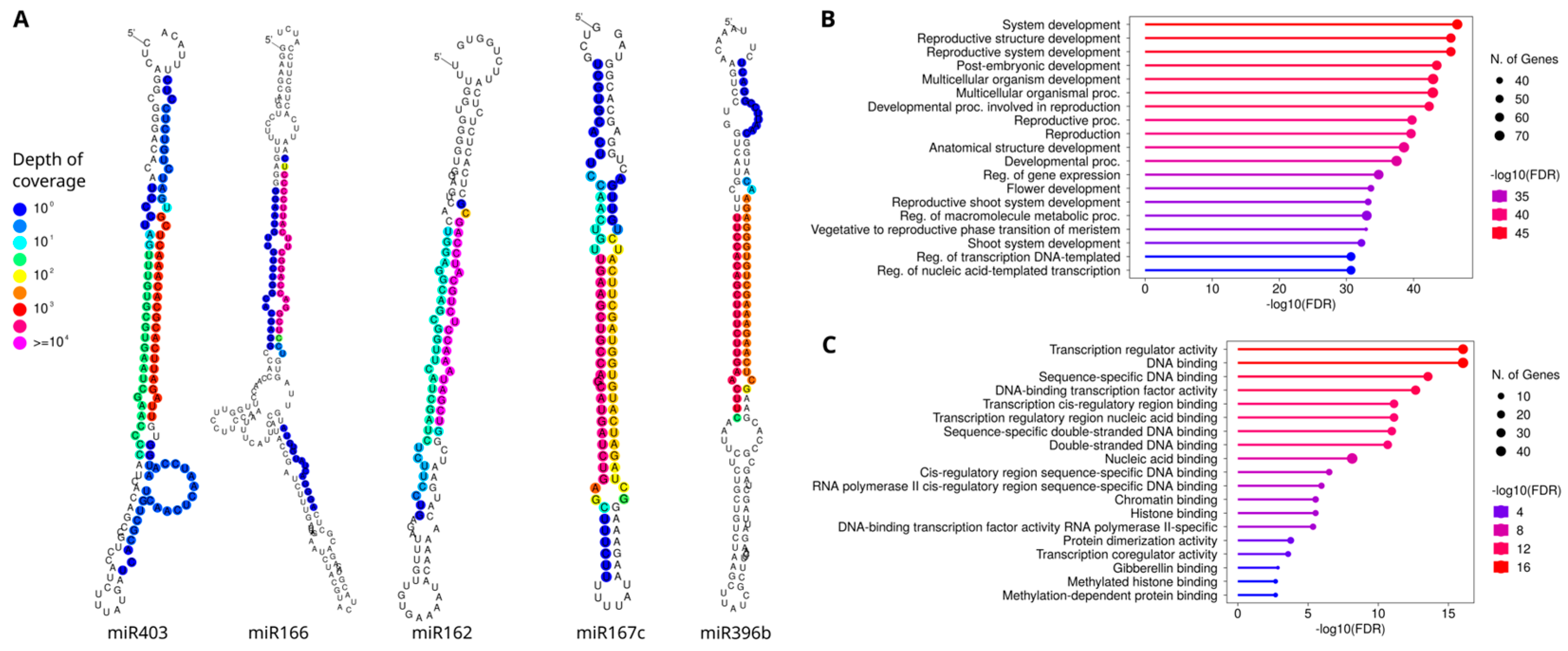
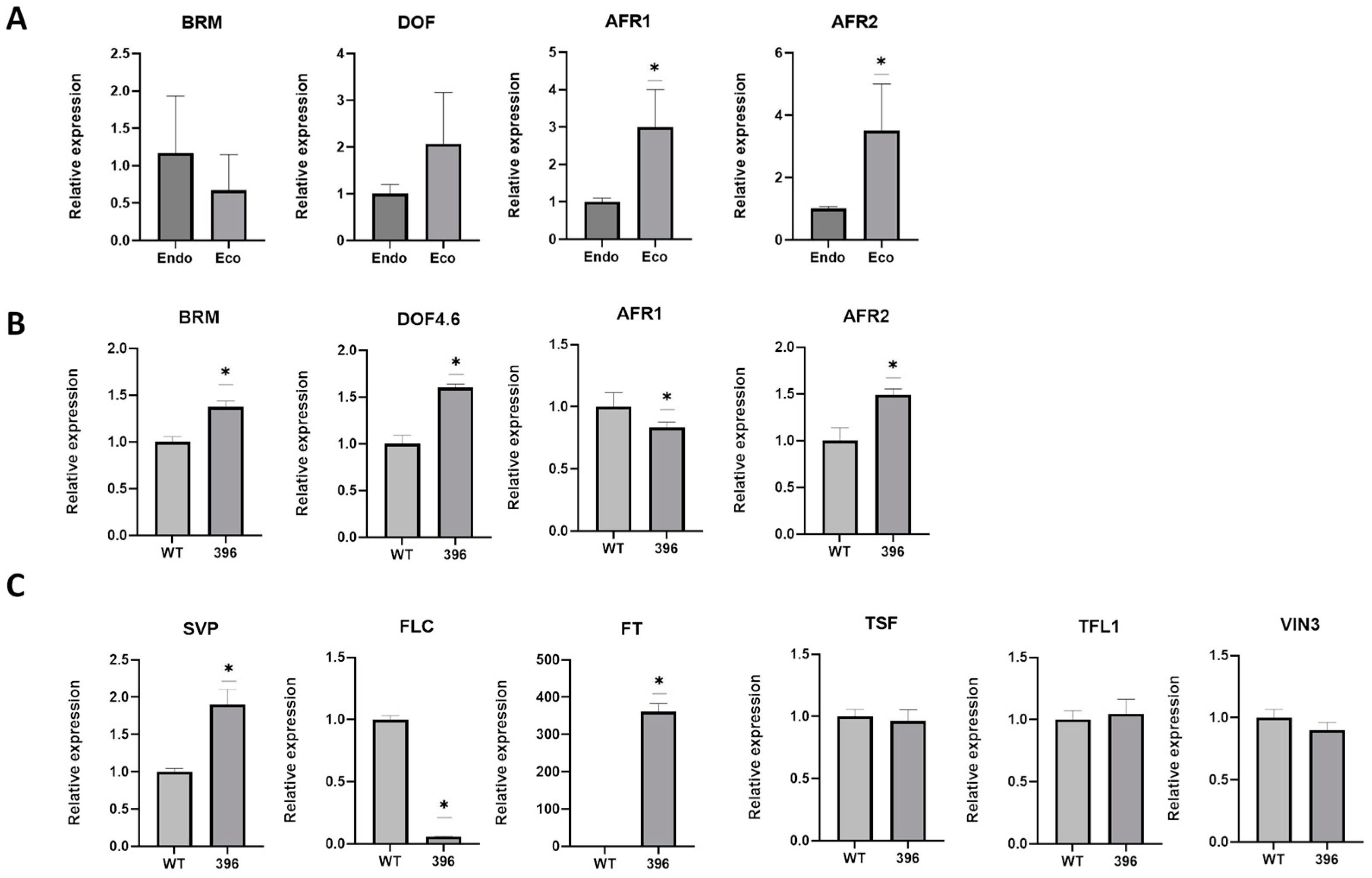
2.2. Insights on the miR396 Activity
3. Discussion
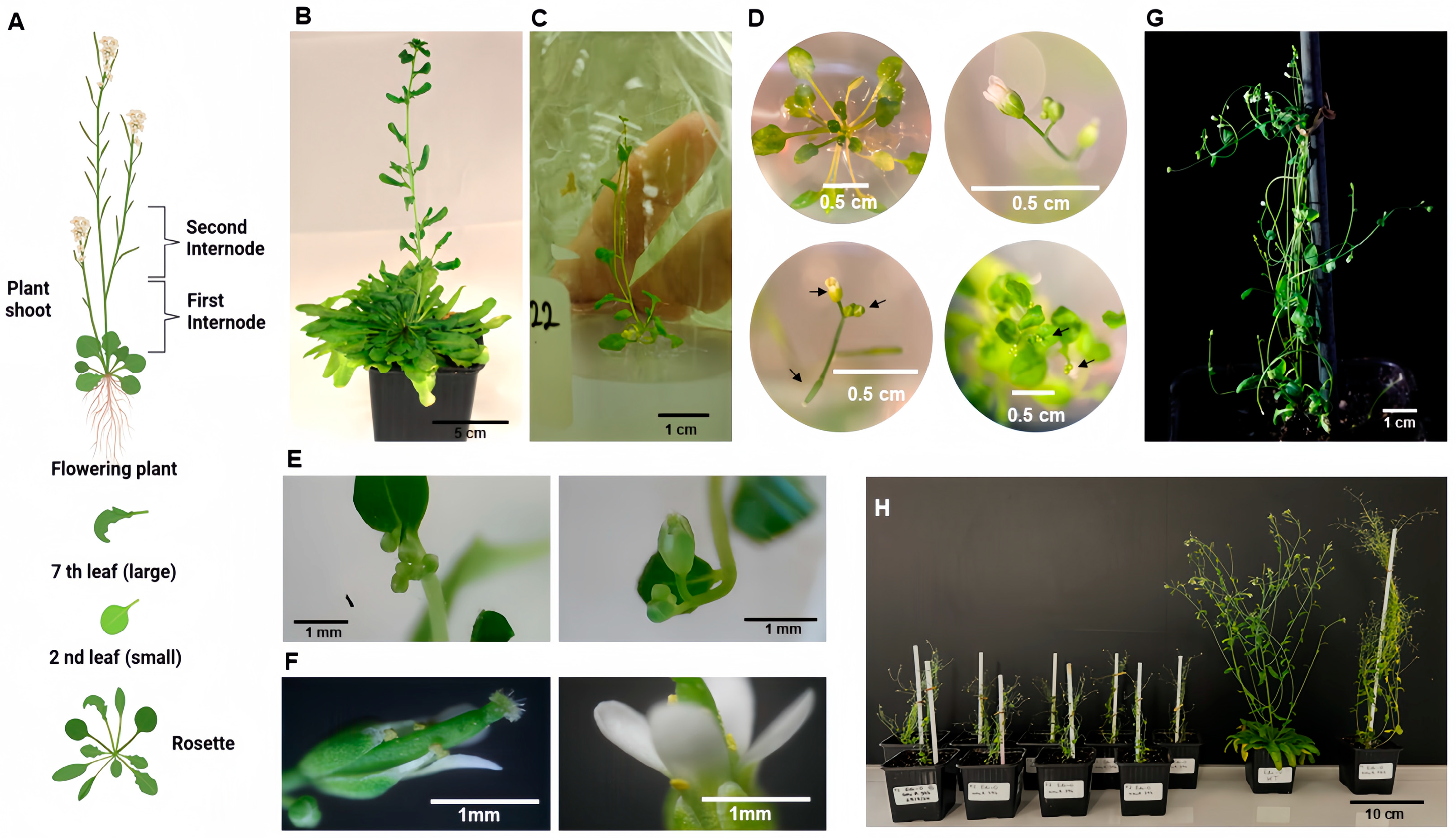
3.1. Ectopic Expression of miR396 in Arabidopsis and Early Flowering
3.2. Other microRNAs
3.3. Final Considerations
4. Materials and Methods
4.1. ‘Regina’ Material, Sampling, and Chilling Requirement Determination Under Forcing Conditions
4.2. ‘Regina’ Total and Small RNA Isolation
4.3. Yield and Quality Analysis of Isolated RNA Fractions
4.4. Library Construction and Sequencing
4.5. Differentially Expressed (DE) Micro RNAs and Target Prediction
4.6. Experimental Determination of Putative Target Genes
4.7. Artificial microRNA (amiRNA) Synthesis
4.8. Expression Vector for Pre-amiRNAs and Agrobacterium Clones
4.9. Genetic Transformation of A. thaliana Ecotype Edi-0
4.10. Genetically Modified Arabidopsis Populations
4.11. Transformed Arabidopsis Total and Small RNA Isolation
4.12. Experimental Determination of microRNAs and Target Genes in Arabidopsis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fadón, E.; Fernandez, E.; Behn, H.; Luedeling, E. A Conceptual Framework for Winter Dormancy in Deciduous Trees. Agronomy 2020, 10, 241. [Google Scholar] [CrossRef]
- Yang, Q.; Gao, Y.; Wu, X.; Moriguchi, T.; Bai, S.; Teng, Y. Bud Endodormancy in Deciduous Fruit Trees: Advances and Prospects. Hortic. Res. 2021, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, Para, and Ecodormancy: Physiological Terminology and Classification for Dormancy Research. HortScience 1987, 22, 701. [Google Scholar] [CrossRef]
- Richardson, E.A.; Seeley, S.D.; Walker, D.R. A Model for Estimating the Completion of Rest for ‘Redhaven’ and ‘Elberta’ Peach Trees1. HortScience 1974, 9, 331–332. [Google Scholar] [CrossRef]
- Erez, A.; Fishman, S.; Gat, Z.; Couvillon, G.A. Evaluation of winter climate for breaking bud rest using the dynamic model. Acta Hortic. 1988, 232, 76–89. [Google Scholar] [CrossRef]
- Fadón, E.; Rodrigo, J.; Luedeling, E. Cultivar-Specific Responses of Sweet Cherry Flowering to Rising Temperatures during Dormancy. Agric. Meteorol. 2021, 307, 108486. [Google Scholar] [CrossRef]
- Conrad, A.O.; Yu, J.; Staton, M.E.; Audergon, J.M.; Roch, G.; Decroocq, V.; Knagge, K.; Chen, H.; Zhebentyayeva, T.; Liu, Z.; et al. Association of the Phenylpropanoid Pathway with Dormancy and Adaptive Trait Variation in Apricot (Prunus armeniaca). Tree Physiol. 2019, 39, 1136–1148. [Google Scholar] [CrossRef]
- Tixier, A.; Gambetta, G.A.; Godfrey, J.; Orozco, J.; Zwieniecki, M.A. Non-Structural Carbohydrates in Dormant Woody Perennials; The Tale of Winter Survival and Spring Arrival. Front. For. Glob. Change 2019, 2, 457838. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, F.; Xie, Q. Balancing Growth and Adaptation to Stress: Crosstalk between Brassinosteroid and Abscisic Acid Signaling. Plant Cell Environ. 2020, 43, 2325–2335. [Google Scholar] [CrossRef]
