Abstract
The genus Vigna Savi (Leguminosae Juss.) comprises approximately 150 species, classified into five subgenera, most of which exhibit a diploid chromosome number of 2n = 22. However, the wild species Vigna lasiocarpa (Benth) Verdc. (V. subg. Lasiospron) is notable for its dysploid chromosome number of 2n = 20. This study aimed to elucidate the chromosomal events involved in the karyotype evolution of V. lasiocarpa (Vla). We used oligopainting probes from chromosomes 1, 2, 3, and 5 of Phaseolus vulgaris L. and two barcode probes from the genome of V. unguiculata (L.) Walp. Additionally, bacterial artificial chromosomes (BACs) from V. unguiculata and P. vulgaris, along with a telomeric probe from Arabidopsis thaliana (L.) Heynh., were hybridized to V. lasiocarpa metaphase chromosomes to characterize Vla3, Vla7/5, and Vla9. Our findings revealed conserved oligo-FISH patterns on chromosomes 2, 6, 8, 10, and 11 between V. unguiculata and V. lasiocarpa. Paracentric and pericentric inversions were identified for Vla3 and Vla9, respectively. Our integrative approach revealed that the dysploid chromosome originated from an “end-to-end fusion” of homoeologous chromosomes 5 and 7. This is the first report on the chromosomal mechanisms underlying descending dysploidy in Vigna, providing new insights into the evolutionary dynamics of the genus.
1. Introduction
Changes in chromosome number and structure are major drivers of species evolution and diversification, leading to disruptions in collinearity and/or synteny among related species [1]. The two primary biological mechanisms responsible for alterations in chromosome number within a karyotype are aneuploidy and dysploidy. Aneuploidy refers to the gain or loss of whole chromosome(s), whereas dysploidy involves changes in chromosome number, either an increase (ascending dysploidy) or decrease (descending dysploidy), resulting from chromosomal rearrangements, such as fissions and fusions, without a net gain or loss of the genetic material [2]. In plants, dysploidy is a common evolutionary event that has shaped the genomes of various species, as evidenced in several angiosperm families, such as Brassicaceae, Asteraceae, and Solanaceae [3].
The paraphyletic Vigna Savi genus belongs to the Leguminosae Juss. family and the Papilionoideae subfamily. It comprises approximately 150 accepted species with a worldwide distribution (https://www.worldfloraonline.org/). Closely related to Phaseolus L., Vigna diverged approximately 10.4 million years ago [4]. This genus includes several socioeconomically and nutritionally important grain legumes, such as cowpea [V. unguiculata (L.) Walp.], mung bean (V. radiata L.), and adzuki bean [V. angularis (Willd.) Ohwi and Ohashi] [5,6,7].
Vigna is divided into five subgenera, each with distinct centers of origin: Lasiospron (Benth.) Maréchal, Mascherpa & Stainier (American); Ceratotropis (Piper) Verdc. (Asian); and Vigna, (R.Wilczek) Verdc., Plectrotropis (Schumach.); and Haydonia (R.Wilczek) Verdc. (African) [8,9,10]. The monophyletic Vigna subg. Lasiospron consists of six wild species native to the wet tropical forests of the Americas, characterized by distinct morphological features compared to other Vigna species [9]. This early-diverging subclade of Vigna subg. Lasiospron has been identified as a sister group to the remaining Old Word Vigna clade (Vigna sensu stricto), with an estimated divergence time of approximately 4–5 million years (My) [8,9]. Most Vigna species have a stable diploid karyotype of 2n = 22 [11,12,13]. However, a reduction in chromosome number has been documented in 6 of the 150 Vigna species (https://taux.evolseq.net/CCDB_web; accessed on 10 February 2025). Among them, Vigna lasiocarpa (Benth.) Verdc. (syn. Phaseolus pilosus Kunth.) stands out as the only dysploid species reported within the small Vigna subg. Lasiospron, possessing a karyotype of 2n = 2x = 20 [9,14].
Since its development, oligonucleotide fluorescent in situ hybridization (oligo-FISH) technique has emerged as a powerful and versatile tool in plant cytogenetics [15]. It has been widely applied in various studies, including comparative karyotype analysis [16], chromosome behavior during meiosis [17], identification of chromosomal territories in nuclei [18], macrosynteny analysis [19], identification of inter- and intraspecific hybrid chromosome [20,21], genome assembly validation [22], and investigation of chromosome number reduction in dysploid species [23]. Our extensive research on Vigna species has revealed that chromosomal inversions and translocations are the primary forces driving evolutionary divergence among Vigna subgenera. We have used various probe sets, including bacterial artificial chromosomes (BACs), 5S and 35S ribosomal DNA (rDNA), and oligo probes [24,25,26,27]. However, dysploid Vigna species have received little attention, making them an excellent target for investigating the evolutionary pathway associated with chromosome reduction.
We performed a cytomolecular mapping in the dysploid species V. lasiocarpa using a well-established methodology based on chromosome painting and barcode oligo-FISH, along with BAC probes from P. vulgaris and V. unguiculata. Our results reveal that “end-to-end chromosome fusion” (EEF) is the primary mechanism driving chromosome reduction in V. lasiocarpa species. Our findings provide valuable insights into the key chromosomal rearrangements underpinning speciation in V. lasiocarpa, enhancing our understanding of the diversity and evolution of this important group of legume crops.
2. Results
2.1. Chromosome Mapping and rDNA Sites in V. lasiocarpa
The two barcode pools developed for V. unguiculata and the painting probes of P. vulgaris chromosomes 1, 2, 3, and 5 were hybridized to the 10 chromosome pairs of V. lasiocarpa (Figure 1a,a’,b,b’). Additionally, the barcode probes and the Pv1 and Pv5 painting probes were used to hybridize the V. unguiculata karyotype (Figure 1c,c’), showing a translocation event involving chromosomes 1 and 5 that was not observed in V. lasiocarpa. The painting probes exhibited strong signals on chromosomes from both Vigna species, except in the pericentromeric and centromeric regions, probably due to sequences divergence in these areas (Figure 1a’–c’).
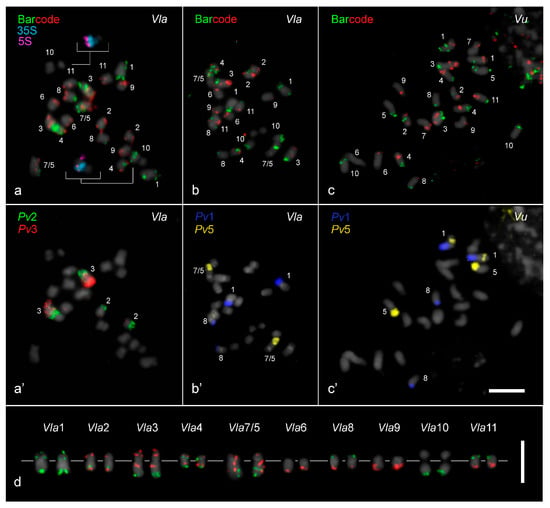
Figure 1.
