Integrated Microbiology and Metabolomics Analysis Reveal Responses of Soil Bacterial Communities and Metabolic Functions to N-Zn Co-Fertilization in the Rhizosphere of Tea Plants (Camellia sinensis L.)
Abstract
1. Introduction
2. Results
2.1. Soil Properties, Microbial Biomass and Enzyme Activities in the Rhizosphere of Tea Plants
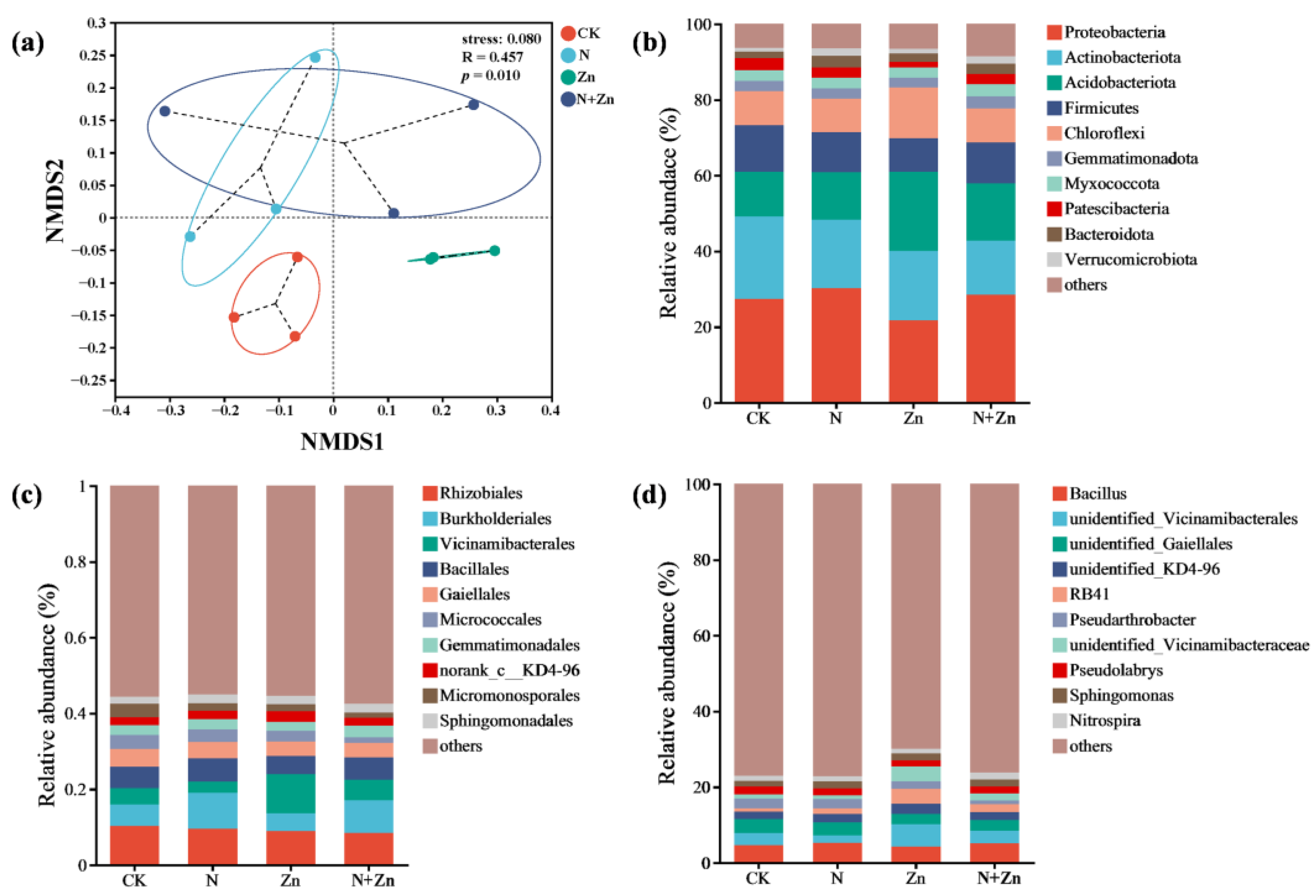
2.2. Soil Rhizobacterial Communities of Tea Plants
2.3. Soil Metabolic Profile in the Rhizosphere of Tea Plants
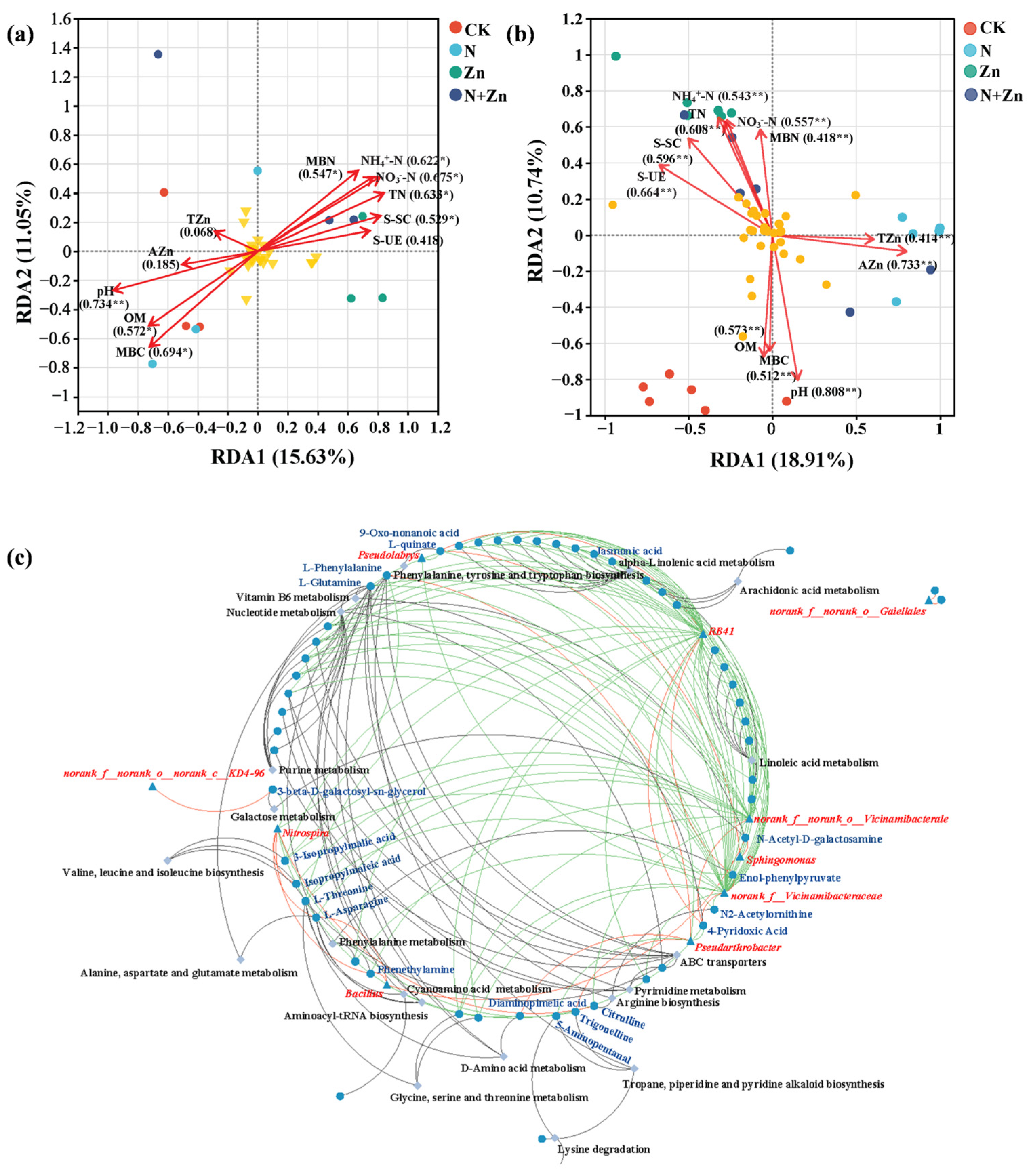
2.4. Correlations Between Soil Properties, Soil Rhizobacterial Community and Rhizosphere Soil Metabolism
3. Discussion