- Beauvieux, R.; Wenden, B.; Dirlewanger, E. Bud Dormancy in Perennial Fruit Tree Species: A Pivotal Role for Oxidative Cues. Front. Plant Sci. 2018, 9, 657. [Google Scholar] [CrossRef]
- Barba-Espín, G.; Hernández, J.A.; Díaz-Vivancos, P. Antioxidant System: The Hub of Bud Dormancy Regulation in Prunus Sp. Sci. Hortic. 2022, 305, 111396. [Google Scholar] [CrossRef]
- Calle, A.; Saski, C.; Wünsch, A.; Grimplet, J.; Gasic, K. Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data. Plants 2022, 11, 2469. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Sun, M.; Chen, S.; Ma, H.; Lin, J.; Sun, Y.; Zhong, M. Molecular Characterization and Gene Expression Analysis of Tomato WOX Transcription Factor Family under Abiotic Stress and Phytohormone Treatment. J. Plant Biochem. Biotechnol. 2021, 30, 973–986. [Google Scholar] [CrossRef]
- Yu, J.; Conrad, A.O.; Decroocq, V.; Zhebentyayeva, T.; Williams, D.E.; Bennett, D.; Roch, G.; Audergon, J.M.; Dardick, C.; Liu, Z.; et al. Distinctive Gene Expression Patterns Define Endodormancy to Ecodormancy Transition in Apricot and Peach. Front. Plant Sci. 2020, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Canton, M.; Forestan, C.; Bonghi, C.; Varotto, S. Meta-Analysis of RNA-Seq Studies Reveals Genes with Dominant Functions during Flower Bud Endo- to Eco-Dormancy Transition in Prunus Species. Sci. Rep. 2021, 11, 13173. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhuo, X.; Zhao, K.; Zheng, T.; Han, Y.; Yuan, C.; Zhang, Q. Transcriptome Profiles Reveal the Crucial Roles of Hormone and Sugar in the Bud Dormancy of Prunus mume. Sci. Rep. 2018, 8, 5090. [Google Scholar] [CrossRef]
- Li, Z.; Lathe, R.S.; Li, J.; He, H.; Bhalerao, R.P. Towards Understanding the Biological Foundations of Perenniality. Trends Plant Sci. 2022, 27, 56–68. [Google Scholar] [CrossRef]
- Sharma, N.; Geuten, K.; Giri, B.S.; Varma, A. The Molecular Mechanism of Vernalization in Arabidopsis and Cereals: Role of Flowering Locus C and Its Homologs. Physiol. Plant 2020, 170, 373–383. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of Flowering Time: All Roads Lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial Activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C Define Distinct Modes of Flowering Regulation in Arabidopsis. Genome Biol. 2015, 16, 31. [Google Scholar] [CrossRef]
- He, Y.; Amasino, R.M. Role of Chromatin Modification in Flowering-Time Control. Trends Plant Sci. 2005, 10, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Shindo, C.; Aranzana, M.J.; Lister, C.; Baxter, C.; Nicholls, C.; Nordborg, M.; Dean, C. Role of FRIGIDA and FLOWERING LOCUS C in Determining Variation in Flowering Time of Arabidopsis. Plant Physiol. 2005, 138, 1163–1173. [Google Scholar] [CrossRef]