Oligo-FISH and chromosome painting based karyotyping of Vigna lasiocarpa (2n = 20) and V. unguiculata (2n = 22). (a) Barcode probes hybridized to V. lasiocarpa chromosomes, (a’) followed by a re-hybridization with P. vulgaris painting probes for chromosomes 2 (Pv2, green) and 3 (Pv3, red), enabling the identification of their putative homoeologs in V. lasiocarpa. (a) Both 5S (pseudo-colored in magenta) and 35S (pseudocolored in light blue) rDNA sites were mapped to a single chromosome Vla10, highlighted in the inserts. (b) The barcode probes hybridized to V. lasiocarpa chromosomes, (b’) followed by a re-hybridization with Pv1 (dark blue) and Pv5 (yellow) probes, allowing the identification of homoeologous chromosomes Vla1, Vla8, and the dysploid Vla7/5 chromosome. (c) Barcode probes hybridized to V. unguiculata chromosomes, (c’) followed by a re-hybridization using Pv1 (dark blue) and Pv5 (yellow) probes. All chromosomes were counter-stained with DAPI (pseudocolored in gray). (d) Karyogram of the barcode pattern in V. lasiocarpa (Vla), with the centromeres aligned along a gray line. Bars in (c’,d) = 5 µm.
Both 5S and 35S rDNA in V. lasiocarpa were each mapped to a single site on chromosome Vla10 (inserts in Figure 1a). The 5S rDNA site was localized at the subterminal region on the long arm, while the 35S rDNA site was localized in a proximal region at the same arm. The chromosome numbering of V. lasiocarpa (Vla) was consistent with the homologous chromosomes of V. unguiculata (Vu) (Figure 1d).
A similar FISH signal pattern for both barcode and painting probes was observed on chromosomes 2, 6, 8, 10, and 11 from V. unguiculata and V. lasiocarpa (Figure 1). For instance, painting probes derived from P. vulgaris chromosomes 2 (Pv2, green) and 3 (Pv3, red), when hybridized to the V. lasiocarpa karyotype, produced the expected labeling pattern for chromosome 2, as previously observed in seven other Vigna species/subspecies [27]. Chromosome Vla2 exhibited signals at the distal region of the short arm labeled with Pv3 (red) and the distal region of the long arm labeled with Pv2 (green), with its pericentromeric region lacking the painting signals (Figure 1a’,c’). It is important to note that the reciprocal translocation Pv2/Pv3 occurred in P. vulgaris and was not shared with Vigna species (Figure 1a’).
In contrast, distinct FISH signal patterns were observed on the homoeologous V. unguiculata chromosomes 1, 3, 4, and 9, and particularly for chromosomes 5 and 7 (Figure 2 and Figure 3b–d). For example, the barcode FISH signal pattern on Vla4 was similar to Vl4, with one red and one green signal on each chromosome arm, but different from Vu4, which exhibited two green signals on the short arm and two red signals on the long arm (Figure 1a–c and Figure 3a). This pericentric inversion appears to be exclusive to V. unguiculata so far. The rearrangements involving the remaining chromosomes, which likely contributed to the genomic diversification and speciation of V. lasiocarpa, are illustrated schematically in Figure 3 and are further detailed in the following sections. The chromosomal positions of the probes in the three species are presented in Supplementary Table S1.
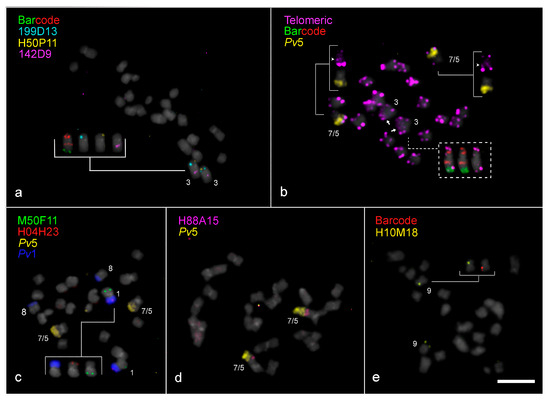
Figure 2.
Chromosomal rearrangements in Vigna lasiocarpa (Vla, 2n = 20) revealed by FISH. (a) A paracentric inversion involving chromosome Vla3, identified by re-hybridization of BACs 199D13 (light blue, from Pv3L), 142D9 (magenta, from Pv3S), and H50P11 (yellow, from Vu3L). From left to right, the insert shows the homoeologous Vla3 hybridized with the barcode probes, followed by the above-mentioned BACs. (b) A metaphase cell of V. lasiocarpa hybridized with a telomeric DNA probe (magenta) and re-hybridized with Pv5 (yellow) and the barcode probes, highlighting the “end-to-end chromosome fusion” in the dysploid Vla7/5. The arrowheads point to the interstitial telomeric site on Vla7/5 (upper inserts), while the arrows point to the interstitial telomeric site in Vla3, indicating a putative inversion. The lower dotted insert shows chromosomes from a different metaphase cell hybridized with telomeric and barcode probes. (c) Reciprocal translocation between chromosomes Pv1 (dark blue) and Pv5 (yellow), confirmed by BAC M050F11 (green, Vu5L) and BAC H04H23 (red, Vu1L), hybridized to Vla1 and Vla8. From left to right, the insert shows homoeologous Vla1 hybridized with the Pv1 oligopainting probe followed by the above-mentioned BACs. (d) Confirmation of the Vla7/5 chromosomal fusion using FISH with Pv5 (yellow) and BAC H088A15 (magenta, Vu7S) probes. (e) A pericentric inversion in chromosome Vla9, identified by FISH with barcode and BAC H010M18 (yellow, Vu9L) probes, showing terminal localization of the BAC signal. All chromosomes were counterstained in DAPI and pseudocolored in gray. Bar in (e) = 5 µm.
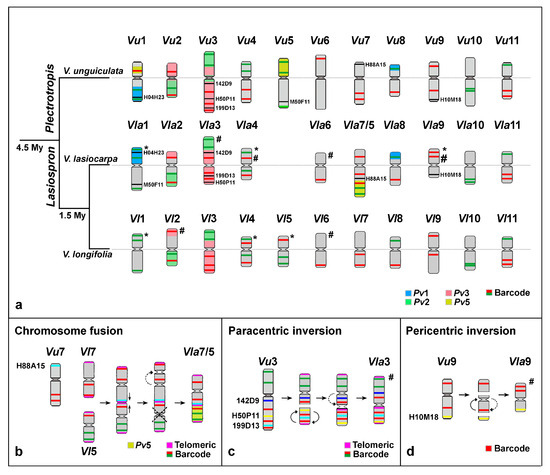
Figure 3.