3.1. Alterations of Rhizosphere Soil N Availability of Tea Plants in Response to Different Combinations of N and Zn Fertilization
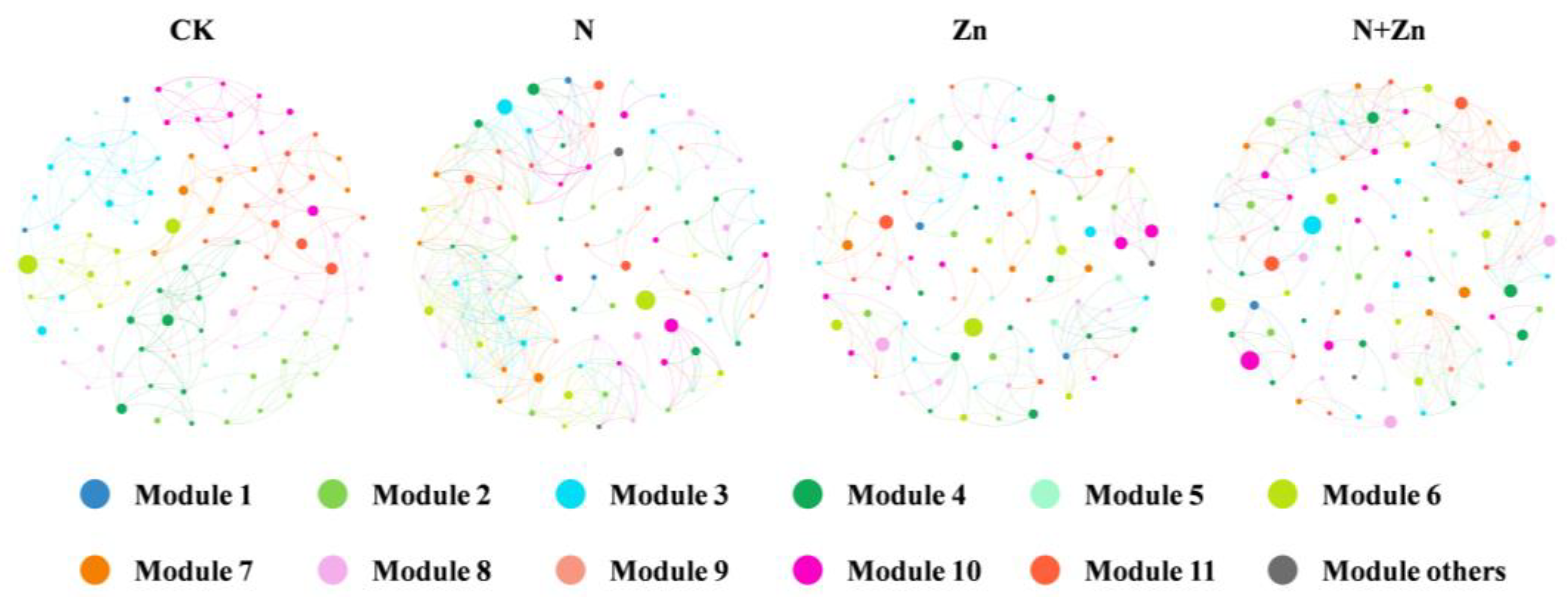
3.2. Modifications of Soil Rhizobacterial Diversity and Communities of Tea Plants in Response to Different Combinations of N and Zn Fertilization
3.3. Responses of Rhizosphere Soil DEMs and Potential Metabolic Pathways of Tea Plants in Response to Different Combinations of N and Zn Fertilization
3.4. Correlations Between Soil Properties, Bacterial Communities and DEMs in the Rhizosphere of Tea Plants in Response to Different Combinations of N and Zn Fertilization
4. Materials and Methods
4.1. Experimental Design
4.2. Soil-Sample Collection and Physicochemical-Properties Analysis
4.3. Measurements of MBC, MBN and Enzyme Activities in Rhizosphere Soil
4.4. Soil DNA Extraction and 16S rRNA Gene Sequencing
4.5. Soil Metabolomics Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yan, P.; Shen, C.; Fan, L.C.; Li, X.; Zhang, L.P.; Zhang, L.; Han, W.Y. Tea planting affects soil acidification and nitrogen and phosphorus distribution in soil. Agric. Ecosyst. Environ. 2018, 254, 20–25. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). Prod. FAOSTAT. 2020. Available online: https://www.fao.org/faostat/en/#home (accessed on 13 May 2023).