- Henderson, I.R.; Dean, C. Control of Arabidopsis Flowering: The Chill before the Bloom. Development 2004, 131, 3829–3838. [Google Scholar] [CrossRef]
- Madrid, E.; Chandler, J.W.; Coupland, G. Gene Regulatory Networks Controlled by FLOWERING LOCUS C That Confer Variation in Seasonal Flowering and Life History. J. Exp. Bot. 2021, 72, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Gazzani, S.; Gendall, A.R.; Lister, C.; Dean, C. Analysis of the Molecular Basis of Flowering Time Variation in Arabidopsis Accessions. Plant Physiol. 2003, 132, 1107–1114. [Google Scholar] [CrossRef]
- Rothkegel, K.; Sánchez, E.; Montes, C.; Greve, M.; Tapia, S.; Bravo, S.; Prieto, H.; Almeida, A.M. DNA Methylation and Small Interference RNAs Participate in the Regulation of MADS-Box Genes Involved in Dormancy in Sweet Cherry (Prunus avium L.). Tree Physiol. 2017, 37, 1739–1751. [Google Scholar] [CrossRef]
- Soto, E.; Sanchez, E.; Nuñez, C.; Montes, C.; Rothkegel, K.; Andrade, P.; Prieto, H.; Almeida, A.M. Small RNA Differential Expression Analysis Reveals MiRNAs Involved in Dormancy Progression in Sweet Cherry Floral Buds. Plants 2022, 11, 2396. [Google Scholar] [CrossRef]
- Martínez de Alba, A.E.; Elvira-Matelot, E.; Vaucheret, H. Gene Silencing in Plants: A Diversity of Pathways. Biochim. Biophys. Acta 2013, 1829, 1300–1308. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Somoza, I.; Weigel, D. MicroRNA Networks and Developmental Plasticity in Plants. Trends Plant Sci. 2011, 16, 258–264. [Google Scholar] [CrossRef]
- Redhu, N.S.; Yashveer, S.; Taunk, J.; Banga, P.; Singh, V.; Tokas, J.; Grewal, S.; Arora, I. Plant MicroRNAs: Unexplored Biogenesis, Prediction Tools and Applications. Gene Rep. 2023, 32, 101799. [Google Scholar] [CrossRef]
- Hong, Y.; Jackson, S. Floral Induction and Flower Formation—The Role and Potential Applications of MiRNAs. Plant Biotechnol. J. 2015, 13, 282–292. [Google Scholar] [CrossRef]
- Waheed, S.; Liang, F.; Zhang, M.; He, D.; Zeng, L. High-Throughput Sequencing Reveals Novel MicroRNAs Involved in the Continuous Flowering Trait of Longan (Dimocarpus longan Lour.). Int. J. Mol. Sci. 2022, 23, 15565. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Saito, T.; Ito, A.; Tuan, P.A.; Xu, Y.; Teng, Y.; Moriguchi, T. Small RNA and PARE Sequencing in Flower Bud Reveal the Involvement of SRNAs in Endodormancy Release of Japanese Pear (Pyrus pyrifolia ‘Kosui’). BMC Genom. 2016, 17, 230. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-Associated MADS-Box Genes and MicroRNAs Jointly Control Dormancy Transition in Pear (Pyrus pyrifolia White Pear Group) Flower Bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Rodriguez, R.E.; Mecchia, M.A.; Palatnik, J.F. Functional Specialization of the Plant MiR396 Regulatory Network through Distinct MicroRNA–Target Interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Li, Z.; Yuan, N.; Hu, Q.; Zhou, M.; Zhao, J.; Li, D.; Luo, H. MiR396 Is Involved in Plant Response to Vernalization and Flower Development in Agrostis stolonifera. Hortic. Res. 2020, 7, 173. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of Cell Proliferation in Arabidopsis thaliana by MicroRNA MiR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular Mechanism of MicroRNA396 Mediating Pistil Development in Arabidopsis. Plant Physiol. 2013, 164, 249. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, X.; Zhao, X.; Ding, D.; Tang, J.; Niu, J. Investigation of MiR396 and Growth-Regulating Factor Regulatory Network in Maize Grain Filling. Acta Physiol. Plant 2015, 37, 28. [Google Scholar] [CrossRef]
- Calle, A.; Grimplet, J.; Le Dantec, L.; Wünsch, A. Identification and Characterization of DAMs Mutations Associated with Early Blooming in Sweet Cherry, and Validation of DNA-Based Markers for Selection. Front. Plant Sci. 2021, 12, 621491. [Google Scholar] [CrossRef]
- Xie, Z.; Kasschau, K.D.; Carrington, J.C. Negative Feedback Regulation of Dicer-Like1 in Arabidopsis by MicroRNA-Guided MRNA Degradation. Curr. Biol. 2003, 13, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Gao, Z.; Wang, F.; Xu, T.; Qi, M.; Liu, Y.; Li, T. MicroRNA162 Regulates Stomatal Conductance in Response to Low Night Temperature Stress via Abscisic Acid Signaling Pathway in Tomato. Front. Plant Sci. 2023, 14, 1045112. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal Regulation of Shoot Development in Arabidopsis thaliana by MiR156 and Its Target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jia, S.; Shen, D.; Liu, J.; Li, J.; Zhao, H.; Han, S.; Wang, Y. Four AUXIN RESPONSE FACTOR Genes Downregulated by MicroRNA167 Are Associated with Growth and Development in Oryza sativa. Funct. Plant Biol. 2012, 39, 736–744. [Google Scholar] [CrossRef]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.N.; Zhao, Y. An Essential Role for MiRNA167 in Maternal Control of Embryonic and Seed Development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef]
- Caruana, J.C.; Dhar, N.; Raina, R. Overexpression of Arabidopsis MicroRNA167 Induces Salicylic Acid-Dependent Defense against Pseudomonas syringae through the Regulation of Its Targets ARF6 and ARF8. Plant Direct 2020, 4, e00270. [Google Scholar] [CrossRef]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. MiR396 Affects Mycorrhization and Root Meristem Activity in the Legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef]
- Liu, H.; Guo, S.; Xu, Y.; Li, C.; Zhang, Z.; Zhang, D.; Xu, S.; Zhang, C.; Chong, K. OsmiR396d-Regulated OsGRFs Function in Floral Organogenesis in Rice through Binding to Their Targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef]
- Chandran, V.; Wang, H.; Gao, F.; Cao, X.L.; Chen, Y.P.; Li, G.B.; Zhu, Y.; Yang, X.M.; Zhang, L.L.; Zhao, Z.X.; et al. MiR396- OsGRF s Module Balances Growth and Rice Blast Disease-Resistance. Front. Plant Sci. 2019, 9, 1999. [Google Scholar] [CrossRef]