Phylogenetic relationships among Vigna unguiculata (Vu), V. lasiocarpa (Vla), and V. longifolia (Vl), highlighting major chromosomal rearrangements associated with the evolution of V. lasiocarpa. (a) Idiograms of V. unguiculata (subgenus Plectotropis), V. lasiocarpa, and V. longifolia (both from subgenus Lasiospron) are plotted along a phylogenetic tree, based on Delgado-Salinas et al. [9] and Horton et al. [10]. The divergence times between the two subgenera and between V. lasiocarpa and V. longifolia are indicated along the branches. The barcode signal patterns and the Pv2 and Pv3 painting in V. unguiculata followed de Oliveira Bustamante et al. [26] and do Vale Martins et al. [25], respectively, while data for V. longifolia followed Dias et al. [27]. The chromosomes are labeled using the species abbreviation and their respective chromosome numbering. Barcode (red and green bars), BACs H50F11 and H04H23 (black bars), and painting probes Pv1, Pv2, Pv3, and Pv5 (light blue, green, red, and yellow, respectively) are mapped onto the idiograms of each Vigna species. Centromeres are aligned by a gray line. Asterisks (*) on Vla1, Vla4, Vla9, Vl1, Vl4, and Vl5 indicate changes in barcode patterns compared to V. unguiculata. Hashtags (#) on Vl2, Vla3, Vla4, Vla6, Vl6, and Vla9 indicate changes in chromosome arm orientation compared to V. unguiculata. (b) Schematic representation of the “end-to-end chromosome fusion” event involving Vla5 and Vla7, evidenced by barcode (red and green), telomeric (magenta), BAC H088A15 (light blue), and the Pv5 (yellow) probes. A black dotted X marks the position of the centromere, which was not visible after the chromosome fusion. The dottle arrow indicates a possible centromere shift following the “chromosome fusion”. (c) Paracentric inversion on Vla3, inferred from barcode and telomeric probe signals (magenta), along with BAC 142D9 (blue), H50P11 (yellow), and 199D13 (light blue), indicating the inversion of an interstitial region on Vla3L. The dottle arrow indicates a possible centromere shift after the rearrangement. The centromere, previously flanked by one green barcode signal and the BAC 142D9 (blue), is now located between one red barcode signal and the proximal telomeric probe. (d) Pericentric inversion on Vla9, with the barcode probe and the BAC H010M18 (yellow) showing both red barcode signals located adjacently on Vla9L as a result of the inversion.
2.2. Translocation Among Vigna Chromosomes 1, 5, and 8
Painting probes from P. vulgaris chromosomes 1 and 5 hybridized to three chromosome pairs (1, 5, and 8) in both V. lasiocarpa and V. unguiculata (Figure 1b’,c’). In V. unguiculata, the Pv1 probe (blue) distally labeled more than half of Vu1L and almost half of Vu8S, while the Pv5 probe (yellow) labeled the entire short arm of Vu5 and an interstitial portion of Vu1S. These findings confirm the presence of a translocation involving chromosomes 1 and 5 in V. unguiculata, which is absent in other Vigna species. We point out that the translocation Pv1/Pv8 has occurred in Phaseolus but not in Vigna species (Figure 1a’ and Figure 3a).
In V. lasiocarpa, the Pv1 probe (blue) hybridized to two chromosome pairs: the entire Vla1S and the distal half of Vla8S. Conversely, the Pv5 probe (yellow) distally labeled almost half of the long arm of the larger chromosome pair (Vla7/5) (Figure 1b’ and Figure 3a). The Pv1/8 translocation was confirmed by the V. lasiocarpa pattern. Additionally, the Vu1/5 reciprocal translocation was absent in V. lasiocarpa. This finding was confirmed by using the BAC H050F11 (Vu5L), which was located on Vla1L, and the BAC H004H23 (Vu1L), located on Vla1S (Figure 2c and Figure 3a). Based on Pv1 and Pv5 chromosome painting and barcode patterns, we revised the previously proposed homoeology of V. longifolia chromosomes 1 and 5, as reported by [27]. The assignments have been corrected, with the former chromosome 1 now designated as Vl5 and the former chromosome 5 as Vl1.
2.3. Chromosomal Rearrangements Associated with the Evolution of V. lasiocarpa
2.3.1. Paracentric Inversion on Vla3 Confirmed by the Presence of an Additional Interstitial Telomeric Signal
A paracentric inversion was observed on Vla3 when compared to Vl3 in addition to a distinct Vla3 orientation. This chromosome exhibited the same barcode pattern as Vu3 and Vl3, with two green and three red signals across Vla3, but with centromere positioned between two red barcode marks, instead of one green and one red mark, as observed in the other Vigna species (Figure 1a,c and Figure 3a). Additionally, the short arm and the proximal region of the long arm of Vla3 were labeled with Pv3 (red), while the distal portion of the long arm was labeled with Pv2 (green), with the centromere embedded in Pv3 sequences. On the other hand, V. longifolia exhibited the same painting pattern, but mapped to opposite chromosome arms, supporting the inversion event inferred in V. lasiocarpa (Figure 3a). Moreover, the oligo-FISH pattern of V. lasiocarpa reflects a morphological change in Vla3, as indicated by the hashtag in Figure 3a. This finding was unexpected, as paracentric inversions typically do not affect chromosome morphology.
To further investigate the structural organization of Vla3, three specific BAC probes were used: 199D13 (from Pv3L), 142D9 (from Pv3S), both syntenic with Vu3, and H50P11 (from Vu3L). BAC 199D13 hybridized to the short arm of Vla3 between two red barcode signals, while BAC 142D9 hybridized to the interstitial region of the long arm between a red and a green barcode signal, consistent with the expected pattern for Vu3 (Figure 3c). On the other hand, BAC H50P11 hybridized to the subterminal region of Vla3S, differing from its position in Vu3L, which was mapped to an interstitial position (Figure 2a and Figure 3c). These results support the occurrence of a paracentric inversion in Vla3, which reorganized the relative positions of these markers along the chromosome.
FISH using a telomeric DNA probe was performed to further understand this structural rearrangement in Vla3. In addition to the expected terminal signals on all V. lasiocarpa chromosome arms, an extra telomeric signal was detected in a proximal region of Vla3S, between the two red signals (Figure 2b and Figure 3c). This interstitial telomeric signal, along with the repositioning of BACs H50P11 and 199D13, provides strong evidence for the occurrence of a paracentric inversion on Vla3. However, we also observed a centromere shift in Vla3 compared to Vu3 and Vl3, with the Vla3 centromere positioned between two red barcode marks, now located near the terminal Vu3L BAC 199D13 instead of the proximal Vu3L BAC 142D9 (Figure 3c and Supplementary Figure S1). This shift altered the chromosome morphology and suggests a centromere repositioning besides the paracentric inversion. The dashed arrow in Figure 3c schematically illustrates this repositioning.
2.3.2. “End-to-End Vla7/5 Chromosome Fusion” as the Major Driver for Descending Dysploidy in V. lasiocarpa
We used a combination of BAC, telomeric DNA, and oligo-FISH probes to further investigate the structure of the distinctively large dysploid chromosome in V. lasiocarpa. In the oligo-FISH barcode analysis, this chromosome showed a unique signal pattern: one red signal on the short arm and two red and two green signals on the long arm. In addition to the expected terminal sites, an interstitial telomeric signal was detected, suggesting a break at the ends of both chromosomes, followed by a reciprocal translocation. This rearrangement may have generated two “fused chromosomes” with the loss of the small chromosome, which was not observed in V. lasiocarpa cells. These alterations could explain the origin of the descending dysploidy. To facilitate our understanding of this chromosomal rearrangement, this reciprocal translocation will hereafter be referred to as “end-to-end chromosome fusion” (EEF). To confirm this rearrangement, we performed additional FISH experiments using a combination of painting and BAC probes (Figure 2d and Figure 3a,b). The distal half of the long arm was entirely labeled with the Pv5 probe (yellow), and BAC H88A15 (Vu7S) hybridized to an interstitial position on the same arm. These findings provide strong evidence that the descending dysploidy in V. lasiocarpa resulted from an EEF involving the homoeologous chromosomes 5 and 7 (Figure 3b).