- National Bureau of Statistics of China (NBSC). Annual Statistical Data. 2020. Available online: http://www.stats.gov.cn/ (accessed on 20 July 2023).
- Wang, Z.T.; Geng, Y.B.; Liang, T. Optimization of reduced chemical fertilizer use in tea gardens based on the assessment of related environmental and economic benefits. Sci. Total Environ. 2020, 713, 136439. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, M.; Li, X.K.; Zhang, Z.; Ren, T.; Cong, R.H.; Ata-Ul-Karim, S.T.; Fahad, S.; Shah, A.N.; Lu, J.W. Nitrogen fertilizer management for enhancing crop productivity and nitrogen use efficiency in a rice-oilseed rape rotation system in China. Front. Plant Sci. 2016, 7, 1496. [Google Scholar] [CrossRef]
- Zecevic, V.; Knezevic, D.; Boskovic, J.; Micanovic, D.; Dozet, G. Effect of nitrogen fertilization on winter wheat quality. Cereal Res. Commun. 2010, 38, 243–249. [Google Scholar] [CrossRef]
- Zhang, X.; Davidson, E.A.; Mauzerall, D.L.; Searchinger, T.D.; Dumas, P.; Shen, Y. Managing nitrogen for sustainable development. Nature 2015, 528, 51–59. [Google Scholar] [CrossRef]
- Tian, J.; Dungait, J.A.J.; Lu, X.K.; Yang, Y.F.; Hartley, I.P.; Zhang, W.; Mo, J.M.; Yu, G.R.; Zhou, J.Z.; Kuzyakov, Y. Long-term nitrogen addition modifies microbial composition and functions for slow carbon cycling and increased sequestration in tropical forest soil. Glob. Change Biol. 2019, 25, 3267–3281. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.Y.; Gerendás, J.; Härdter, R.; Sattelmacher, B. Effect of nitrogen form and root-zone pH on growth and nitrogen uptake of tea (Camellia sinensis) plants. Ann. Bot. 2007, 99, 301–310. [Google Scholar] [CrossRef]
- Quemada, M.; Lassaletta, L.; Jensen, L.S.; Godinot, O.; Brentrup, F.; Buckley, C.; Foray, S.; Hvid, S.K.; Oenema, J.; Richards, K.G.; et al. Exploring nitrogen indicators of farm performance among farm types across several European case studies. Agr. Syst. 2020, 177, 102689. [Google Scholar] [CrossRef]
- Cakmak, I.; Kutman, U.B. Agronomic biofortification of cereals with zinc: A review. Eur. J. Soil Sci. 2018, 69, 172–180. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Wang, Y.; Ding, Z.T.; Wang, H.; Song, L.B.; Jia, S.S.; Ma, D.X. Zinc stress affects ionome and metabolome in tea plants. Plant Physiol. Biochem. 2017, 111, 318–328. [Google Scholar] [CrossRef]
- Glass, J.B.; Orphan, V.J. Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol. 2012, 3, 61. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.L.; Li, R.; Zhang, X.Y.; Chen, Y.Q.; Ni, D.J.; Wang, M.L. Zinc/iron-regulated transporter-like protein CsZIP4 enhances zinc and nitrogen uptake and alleviates zinc stresses with nitrogen supply in Camellia sinensis. J. Agric. Food Chem. 2024, 72, 21193–21207. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, H.C.; Sun, L.G.; Qi, G.F.; Chen, S.; Zhao, X.Y. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef]
- Peng, X.; Yan, X.; Zhou, H.; Zhang, Y.Z.; Sun, H. Assessing the contributions of sesquioxides and soil organic matter to aggregation in an ultisol under long-term fertilization. Soil Tillage Res. 2015, 146, 89–98. [Google Scholar] [CrossRef]
- Ma, L.F.; Yang, X.D.; Shi, Y.Z.; Yi, X.Y.; Ji, L.F.; Cheng, Y.; Ni, K.; Ruan, J.Y. Response of tea yield, quality and soil bacterial characteristics to long-term nitrogen fertilization in an eleven-year field experiment. Appl. Soil Ecol. 2021, 166, 103976. [Google Scholar] [CrossRef]