- Szczygieł-Sommer, A.; Gaj, M.D. The MiR396-GRF Regulatory Module Controls the Embryogenic Response in Arabidopsis via an Auxin-Related Pathway. Int. J. Mol. Sci. 2019, 20, 5221. [Google Scholar] [CrossRef]
- Liebsch, D.; Palatnik, J.F. MicroRNA MiR396, GRF Transcription Factors and GIF Co-Regulators: A Conserved Plant Growth Regulatory Module with Potential for Breeding and Biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Z.; Bai, J.; Tao, X.; Wang, L.; Zhang, H.; Zhu, J.K. Disruption of MIR396e and MIR396f Improves Rice Yield under Nitrogen-Deficient Conditions. Natl. Sci. Rev. 2020, 7, 102–112. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-Directed Phasing during Trans-Acting SiRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Garighan, J.; Dvorak, E.; Estevan, J.; Loridon, K.; Huettel, B.; Sarah, G.; Farrera, I.; Leclercq, J.; Grynberg, P.; Coiti Togawa, R.; et al. The Identification of Small RNAs Differentially Expressed in Apple Buds Reveals a Potential Role of the Mir159-MYB Regulatory Module during Dormancy. Plants 2021, 10, 2665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, X.; Yuan, Y.; Zhu, S.; Liu, C.; Zhang, Y.; Gai, S. PsmiR159b-PsMYB65 Module Functions in the Resumption of Bud Growth after Endodormancy by Affecting the Cell Cycle in Tree Peony. Hortic. Res. 2024, 11, uhae052. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, Y.; He, Y. Photoperiodic Regulation of Flowering Time through Periodic Histone Deacetylation of the Florigen Gene FT. PLoS Biol. 2013, 11, e1001649. [Google Scholar] [CrossRef]
- Thouly, C.; Le Masson, M.; Lai, X.; Carles, C.C.; Vachon, G. Unwinding BRAHMA Functions in Plants. Genes 2020, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Liu, X.; Huan, L.; Sun, M.; Liu, L.; Chen, X.; Gao, D.; Li, L. Genome-Wide Analysis of Dof Family Genes and Their Expression during Bud Dormancy in Peach (Prunus persica). Sci. Hortic. 2017, 214, 18–26. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J.; Dennis, E.S. The FLF MADS Box Gene: A Repressor of Flowering in Arabidopsis Regulated by Vernalization and Methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C Encodes a Novel MADS Domain Protein That Acts as a Repressor of Flowering. Plant Cell 1999, 11, 949. [Google Scholar] [CrossRef]
- Zhu, P.; Lister, C.; Dean, C. Cold-Induced Arabidopsis FRIGIDA Nuclear Condensates for FLC Repression. Nature 2021, 599, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.K.; Schmitt, J.; Runcie, D.E. Exploring the Molecular Regulation of Vernalization-Induced Flowering Synchrony in Arabidopsis. New Phytol. 2024, 242, 947–959. [Google Scholar] [CrossRef]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 Redundantly Control AGAMOUS during Early Stages of Flower Development in Arabidopsis. Plant Cell 2006, 18, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Thong, Z.; Yu, H. Coming into Bloom: The Specification of Floral Meristems. Development 2009, 136, 3379–3391. [Google Scholar] [CrossRef] [PubMed]