The centromere in the dysploid chromosome was located between the two red barcode signals of the Vla7, rather than adjacent to both signals as observed in the ortholog chromosomes of Vigna species (Figure 3a,b). Additionally, the centromeric region expected for Vla5 was not detected in the fused chromosome (Figure 3b).
2.3.3. Exclusive Pericentric Inversion on Vla9
In Vla9, a distinct barcode FISH pattern was observed when compared to Vu9, characterized by two red signals located on the same arm, whereas in Vu9, these signals are positioned on opposite arms (Figure 1a–c, Figure 2e and Figure 3d). This pattern is indicative of a pericentric inversion involving one of the red barcode signals in Vla9. To further investigate this rearrangement, we performed FISH using the BAC H10M18 (from Vu9L) in combination with the red barcode probe. While H10M18 hybridized to the Vla9S, both red barcode signals were detected in adjacent positions on Vla9L, supporting the inversion hypothesis. This represents the first report of a pericentric inversion on chromosome 9 in a Vigna species, indicating that it may be a lineage-specific chromosomal rearrangement unique to V. lasiocarpa.
3. Discussion
3.1. Chromosomal Rearrangements Associated with Descending Dysploidy and Evolution of V. lasiocarpa
Our research provides novel insights into the evolutionary mechanisms underlying descending dysploidy in V. lasiocarpa, a species belonging to Vigna subg. Lasiospron, one of the most basal subgenera within the Vigna lineage [10]. We identified three major chromosomal rearrangements associated with the diversification and speciation of this dysploid species: (1) “an end-to-end fusion of chromosomes” 5 and 7; (2) a paracentric inversion on chromosome 3; and (3) a pericentric inversion on chromosome 9. These structural changes appear to be exclusive to V. lasiocarpa and may represent lineage-specific events that occurred after the divergence of Vigna subg. Lasiospron and Vigna subg. Plectrotropis, which is estimated to have occurred approximately 4.5 Mya [9].
Inversions are among the most common chromosomal rearrangements in plants and have been associated with changes in gene expression, suppression of recombination, and genomic diversification [28]. Although their precise biological roles remain unclear, inversions are widely recognized as important drivers of genome evolution and speciation. For instance, several inversions, typically ranging from 100 to 200 kb, along with translocations, have contributed to the divergence between wild and cultivated soybean varieties [29]. Our recent findings reinforce the role of inversions and translocations in shaping the genome architecture and evolutionary diversification of Vigna species [25,27]. In the present work, a paracentric inversion on Vla3 with one interstitial and one terminal breakpoint was inferred based on the presence of additional Interstitial Telomeric Repeats (ITRs) and was confirmed through FISH using BAC probes from P. vulgaris and V. unguiculata. Previous studies have also reported rearrangements on chromosome 3 among Vigna and Phaseolus species [25,26,27,30], suggesting that this chromosome may represent a hotspot for rearrangements. Notably, in Phaseolus, ITRs associated with inversion events have already been described. In P. microcarpus, for example, an additional ITR signal was identified on chromosome 3, derived from a pericentric inversion involving the entire short arm and part of the long arm [31].
We also identified a telomeric probe signal at an interstitial position on the dysploid chromosome Vla7/5. The presence of ITRs, particularly in centromeric or pericentromeric regions, is a reliable marker of end-to-end fusions between non-homologous chromosomes [32,33,34,35]. In V. lasiocarpa, the fusion between chromosomes 5 and 7 was confirmed through FISH using a combination of a BAC probe from Vu7 and an oligopainting probe corresponding to Vu5, supporting the conclusion that this descending dysploidy arose via non-homologous chromosome fusion.
Dysploidy has played a key role in shaping chromosome number evolution across various plant species, as observed in the case of V. lasiocarpa in the present study. Mandáková et al. [36] demonstrated that end-to-end chromosome fusions are the primary mechanism driving independent descending dysploidy in the Microlepidieae (MICR) lineage. A comparable pattern is observed in the closely related Phaseolus genus, in which three dysploid species (P. macvaughii, P. leptostachyus, and P. micranthus) with 2n = 20 chromosomes have been identified within the Leptostachyus group [37,38]. Recent studies have revealed a high rate of chromosomal rearrangements in P. leptostachyus and P. macvaughii, including 16 rearrangements in P. leptostachyus, one of which involved a nested chromosome fusion [23]. The high frequency of chromosomal changes in the Leptostachyus group may be related to the abundance of repetitive elements, which are thought to have played a major role in the genomic restructuring and diversification of Phaseolus and related genera [39,40]. These observations highlight the importance of further investigation into the distribution and composition of repetitive DNA across Vigna genomes, particularly in dysploid species, to better understand how these elements influence genome architecture and evolution.
3.2. Positional Centromere Changes on V. lasiocarpa Chromosomes
The centromere plays a fundamental role in chromosome segregation and transmission by serving as the assembly site for the kinetochore, which mediates attachment to the mitotic spindle fibers, thereby ensuring accurate chromosome movement [41]. Despite this essential function, centromeres in eukaryotes display remarkable variation in size, structure, and sequence composition, a phenomenon known as centromere paradox. Their formation and identity are largely governed by epigenetic mechanisms [42,43]. Moreover, changes in centromere position without disrupting gene collinearity are frequently observed in plant genomes and can arise through different mechanisms, including (1) a pericentric inversion with breaks in the opposite arms—one near the centromere and the other distantly located—followed by a paracentric inversion involving the distal breakpoint and a new break adjacent to centromere; (2) a three-break-event, in which the centromeric fragment is inserted at a third breakpoint, distinct from the original centromere position; and (3) de novo centromere formation at a novel chromosomal location, generally accompanied by inactivation of the original centromere [44]. These mechanisms are referred to as centromere repositioning [44,45]. In the present study, we identified positional changes in the centromere of chromosomes Vla3, Vla9, and Vla7/5.
In cases of descending dysploidy, an end-to-end fusion (EEF) typically occurs between two non-homologous chromosomes. This fusion may be mediated by two DSBs at the terminal regions of both chromosomes, followed by a reciprocal ligation that results in the formation of a dicentric chromosome. To ensure proper chromosome segregation and maintain genome stability, one of the two centromeres must be subsequently silenced or eliminated [3,46]. We propose that such a mechanism underlies the formation of the dysploid chromosome 7/5 in V. lasiocarpa, in which the centromere of the ancestral chromosome 5 appears to have been inactivated, followed by a potential repositioning of the remaining functional centromere, stabilizing the fused chromosome.