- Liu, Y.M.; Cao, W.Q.; Chen, X.X.; Yu, B.G.; Lang, M.; Chen, X.P.; Zou, C.Q. The responses of soil enzyme activities, microbial biomass and microbial community structure to nine years of varied zinc application rates. Sci. Total Environ. 2020, 737, 140245. [Google Scholar] [CrossRef]
- Lv, H.H.; Ji, C.C.; Zhang, L.; Jiang, C.C.; Cai, H.M. Zinc application promotes nitrogen transformation in rice rhizosphere soil by modifying microbial communities and gene expression levels. Sci. Total Environ. 2022, 849, 157858. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.N.; Yao, S.; Yang, X.L.; Jiang, X. Correlations between soil metabolomics and bacterial community structures in the pepper rhizosphere under plastic greenhouse cultivation. Sci. Total Environ. 2020, 728, 138439. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Wang, Z.N.; Ma, Y.; Luo, J.Y.; Gao, X.K.; Ning, J.; Mei, X.D.; She, D.M. Influence of the neonicotinoid insecticide thiamethoxam on soil bacterial community composition and metabolic function. J. Hazard. Mater. 2021, 405, 124275. [Google Scholar] [CrossRef]
- Wu, C.C.; Ma, Y.J.; Wang, D.; Shan, Y.P.; Song, X.P.; Hu, H.Y.; Ren, X.L.; Ma, X.Y.; Cui, J.J.; Ma, Y. Integrated microbiology and metabolomics analysis reveal plastic mulch film residue affects soil microorganisms and their metabolic functions. J. Hazard. Mater. 2022, 423, 127258. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Yuan, M.S.; Tang, L.; Shen, Y.F.; Yu, Q.; Li, S.Q. Integrated microbiology and metabolomics analysis reveal responses of soil microorganisms and metabolic functions to phosphorus fertilizer on semiarid farm. Sci. Total Environ. 2022, 817, 152878. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.T.; Aksenov, A.A.; Nothias, L.F.; Foulds, J.R.; Quinn, R.A.; Badri, M.H.; Swenson, T.L.; Van Goethem, M.W.; Northen, T.R.; Vazquez-Baeza, Y.; et al. Learning representations of microbe-metabolite interactions. Nat. Methods 2019, 16, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Li, F.; Zeng, Q.; Li, C.Y.; Shen, W.; Gao, X.; Li, X.Y.; Zhao, W.C.; Dong, J.; Li, J.G.; et al. Influence of fertilization methods and types on wheat rhizosphere microbiome community and functions. J. Agric. Food Chem. 2024, 72, 7794–7806. [Google Scholar] [CrossRef] [PubMed]
- Bremner, J.M.; Mulvaney, R.L. Urease activity in soils. In Soil Enzyme; Burns, R.G., Ed.; Academic Press: London, UK, 1978; pp. 149–197. [Google Scholar]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational improvements reveal great bacteria diversity and high metal toxicity in soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef]
- Shi, S.J.; Richardson, A.E.; O’Callaghan, M.; DeAngelis, K.M.; Jones, E.E.; Stewart, A.; Firestone, M.K.; Condron, L.M. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 2011, 77, 600–610. [Google Scholar] [CrossRef]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.W.; McHardy, A.C.; Schulze-Lefert, P.; AgBiome, T. Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 2018, 24, 155–167. [Google Scholar] [CrossRef]
- Ren, C.L.; Teng, Y.R.; Chen, X.Y.; Shen, Y.J.; Xiao, H.; Wang, H.Y. Impacts of earthworm introduction and cadmium on microbial communities composition and function in soil. Environ. Toxicol. Pharmacol. 2021, 83, 103606. [Google Scholar] [CrossRef]