- Chorostecki, U.; Crosa, V.A.; Lodeyro, A.F.; Bologna, N.G.; Martin, A.P.; Carrillo, N.; Schommer, C.; Palatnik, J.F. Identification of New MicroRNA-Regulated Genes by Conserved Targeting in Plant Species. Nucleic Acids Res. 2012, 40, 8893–8904. [Google Scholar] [CrossRef]
- Yang, C.Y.; Huang, Y.H.; Lin, C.P.; Lin, Y.Y.; Hsu, H.C.; Wang, C.N.; Liu, L.Y.D.; Shen, B.N.; Lin, S.S. MicroRNA396-Targeted SHORT VEGETATIVE PHASE Is Required to Repress Flowering and Is Related to the Development of Abnormal Flower Symptoms by the Phyllody Symptoms1 Effector. Plant Physiol. 2015, 168, 1702–1716. [Google Scholar] [CrossRef]
- Hou, N.; Cao, Y.; Li, F.; Yuan, W.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Epigenetic Regulation of MiR396 Expression by SWR1-C and the Effect of MiR396 on Leaf Growth and Developmental Phase Transition in Arabidopsis. J. Exp. Bot. 2019, 70, 5217–5229. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, M.P.; Lahiji, H.S.; Golfazani, M.M.; Biglouei, M.H. New Insights on the Regulatory Network of Drought-Responsive Key Genes in Arabidopsis thaliana. Genetica 2023, 151, 29–45. [Google Scholar] [CrossRef]
- Liang, G.; Li, Y.; He, H.; Wang, F.; Yu, D. Identification of MiRNAs and MiRNA-Mediated Regulatory Pathways in Carica papaya. Planta 2013, 238, 739–752. [Google Scholar] [CrossRef]
- Várallyay, É.; Válóczi, A.; Ágyi, Á.; Burgyán, J.; Havelda, Z. Plant Virus-Mediated Induction of MiR168 Is Associated with Repression of ARGONAUTE1 Accumulation. EMBO J. 2010, 29, 3507–3519. [Google Scholar] [CrossRef]
- Harvey, J.J.W.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An Antiviral Defense Role of AGO2 in Plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Mallory, A.C.; Bartel, D.P. AGO1 Homeostasis Entails Coexpression of MIR168 and AGO1 and Preferential Stabilization of MiR168 by AGO1. Mol. Cell 2006, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xian, Z.; Huang, W.; Li, Z. Evidence for the Biological Function of MiR403 in Tomato Development. Sci. Hortic. 2015, 197, 619–626. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.; Hen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-Directed Regulation of the Auxin Response Factors GmARF8a and GmARF8b Is Required for Soybean Nodulation and Lateral Root Development. Plant Physiol. 2015, 168, 101–116. [Google Scholar] [CrossRef]
- Jin, L.; Yarra, R.; Zhou, L.; Zhao, Z.; Cao, H. MiRNAs as Key Regulators via Targeting the Phytohormone Signaling Pathways during Somatic Embryogenesis of Plants. 3 Biotech 2020, 10, 495. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Guo, Q.; Zhang, X.; Zhou, L.; Zhang, Y.; Zhang, C. Conservation and Diversity of MiR166 Family Members From Highbush Blueberry (Vaccinium corymbosum) and Their Potential Functions in Abiotic Stress. Front. Genet. 2022, 13, 919856. [Google Scholar] [CrossRef]