On the other hand, the centromere shift on chromosome Vla9 is consistent with an unequal pericentric inversion. In turn, the centromere relocation on Vla3 was not a result of the paracentric inversion but rather appears to have resulted from a de novo centromere formation accompanied by the silencing of the ancestral centromere. Previous studies have reported centromere repositioning through de novo centromere formation in the Vigna genus [25,27]. This phenomenon appears to have occurred across multiple chromosomes in Vigna and Phaseolus species, based on analyses of genomic blocks (GBs) from V. unguiculata subsp. unguiculata, V. unguiculata subsp. sesquipedalis, and P. vulgaris, compared to the Ancestral Phaseoleae Karyotype (APK) [27,47]. Centromere repositioning has also been documented in other plant groups, such as the Brassicaceae family, particularly within the Arabideae tribe, where a high frequency of repositioning events has been observed. They are referred to as evolutionary new centromeres (ENCs) and can influence the epigenetic regulation of nearby genes [45].
Epigenetic mechanisms, such as CENH3/CENPA deposition, are believed to facilitate centromeric flexibility, enabling repositioning events in response to evolutionary and genomic pressures [48]. These processes not only contribute to chromosomal stability but may also play a significant role in plant speciation. To advance our understanding of centromere dynamics in Vigna, further functional and comparative studies, such as chromatin immunoprecipitation sequencing (ChIP-seq) or fine-scale centromere mapping, are essential to unravel the mechanisms and evolutionary implications of centromere repositioning in this genus.
3.3. Comparative Analysis Between Two Species of the American subg. Lasiospron: V. longifolia and V. lasiocarpa
Two of the six species from the American Lasiospron subgenus analyzed in this study, V. longifolia and V. lasiocarpa, are native to the Amazon basin, Restinga, and the Paraguay-Paraná river system [9]. These species are phylogenetically and morphologically closely related, both characterized by nearly cylindrical pods with thickened walls and subglobose seeds. This contrasts with other American Vigna species, which typically bear laterally flattened pods and seeds [9,10]. Despite their similarities, slight morphological differences distinguish V. lasiocarpa from V. longifolia. For instance, V. lasiocarpa generally presents five to seven floral nodes per inflorescence and a short-hooked style, whereas V. longifolia typically has two to three floral nodes and a poorly developed style [9].
Cytogenetically, chromosomal changes have occurred between V. lasiocarpa and V. longifolia, as evidenced by breaks in collinearity and macrosynteny involving five chromosomes. Additionally, V. lasiocarpa has undergone evolutionary processes leading to descending dysploidy (2n = 20), in contrast to V. longifolia and most Vigna species, which retain the ancestral chromosome number of 2n = 22 [49,50,51,52]. Our previous cytogenetic analyses revealed that V. longifolia possesses a conserved karyotype compared to other Vigna species, with only minor differences in the painting patterns of chromosomes 2 and 3 [27]. In contrast, the major chromosomal rearrangements observed in V. lasiocarpa, including a paracentric inversion on chromosome 3, a pericentric inversion on chromosome 9, and an “end-to-end fusion” between chromosomes 5 and 7, are likely species-specific events that occurred after the divergence between V. lasiocarpa and V. longifolia, estimated at approximately 1.5 Mya [9].
Taken together with our previous studies involving species from different origins and subgenera, our findings provide additional evidence that deepens our understanding of the mechanisms shaping Vigna karyotypes and their implications for biodiversity and speciation. Therefore, further research on other understudied species is needed to develop a broader overview of the karyotype evolution and genome organization in these important Vigna legumes.
4. Materials and Methods
4.1. Plant Material and Chromosome Preparation
Seeds of V. unguiculata cv. BR-14 Mulato and V. lasiocarpa (PI 306376) were obtained from Embrapa Meio-Norte (Teresina, Piauí, Brazil) and the U.S. National Plant Germplasm System (NGPS, Odessa, TX, USA), respectively, and were multiplied at the Instituto Agronômico de Pernambuco (IPA, Recife, Brazil).
Root tips from germinated seeds were pretreated with 8-hydroxyquinoline solution (2 mM) for 5 h at 18 °C, then fixed in a solution of 3:1 (v/v) methanol: 1 glacial acetic acid for 2–22 h at room temperature (RT) and stored at −20 °C until use. For chromosome preparation, the root tips were washed twice in distilled water and digested in an enzymatic solution consisting of 2% pectolyase (w/v), 4% cellulase (w/v), and 20% pectinase (v/v) for 2 h at 37 °C in a moist chamber. After the digestion, the slides were prepared using the air-drying protocol [53] with minor modifications: after air-drying, the slides were incubated in 45% acetic acid for 2 min (RT) and dried for 5 min at 37 °C.
4.2. FISH Probe Sets
To identify the individual chromosomes and the chromosomal rearrangements between V. lasiocarpa and V. unguiculata, we performed the FISH technique using various oligo probes from V. unguiculata and P. vulgaris (oligobarcode and chromosome painting, respectively), as well as BAC probe sets from both species, rDNA, and telomeric probes.
For chromosome painting, the probes were designed from P. vulgaris genome (Pvulgaris_442_v2.0; available at https://phytozome.jgi.doe.gov/), for chromosomes 2 and 3 (Pv2 and Pv3), as described by do Vale Martins et al. [25], and for chromosomes 1 and 5 (Pv1 and Pv5), as described by Montenegro et al. [54]. For the barcode probes, we used two libraries designed from the V. unguiculata reference genome (‘IT97K-499-35’, [55]), as described by de Oliveira Bustamante et al. [26]. Barcode libraries 1 (red) and 2 (green) correspond to 16 and 14 FISH signals, respectively, covering the 11 chromosome pairs of different Vigna species [27]. The oligo probes were synthesized by Arbor Biosciences (Ann Arbor, MI, USA), amplified, and indirectly labeled with biotin or dual biotin (www.idtdna.com, Pv1, Pv2, and library 2) and digoxigenin (www.idtdna.com, Pv3, Pv5, and library 1) according to published protocols [18,56].
Five BAC clones of V. unguiculata chromosomes (Vu) were selected: Vu1L (H004H23, long arm of Vu1), Vu3L (H050P11), Vu5L (M050F11), Vu7S (H088A15; short arm of Vu7), and Vu9L (H010M18). These BACs were previously mapped in V. unguiculata mitotic chromosomes by do Vale Martins et al. [25]. Additionally, we selected BACs from Pv3S and Pv3L of P. vulgaris (142D9 and 199D13, respectively), which were previously mapped in P. vulgaris by Pedrosa-Harand et al. [57]. The BAC synteny analysis between V. unguiculata and P. vulgaris was deeply described by de Oliveira Bustamante et al. [26]. All BACs were tested and used to confirm the chromosomal rearrangements between V. unguiculata and V. lasiocarpa. Moreover, we used the D2 sequence, a 400 bp fragment of two 5S rDNA repeat units from Lotus japonicus (Regel) K. Larsen [58], and the R2 sequence, a 6.5 kb fragment of 18S-5.8S-25S rDNA repeat unit from Arabidopsis thaliana [59]. The Arabidopsis thaliana plasmid clone, pAtT4 [60], served as a telomeric (5′-CCCTAAA-3′) DNA probe. BAC, 5S, 35S rDNA, and pAtT4 clones were isolated using the Plasmid Mini Kit (Qiagen). The BAC clones were directly labeled with Cy3-dUTP (GE Healthcare). The telomeric and 35S rDNA probes were indirectly labeled with digoxigenin-11-dUTP (Roche Diagnostics), while the 5S rDNA probes were indirectly labeled with biotin-11-dUTP (Roche Diagnostics, Basel, Switzerland), all by nick translation (ThermoFisher Scientific, Waltham, MA, USA). We followed the manufacturer’s instructions for extraction and labeling procedures.