- Jiao, S.; Wang, J.M.; Wei, G.H.; Chen, W.M.; Lu, Y.H. The dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere 2019, 235, 248–259. [Google Scholar] [CrossRef]
- Cardoni, M.; Fernandez-Gonzalez, A.J.; Valverde-Corredor, A.; Fernandez-Lopez, M.; Mercado-Blanco, J. Co-occurrence network analysis unveils the actual differential impact on the olive root microbiota by two Verticillium wilt biocontrol Rhizobacteria. Environ. Microbiome 2023, 18, 21. [Google Scholar] [CrossRef]
- Yuan, Q.S.; Wang, P.F.; Wang, C.; Chen, J.; Wang, X.; Liu, S. Indicator species and co-occurrence pattern of sediment bacterial community in relation to alkaline copper mine drainage contamination. Ecol. Indic. 2020, 120, 106884. [Google Scholar] [CrossRef]
- Zhang, F.G.; Meng, X.H.; Yang, X.M.; Ran, W.; Shen, Q.R. Quantification and role of organic acids in cucumber root exudates in Trichoderma harzianum T-E5 colonization. Plant Physiol. Biochem. 2014, 83, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Zhang, X.J.; Li, X.J.; Shen, J.Z.; Sun, L.T.; Zaman, S.; Wang, Y.; Ding, Z.T. Different changes of bacterial diversity and soil metabolites in tea plants-legume intercropping systems. Front. Plant Sci. 2023, 14, 1110623. [Google Scholar] [CrossRef] [PubMed]
- Li, X.N.; Qu, C.S.; Bian, Y.R.; Gu, C.G.; Jiang, X.; Song, Y. New insights into the responses of soil microorganisms to polycyclic aromatic hydrocarbon stress by combining enzyme activity and sequencing analysis with metabolomics. Environ. Pollut. 2019, 255, 113312. [Google Scholar] [CrossRef]
- Diaz, C.; Purdy, S.; Christ, A.; Morot-Gaudry, J.F.; Wingler, A.; Masclaux-Daubresse, C.L. Characterization of markers to determine the extent and variability of leaf senescence in Arabidopsis. A metabolic profiling approach. Plant Physiol. 2005, 138, 898–908. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Chen, W.H.; Yuan, W.; Wang, J.; Wang, Z.Y.; Zhou, Z.P.; Liu, S.P. No-tillage combined with appropriate amount of straw returning increased soil biochemical properties. Sustainability 2022, 14, 4875. [Google Scholar] [CrossRef]
- Tang, S.; Zhou, J.J.; Pan, W.K.; Sun, T.; Liu, M.J.; Tang, R.; Li, Z.J.; Ma, Q.X.; Wu, L.H. Effects of combined application of nitrogen, phosphorus, and potassium fertilizers on tea (Camellia sinensis) growth and fungal community. Appl. Soil Ecol. 2023, 181, 104611. [Google Scholar] [CrossRef]
- Chen, D.M.; Lan, Z.C.; Bai, X.; Grace, J.B.; Bai, Y.F. Evidence that acidification induced declines in plant diversity and productivity are mediated by changes in below-ground communities and soil properties in a semi-arid steppe. J. Ecol. 2013, 101, 1322–1334. [Google Scholar] [CrossRef]
- Schinner, F.; VonMersi, W. Xylanase-, CM-cellulase- and invertase activity in soil: An improved method. Soil Biol. Biochem. 1990, 22, 511–515. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Bao, S.D. Soil Agricultural Chemistry Analysis Method, 3rd ed.; China Agriculture Press: Beijing, China, 2008. [Google Scholar]
- Schneider, A.R.; Gommeaux, M.; Duclercq, J.; Fanin, N.; Conreux, A.; Alahmad, A.; Lacoux, J.; Roger, D.; Spicher, F.; Ponthieu, M.; et al. Response of bacterial communities to Pb smelter pollution in contrasting soils. Sci. Total Environ. 2017, 605, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, G.B.; Xue, S.; Song, Z.L. Rhizosphere soil microbial activity under different vegetation types on the Loess Plateau, China. Geoderma 2011, 161, 115–125. [Google Scholar] [CrossRef]