- Fadón, E.; Herrero, M.; Rodrigo, J. Flower Development in Sweet Cherry Framed in the BBCH Scale. Sci. Hortic. 2015, 192, 141–147. [Google Scholar] [CrossRef]
- Sánchez, E.; Tricon, D.; Mora, R.; Quiroz, D.; Decroocq, V.; Prieto, H. A Fast and Efficient Protocol for Small RNA Extraction in Japanese Plum and Other Prunus Species. Electron. J. Biotechnol. 2017, 30, 103–109. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-Scale Genome Assembly of Sweet Cherry (Prunus avium L.) Cv. Tieton Obtained Using Long-Read and Hi-C Sequencing. Hortic. Res. 2020, 7, 122. [Google Scholar] [CrossRef]
- Johnson, N.R.; Yeoh, J.M.; Coruh, C.; Axtell, M.J. Improved Placement of Multi-Mapping Small RNAs. G3 2016, 6, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Lunardon, A.; Johnson, N.R.; Hagerott, E.; Phifer, T.; Polydore, S.; Coruh, C.; Axtell, M.J. Integrated Annotations and Analyses of Small RNA-Producing Loci from 47 Diverse Plants. Genome Res. 2020, 30, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Yan, G.; Zhou, Y.; Zhang, K. Over-Expression of the PaAP1 Gene from Sweet Cherry (Prunus avium L.) Causes Early Flowering in Arabidopsis thaliana. J. Plant Physiol. 2013, 170, 315–320. [Google Scholar] [CrossRef]
- Castro, Á.; Quiroz, D.; Sánchez, E.; de los Ángeles Miccono, M.; Aguirre, C.; Ramírez, A.; Montes, C.; Prieto, H. Synthesis of an Artificial Vitis Vinifera MiRNA 319e Using Overlapping Long Primers and Its Application for Gene Silencing. J. Biotechnol. 2016, 233, 200–210. [Google Scholar] [CrossRef]
- Nakagawa, T.; Ishiguro, S.; Kimura, T. Gateway Vectors for Plant Transformation. Plant Biotechnol. 2009, 26, 275–284. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Edwards, K.; Johnstone, C.; Thompson, C. A Simple and Rapid Method for the Preparation of Plant Genomic DNA for PCR Analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Urtubia, C.; Devia, J.; Castro, Á.; Zamora, P.; Aguirre, C.; Tapia, E.; Barba, P.; Dell’Orto, P.; Moynihan, M.R.; Petri, C.; et al. Agrobacterium-Mediated Genetic Transformation of Prunus salicina. Plant Cell Rep. 2008, 27, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.J.; Silva, J.; Pinto, S.C.; Coimbra, S. I Choose You: Selecting Accurate Reference Genes for QPCR Expression Analysis in Reproductive Tissues in Arabidopsis thaliana. Biomolecules 2023, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem–Loop RT–PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
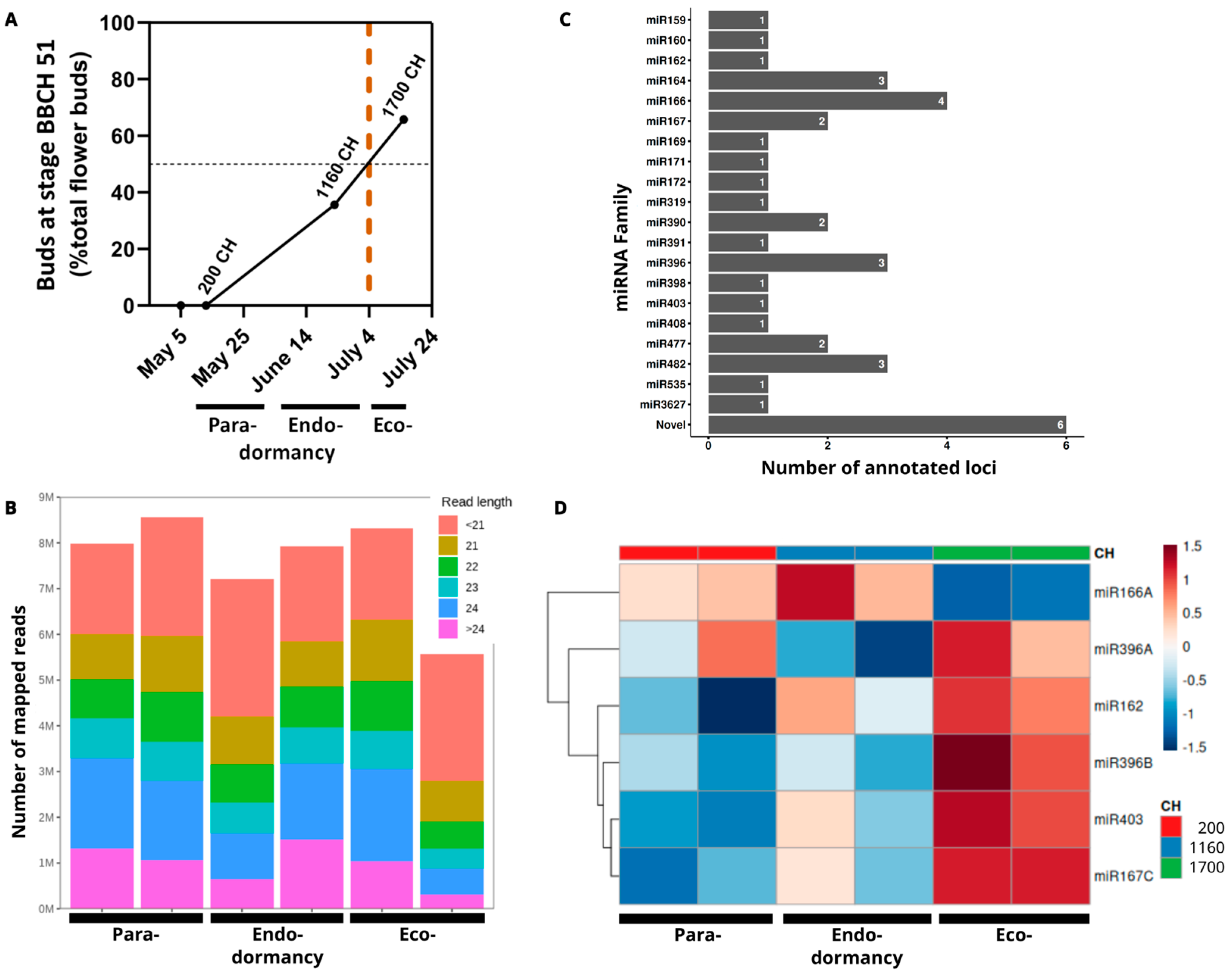
| Condition | Raw Reads | HQ Reads | Mapping to Reference Genome (%) | 21-nt | 22-nt | 23-nt | 24-nt |
|---|---|---|---|---|---|---|---|
| Para- dormancy (200 CH) | 24,078,331 | 18,843,039 | 87.8 | 2,215,658 | 1,932,853 | 1,727,362 | 3,717,064 |