4.3. FISH and Image Processing
For the FISH procedure, we followed Braz et al. [56] with minor modifications. The hybridization mix (10 µL/slide) contained 50% formamide, 10% dextran sulfate, and 2× saline sodium citrate (SSC). Depending on the type of probe, we used different concentrations. For chromosome painting, we added 400 ng of Pv2 and 350 ng of Pv3 or 300 ng of Pv1 and 350 ng of Pv5 to the hybridization mix. For the oligo-FISH barcode, we added 200 ng of library 1 and 250 ng of library 2 to the hybridization mix, while for BAC clones, 5S and 35S rDNA, and telomeric probes, we added 10 ng/µL. The final hybridization mix was directly applied to the slides for 7 min at 75 °C and hybridized for 18–36 h at 37 °C. After incubation, the slides were washed with 2× SCC and subsequently with 0.1× SSC at 42 °C, in the absence of formamide (76% stringency). The slides were then incubated for 30 min at 37 °C in 50 µL of 1× TNB (1 M Tris HCl pH 7.5, 3 M NaCl and blocking reagent, Sigma-Aldrich, St. Louis, MO, USA) or 5% BSA. For probe detection, the antibody mix consisted of 0.1–0.2 µL of rhodamine sheep anti-DIG (Roche) and 0.2 µL of Alexa Fluor 488 Streptavidin (Invitrogen, Waltham, MA, USA) diluted up to 20 µL with 1× TNB or 1% BSA and incubated for 1 h at 37 °C. Subsequently, the slides were washed three times with 4× SCC/1% Tween-20, 5 min each at 42 °C, and counterstained with 2 µg/mL DAPI (4,6-diamidino-2-phenylindole) in Vectashield (Vector Laboratories, Newark, CA, USA) antifade solution. To detect different DNA sequences using the same slide, we performed the rehybridization procedure according to Heslop-Harrison et al. [61].
Metaphase images were captured using a CytoVision Station (Leica, Wetzlar, German) with a MV4 camera (Jai, Valby, Denmark) with CytoVision Version 7.7/FISH software upgrade, connected to a Leica DM2500 microscope. The images were adjusted for brightness and contrast using Adobe Photoshop CS6 software. The chromosomes were identified based on their orthology with V. unguiculata [25,55]. The measurements were performed for both chromatids of five chromosomes from metaphase plates. The positions of each barcode, rDNA, telomeric, and BAC signal were determined using DRAWID 0.26 software [62]. The idiograms of V. lasiocarpa and V. unguiculata were performed using Adobe Photoshop CS6 software. Vigna longifolia idiogram was adapted from Dias et al. [27].
5. Conclusions
This study presents a comprehensive cytogenetic analysis of a dysploid Vigna karyotype, offering new insights into the chromosomal structure and evolutionary dynamics of V. lasiocarpa. By combining oligo- and BAC-FISH with telomeric DNA probes, we identified key chromosomal rearrangements, including inversions and an “end-to-end chromosome fusion”, supporting the descending dysploidy in this species. Specifically, we propose that a fusion between chromosomes 5 and 7 gave rise to the reduced chromosome number observed in V. lasiocarpa (2n = 20). This study not only enhances our understanding of the chromosomal structure of V. lasiocarpa but also provides insights into the evolutionary mechanisms that have shaped genomic diversity within the Vigna genus.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants14121872/s1: Table S1: List of probes and their chromosomal positions, including barcode1, BAC clones1, and 5S and 35S rDNA1,2, tested and used for comparative FISH mapping among V. unguiculata, V. lasiocarpa and V. longifolia, organized from the terminal region of the short arm to the terminal region of the long arm. These probe sets were used to characterize the V. lasiocarpa karyotype and to identify chromosomal rearrangements and homoeology with V. unguiculata and V. longifolia. The probes’ positions on V. longifolia chromosomes are based on Dias et al. [27]. No BACs probes were hybridized to the chromosomes of this species. Hashtags (#) on Vl2, Vla3, Vla4, Vla6, Vl6, and Vla9 indicate changes in chromosome arm orientation compared to V. unguiculata. Figure S1: Oligobarcode probes (red and green) hybridized to metaphase chromosomes of Vigna unguiculata (a, Vu, 2n = 22), V. lasiocarpa (b, Vla, 2n = 20), and V. longifolia (c, Vl, 2n = 22). (d) Karyogram showing the barcode pattern on chromosome 3 of the three species, with the centromeres aligned along a thin white line. All chromosomes were counter-stained with DAPI (pseudocolored in gray). Bars in (c,d) = 5 µm.
Author Contributions
All authors contributed to the study conception and design. Seed multiplication was made by A.F.d.C., L.S., J.H.S., S.D., and A.R.d.S.O. Material preparation, data collection, and analysis were performed by M.C.N. The first draft of the manuscript was written by L.S., J.H.S., S.D., A.M.B.-I., J.J., L.d.V.M., and A.C.B.-V. The experiments were conceptualized and supervised by A.C.B.-V. The resources for this research were provided by A.M.B.-I., J.J. and A.C.B.-V. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) Finance Code 001, and by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) Grant No. 315500/2023-9, 406048/2022-3, 313581/2020-7, and 442019/2019-0.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Acknowledgments
We thank Embrapa Meio-Norte (Teresina, Piauí, Brazil) and Germplasm Resources Information Network (GRIN Global, USA) for providing the V. unguiculata and V. lasiocarpa seeds.