- Guan, S.Y. Soil Enzyme and Its Research Method; Agricultural Press: Beijing, China, 1986. [Google Scholar]
- Liu, C.S.; Zhao, D.F.; Ma, W.J.; Guo, Y.D.; Wang, A.J.; Wang, Q.L.; Lee, D.J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biot. 2016, 100, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media: ICWSM 2009, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]




| CK | N | Zn | N+Zn | |
|---|---|---|---|---|
| pH (1:2.5) | 6.17 ± 0.057 a | 5.50 ± 0.018 c | 5.94 ± 0.025 b | 5.63 ± 0.054 c |
| Organic matter (g/kg) | 19.5 ± 0.244 a | 14.0 ± 0.033 c | 16.7 ± 0.143 b | 12.2 ± 0.303 d |
| CEC (cmol(+)/kg) | 8.95 ± 0.748 a | 8.06 ± 0.232 a | 9.47 ± 0.729 a | 8.81 ± 0.315 a |
| Total N (g/kg) | 0.740 ± 0.025 b | 1.17 ± 0.040 a | 0.723 ± 0.067 b | 1.13 ± 0.017 a |
| NO3−-N (mg/kg) | 8.12 ± 0.094 b | 19.5 ± 0.579 a | 8.15 ± 0.101 b | 20.3 ± 0.534 a |
| NH4+-N (mg/kg) | 0.290 ± 0.012 b | 0.970 ± 0.040 a | 0.270 ± 0.017 b | 0.953 ± 0.083 a |
| Available P (mg/kg) | 22.0 ± 0.687 d | 24.7 ± 0.444 c | 30.0 ± 0.852 b | 40.9 ± 0.632 a |
| Available K (mg/kg) | 62.4 ± 0.590 b | 70.4 ± 0.993 a | 70.4 ± 0.995 a | 69.3 ± 0.973 a |
| Total Zn (mg/kg) | 124 ± 0.273 b | 120 ± 0.705 b | 142 ± 3.83 a | 145 ± 3.76 a |
| DTPA-Zn (mg/kg) | 2.19 ± 0.119 c | 1.12 ± 0.039 d | 8.04 ± 0.165 a | 3.81 ± 0.096 b |
| Topological Properties | CK | N | Zn | N+Zn |
|---|---|---|---|---|
| Number of Nodes | 98 | 99 | 98 | 99 |
| Number of Edges | 235 | 347 | 230 | 315 |
| Number of Positive Edges | 111 (47.23%) | 183 (52.74%) | 137 (59.57%) | 172 (54.60%) |
| Number of Negative Edges | 124 (52.77%) | 164(47.26%) | 93 (40.43%) | 143 (45.40%) |
| Average degree | 4.796 | 7.01 | 4.694 | 6.364 |
| Average clustering coefficient | 0.736 | 0.783 | 0.745 | 0.75 |
| Network diameter | 15 | 13 | 13 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, M.; Shi, Y.; Qi, D.; Wang, Q.; Zhang, H.; Feng, Y.; He, Z.; Dong, C.; Yang, X.; Yuan, C. Integrated Microbiology and Metabolomics Analysis Reveal Responses of Soil Bacterial Communities and Metabolic Functions to N-Zn Co-Fertilization in the Rhizosphere of Tea Plants (Camellia sinensis L.). Plants 2025, 14, 1811. https://doi.org/10.3390/plants14121811
Lu M, Shi Y, Qi D, Wang Q, Zhang H, Feng Y, He Z, Dong C, Yang X, Yuan C. Integrated Microbiology and Metabolomics Analysis Reveal Responses of Soil Bacterial Communities and Metabolic Functions to N-Zn Co-Fertilization in the Rhizosphere of Tea Plants (Camellia sinensis L.). Plants. 2025; 14(12):1811. https://doi.org/10.3390/plants14121811
Chicago/Turabian StyleLu, Min, Yali Shi, Dandan Qi, Qiong Wang, Haowen Zhang, Ying Feng, Zhenli He, Chunwang Dong, Xiaoe Yang, and Changbo Yuan. 2025. "Integrated Microbiology and Metabolomics Analysis Reveal Responses of Soil Bacterial Communities and Metabolic Functions to N-Zn Co-Fertilization in the Rhizosphere of Tea Plants (Camellia sinensis L.)" Plants 14, no. 12: 1811. https://doi.org/10.3390/plants14121811
APA StyleLu, M., Shi, Y., Qi, D., Wang, Q., Zhang, H., Feng, Y., He, Z., Dong, C., Yang, X., & Yuan, C. (2025). Integrated Microbiology and Metabolomics Analysis Reveal Responses of Soil Bacterial Communities and Metabolic Functions to N-Zn Co-Fertilization in the Rhizosphere of Tea Plants (Camellia sinensis L.). Plants, 14(12), 1811. https://doi.org/10.3390/plants14121811