| Endo- dormancy (1160 CH) | 25,413,001 | 17,044,809 | 88.7 | 2,035,231 | 1,723,603 | 1,470,882 | 2,667,987 |
| Eco- dormancy (1700 CH) | 23,798,208 | 15,740,688 | 88.1 | 2,242,584 | 1,676,026 | 1,260,979 | 2,605,945 |
| miRNA | Target | Target Function | Species | References |
|---|---|---|---|---|
| miR162 | DCL1 | Regulate miRNA biogenesis. Involved in the low night temperature responsive pathway by indirectly regulating stomatal conductance and photosynthesis | Arabidopsis and Solanum | [41,42] |
| miR167 | ARF | Development of male organ, roots, stems, leaves and flowers, flowering time, embryonic development, seed development and stress | Arabidopsis and Oryza | [43,44,45,46] |
| miR396 | GRF | Cell proliferation in leaves, disease resistance, somatic embryogenesis, grain size, and panicle branching | Arabidopsis, Medicago, and Oryza | [35,47,48,49,50,51,52] |
| miR403 | AGO2 | miRNA metabolism | Arabidopsis | [53] |
| Genotype/Plant * | Flowering Day (After Sowing) | Rosette Leaves at Flowering | Stemloop PCR/Sequencing ** |
|---|---|---|---|
| WT (Edi-0)/p1 | 207 | 136 | −/n.a. |
| WT (Edi-0)/p2 | 209 | 140 | −/n.a. |
| WT (Edi-0)/p3 | 209 | 124 | −/n.a. |
| WT (Edi-0)/p4 | 210 | 110 | −/n.a. |
| WT (Edi-0)/p5 | 210 | 133 | −/n.a. |
| 396 T2 a/p1 | 49 | 3 | +/amiR396 |
| 396 T2/p2 | 61 | 4 | +/amiR396 |
| 396 T2/p3 | 88 | 2 | +/amiR396 |
| 396 T2/p4 | 60 | 0 | +/amiR396 |
| 396 T2/p5 | 61 | 9 | +/amiR396 |
| 162 T1/p1 | 146 | 40 | +/amiR162 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaete-Loyola, J.; Olivares, F.; Saavedra, G.M.; Zúñiga, T.; Mora, R.; Ríos, I.; Valdovinos, G.; Barrera, M.; Almeida, A.M.; Prieto, H. Artificial Sweet Cherry miRNA 396 Promotes Early Flowering in Vernalization-Dependent Arabidopsis Edi-0 Ecotype. Plants 2025, 14, 899. https://doi.org/10.3390/plants14060899
Gaete-Loyola J, Olivares F, Saavedra GM, Zúñiga T, Mora R, Ríos I, Valdovinos G, Barrera M, Almeida AM, Prieto H. Artificial Sweet Cherry miRNA 396 Promotes Early Flowering in Vernalization-Dependent Arabidopsis Edi-0 Ecotype. Plants. 2025; 14(6):899. https://doi.org/10.3390/plants14060899
Chicago/Turabian StyleGaete-Loyola, José, Felipe Olivares, Gabriela M. Saavedra, Tiare Zúñiga, Roxana Mora, Ignacio Ríos, Gonzalo Valdovinos, Marion Barrera, Andrea Miyasaka Almeida, and Humberto Prieto. 2025. "Artificial Sweet Cherry miRNA 396 Promotes Early Flowering in Vernalization-Dependent Arabidopsis Edi-0 Ecotype" Plants 14, no. 6: 899. https://doi.org/10.3390/plants14060899
APA StyleGaete-Loyola, J., Olivares, F., Saavedra, G. M., Zúñiga, T., Mora, R., Ríos, I., Valdovinos, G., Barrera, M., Almeida, A. M., & Prieto, H. (2025). Artificial Sweet Cherry miRNA 396 Promotes Early Flowering in Vernalization-Dependent Arabidopsis Edi-0 Ecotype. Plants, 14(6), 899. https://doi.org/10.3390/plants14060899