Conflicts of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| FISH | Fluorescence in situ hybridization |
| BAC | Bacterial artificial chromosome |
| EMBRAPA | Empresa Brasileira de Pesquisa Agropecuária |
| NGPS | National Plant Germplasm System |
| RT | Room temperature |
| EEF | End-to-end fusion |
| rDNA | Ribosomal DNA |
| IPA | Instituto Agronômico de Pernambuco |
| APK | Ancestral Phaseoleae karyotype |
| GB | Genomic blocks |
| EC | Evolutionary new centromeres |
| DSB | Double-strand breaks |
| ITR | Interstitial telomeric repeats |
| SSC | Saline sodium citrate |
| ChIP-seq | Chromatin immunoprecipitation sequencing |
| Pv | Phaseolus vulgaris |
| Vu | Vigna unguiculata |
| Vl | Vigna longifolia |
| Vla | Vigna lasiocarpa |
References
- Lysak, M.A. Celebrating Mendel, McClintock, and Darlington: On end-to-end chromosome fusions and nested chromosome fusions. Plant Cell 2022, 34, 2475–2491. [Google Scholar] [CrossRef] [PubMed]
- de Storme, N.; Mason, A. Plant speciation through chromosome instability and ploidy change: Cellular mechanisms, molecular factors and evolutionary relevance. Curr. Plant Biol. 2014, 1, 10–33. [Google Scholar] [CrossRef]
- Mandáková, T.; Lysak, M.A. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 2018, 42, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, W.; Lin, L.; Zhu, X.; Li, J.; Zhu, X.; Chen, Z. Diversification of the Phaseoloid legumes: Effects of climate change, range expansion and habit shift. Front. Plant Sci. 2013, 4, 386. [Google Scholar] [CrossRef]
- Boukar, O.; Belko, N.; Chamarthi, S.; Togola, A.; Batieno, J.; Owusu, E.; Haruna, M.; Diallo, S.; Umar, M.L.; Olufajo, O.; et al. Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed. 2019, 138, 415–424. [Google Scholar] [CrossRef]
- Somta, P.; Laosatit, K.; Yuan, X.; Chen, X. Thirty years of mungbean genome research: Where do we stand and what have we learned? Front. Plant Sci. 2022, 13, 944721. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, X.; Shen, H.; Zhao, R.; Li, Z.; Shen, X.; Wang, F.; Chen, K.; Zhou, Y.; Li, B.; et al. Nutritional composition, efficacy, and processing of Vigna angularis (Adzuki Bean) for the human diet: An overview. Molecules 2022, 27, 6079. [Google Scholar] [CrossRef]
- Delgado-Salinas, A.; Thulin, M.; Pasquet, R.; Weeden, N.; Lavin, M. Vigna (Leguminosae) Sensu lato: The names and identities of the American segregate genera. Am. J. Bot. 2011, 98, 1694–1715. [Google Scholar] [CrossRef]
- Delgado-Salinas, A.; Lavin, M.; Snak, C.; Lewis, G.P. Systematics of Vigna subgenus Lasiospron (Leguminosae: Papilionoideae: Phaseolinae). Syst. Bot. 2022, 47, 97–124. [Google Scholar] [CrossRef]
- Horton, D.M.; Feleke, Y.; Pasquet, R.S.; Javadi, F.; Melville, K.A.; Delgado-Salinas, A.; Thulin, M.; Mithen, R.F.; Gepts, P.; Egan, A.N. Phylogenetic systematics of Vigna sensu stricto in the context of Physostigma and allies. Am. J. Bot. 2024, 111, e16381. [Google Scholar] [CrossRef]
- Forni-Martins, E.R. New chromosome number in the genus Vigna Savi (Leguminosae-Papilionoideae). Bull. Jard. Bot. Natl. Belg. /Bull. Natl. Plantentuin Belg. 1986, 56, 129. [Google Scholar] [CrossRef]
- Venoral, G.; Blangifortil, S.; Cremonini, R. Karyotype analysis of twelve species belonging to genus Vigna. Cytologia 1999, 64, 117–127. [Google Scholar] [CrossRef]
- Shamurailatpam, A.; Madhavan, L.; Yadav, S.R.; Bhat, K.V.; Rama Rao, S. Heterochromatin distribution and comparative karyo-morphological studies in Vigna umbellata Thunberg, 1969 and V. aconitifolia Jacquin, 1969 (Fabaceae) accessions. Comp. Cytogenet. 2015, 9, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Galasso, I.; Saponetti, L.S.; Pignone, D. Cytotaxonomic studies in Vigna. III. Chromosomal distribution and reacting properties of the heterochromatin in five wild species of the section Vigna. Caryologia 1996, 49, 311–319. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in Situ Hybridization in plants: Recent developments and future applications. Chromosome Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Braz, G.T.; He, L.; Zhao, H.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Torres, G.A.; Jiang, J. Comparative oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2018, 208, 513–523. [Google Scholar] [CrossRef]
- He, L.; Braz, G.T.; Torres, G.A.; Jiang, J. Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum Species. Chromosoma 2018, 127, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef]
- Zhao, Q.; Meng, Y.; Wang, P.; Qin, X.; Cheng, C.; Zhou, J.; Yu, X.; Li, J.; Lou, Q.; Jahn, M.; et al. Reconstruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J. 2021, 107, 1243–1259. [Google Scholar] [CrossRef]
- do Vale Martins, L.; Yu, F.; Zhao, H.; Dennison, T.; Lauter, N.; Wang, H.; Deng, Z.; Thompson, A.; Semrau, K.; Rouillard, J.-M.; et al. Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat. Commun. 2019, 10, 4604. [Google Scholar] [CrossRef]
- Wang, K.; Cheng, H.; Han, J.; Esh, A.; Liu, J.; Zhang, Y.; Wang, B. A comprehensive molecular cytogenetic analysis of the genome architecture in modern sugarcane cultivars. Chromosome Res. 2022, 30, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Doležalová, A.; Sládeková, L.; Šimoníková, D.; Holušová, K.; Karafiátová, M.; Varshney, R.K.; Doležel, J.; Hřibová, E. Karyotype differentiation in cultivated chickpea revealed by oligopainting fluorescence in situ hybridization. Front. Plant Sci. 2022, 12, 791303. [Google Scholar] [CrossRef]
- Nascimento, T.; Pedrosa-Harand, A. High rates of structural rearrangements have shaped the chromosome evolution in dysploid Phaseolus beans. Theor. Appl. Genet. 2023, 136, 215. [Google Scholar] [CrossRef]
- Oliveira, A.R.D.S.; Martins, L.D.V.; Bustamante, F.D.O.; Muñoz-Amatriaín, M.; Close, T.; Da Costa, A.F.; Benko-Iseppon, A.M.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosome Res. 2020, 28, 293–306. [Google Scholar] [CrossRef] [PubMed]
- do Vale Martins, L.; de Oliveira Bustamante, F.; da Silva Oliveira, A.R.; da Costa, A.F.; de Lima Feitoza, L.; Liang, Q.; Zhao, H.; Benko-Iseppon, A.M.; Muñoz-Amatriaín, M.; Pedrosa-Harand, A.; et al. BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 2021, 130, 133–147. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Bustamante, F.; do Nascimento, T.H.; Montenegro, C.; Dias, S.; do Vale Martins, L.; Braz, G.T.; Benko-Iseppon, A.M.; Jiang, J.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Oligo-FISH barcode in beans: A new chromosome identification system. Theor. Appl. Genet. 2021, 134, 3675–3686. [Google Scholar] [CrossRef]
- Dias, S.; de Oliveira Bustamante, F.; do Vale Martins, L.; Da Costa, V.A.; Montenegro, C.; Oliveira, A.R.D.S.; de Lima, G.S.; Braz, G.T.; Jiang, J.; Da Costa, A.F.; et al. Translocations and inversions: Major chromosomal rearrangements during Vigna (Leguminosae) evolution. Theor. Appl. Genet. 2024, 137, 29. [Google Scholar] [CrossRef]
- Hu, H.; Scheben, A.; Wang, J.; Li, F.; Li, C.; Edwards, D.; Zhao, J. Unravelling inversions: Technological advances, challenges, and potential impact on crop breeding. Plant Biotechnol. J. 2024, 22, 544–554. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef]
- Vasconcelos, E.V.; de Andrade Fonsêca, A.F.; Pedrosa-Harand, A.; de Andrade Bortoleti, K.C.; Benko-Iseppon, A.M.; Da Costa, A.F.; Brasileiro-Vidal, A.C. Intra- and Interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosome Res. 2015, 23, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Fonsêca, A.; Pedrosa-Harand, A. karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 2013, 56, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Brandes, A.; Schubert, I. Telomere sequence localization and karyotype evolution in higher plants. Plant Syst. Evol. 1995, 196, 227–241. [Google Scholar] [CrossRef]
- Sousa, A.; Renner, S.S. Interstitial telomere-like repeats in the monocot family Araceae: Chromosome evolution in the Araceae. Bot. J. Linn. Soc. 2015, 177, 15–26. [Google Scholar] [CrossRef]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 207–216. [Google Scholar] [CrossRef]
- Wang, X.; Jin, D.; Wang, Z.; Guo, H.; Zhang, L.; Wang, L.; Li, J.; Paterson, A.H. Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytol. 2015, 205, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Mandáková, T.; Pouch, M.; Harmanová, K.; Zhan, S.H.; Mayrose, I.; Lysak, M.A. Multispeed genome diploidization and diversification after an ancient allopolyploidization. Mol. Ecol. 2017, 26, 6445–6462. [Google Scholar] [CrossRef]
- Mercado-Ruaro, P.; Delgado-Salinas, A. Karyological studies in several mexican species of Phaseolus L. and Vigna Savi (Phaseolinae, Fabaceae). Adv. Legume Syst. 1996, 8, 83–87. [Google Scholar]
- Mercado-Ruaro, P.; Delgado-Salinas, A. Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am. J. Bot. 1998, 85, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Vasconcelos, E.; dos Santos, K.G.B.; Vaio, M.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Diversity of repetitive sequences within compact genomes of Phaseolus L. Beans and Allied Genera Cajanus L. and Vigna Savi. Chromosome Res. 2020, 28, 139–153. [Google Scholar] [CrossRef]
- Ferraz, M.E.; Fonsêca, A.; Pedrosa-Harand, A. Multiple and independent rearrangements revealed by comparative Cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosome Res. 2020, 28, 395–405. [Google Scholar] [CrossRef]
- Allshire, R.C.; Karpen, G.H. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 2008, 9, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef]
- Keçeli, B.N.; Jin, C.; van Damme, D.; Geelen, D. Conservation of centromeric histone 3 interaction partners in plants. J. Exp. Bot. 2020, 71, 5237–5246. [Google Scholar] [CrossRef]
- Schubert, I. What is behind “Centromere repositioning”? Chromosoma 2018, 127, 229–234. [Google Scholar] [CrossRef]
- Mandáková, T.; Hlousková, P.; Koch, M.A.; Lysak, M.A. Genome evolution in Arabideae was marked by frequent centromere repositioning. Plant Cell 2020, 32, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Steckenborn, S.; Marques, A. Centromere diversity and its evolutionary impacts on plant karyotypes and Plant Reproduction. New Phytol. 2025, 245, 1879–1886. [Google Scholar] [CrossRef]
- Montenegro, C.; do Vale Martins, L.; Bustamante, F.D.O.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Comparative cytogenomics reveals genome reshuffling and centromere repositioning in the legume tribe Phaseoleae. Chromosome Res. 2022, 30, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Leo, L.; Marchetti, M.; Giunta, S.; Fanti, L. Epigenetics as an evolutionary tool for centromere flexibility. Genes 2020, 11, 809. [Google Scholar] [CrossRef]
- Marechal, R. Donnees Cytologiques Sur Les Especes de La Sous-Tribu Des Papilionaceae—Phaseoleae—Phaseolinae Premiere Serie. Bull. Jard. Bot. Natl. Belg. 1969, 39, 125. [Google Scholar] [CrossRef]
- Senff, M.; Hickenbick, M.M.; Paim, N.R. Cytogenetic studies in species of the genus Vigna Savi (Leguminosae-Papilionoideae). Rev. Bras. Genét. 1992, 15, 2. [Google Scholar]
- Senff, M.; Schifino-Wittmann, M.T.S.-W.M.; Paim, N.R. Cytogenetic studies of populations of Arachis, Desmodium and Vigna Species (Leguminosae, Papilionoideae) from Rio Grande Do Sul. Braz. J. Genet. 1995, 18, 4. [Google Scholar]
- Dias, S.; Souza, R.C.; Vasconcelos, E.V.; Vasconcelos, S.; da Silva Oliveira, A.R.; do Vale Martins, L.; de Oliveira Bustamante, F.; da Costa, V.A.; Souza, G.; da Costa, A.F.; et al. Cytomolecular diversity among Vigna Savi (Leguminosae) subgenera. Protoplasma 2024, 261, 859–875. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, C.R.; Saraiva, L.S. An Air Drying technique for maize chromosomes without enzymatic maceration. Biotech. Histochem. 1993, 68, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, C.; Ibiapino, A.; Nascimento, T.; Costa, A.F.D.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Cytogenomic and phylogenomic evidence for new Infrageneric relationships in Macroptilium (Benth.) beans. Ann. Bot. 2024; submitted. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Braz, G.T.; Yu, F.; do Vale Martins, L.; Jiang, J. Fluorescent in situ hybridization using oligonucleotide-based probes. In In Situ Hybridization Protocols, 5th ed.; Nielsen, B.S., Jones, J., Eds.; Springer: New York, NY, USA, 2020; pp. 71–83. [Google Scholar]
- Pedrosa-Harand, A.; Kami, J.; Gepts, P.; Geffroy, V.; Schweizer, D. Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosome Res. 2009, 17, 405–417. [Google Scholar] [CrossRef]
- Pedrosa, A.; Sandal, N.; Stougaard, J.; Schweizer, D.; Bachmair, A. Chromosomal map of the model legume Lotus japonicus. Genetics 2002, 161, 1661–1672. [Google Scholar] [CrossRef]
- Wanzenböck, E.; Schöfer, C.; Schweizer, D.; Bachmair, A. Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J. 1997, 11, 1007–1016. [Google Scholar] [CrossRef]
- Richards, E.J.; Ausubel, F.M. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 1988, 53, 127–136. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Schwarzacher, T.; Anamthawat-Jónsson, K.; Leitch, A.R.; Shi, M.; Leitch, I.J. In Situ Hybridization with automated chromosome denaturation. Tecnhique 1991, 3, 109–115. [Google Scholar]
- Kirov, I.; Khrustaleva, L.; van Laere, K.; Soloviev, A.; Meeus, S.; Romanov, D.; Fesenko, I. DRAWID: User-friendly java software for chromosome measurements and idiogram drawing. Comp. Cytogenet. 2017, 11, 747–757. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).