Single- and Multi-Locus GWAS Unravels Novel Genomic Regions Related to Low-Phosphate Stress in Cotton Seedlings
Abstract
1. Introduction
2. Results
2.1. Phenotypic Analysis of Related Traits Under Low-Phosphate Stress
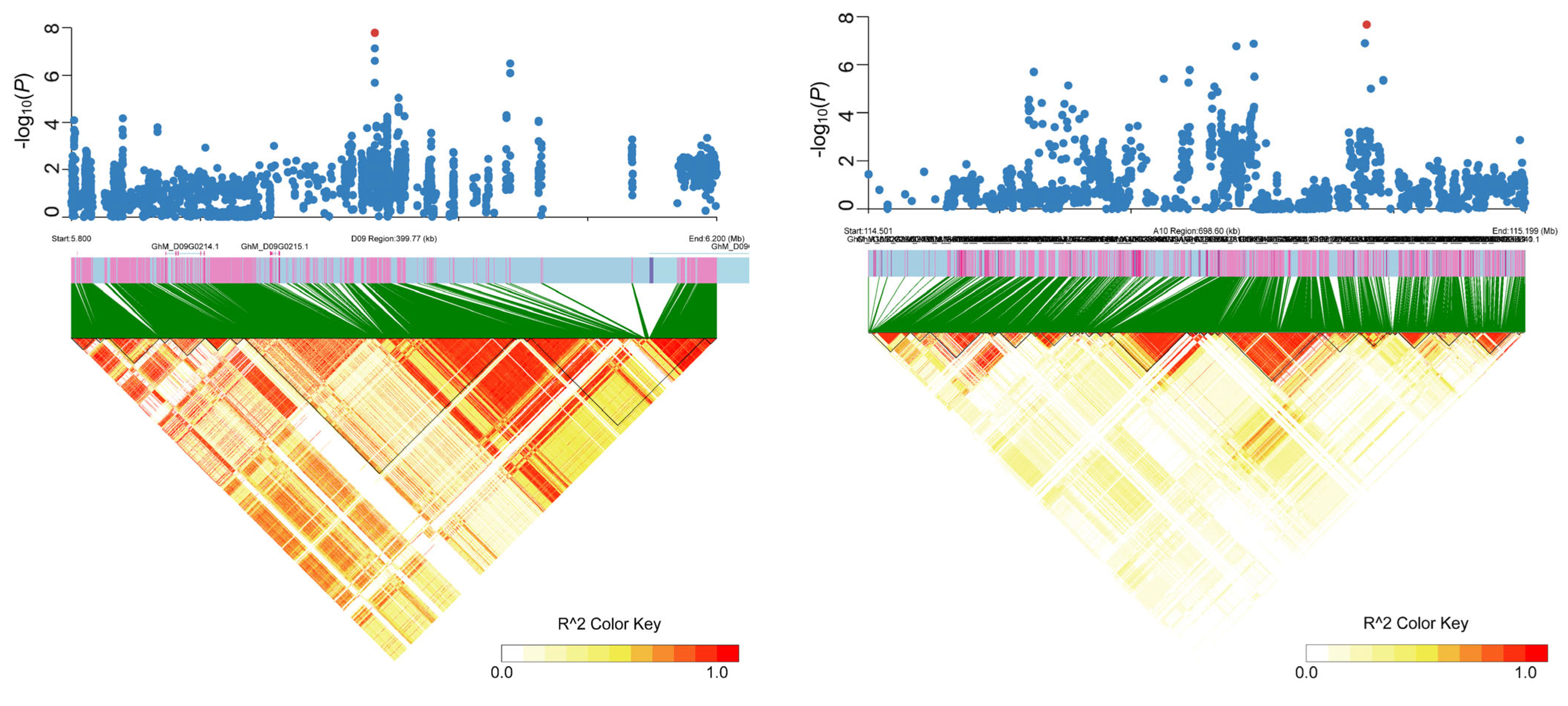
2.2. Identification of Genetic Loci for Related Traits
2.3. Candidate Gene Prediction
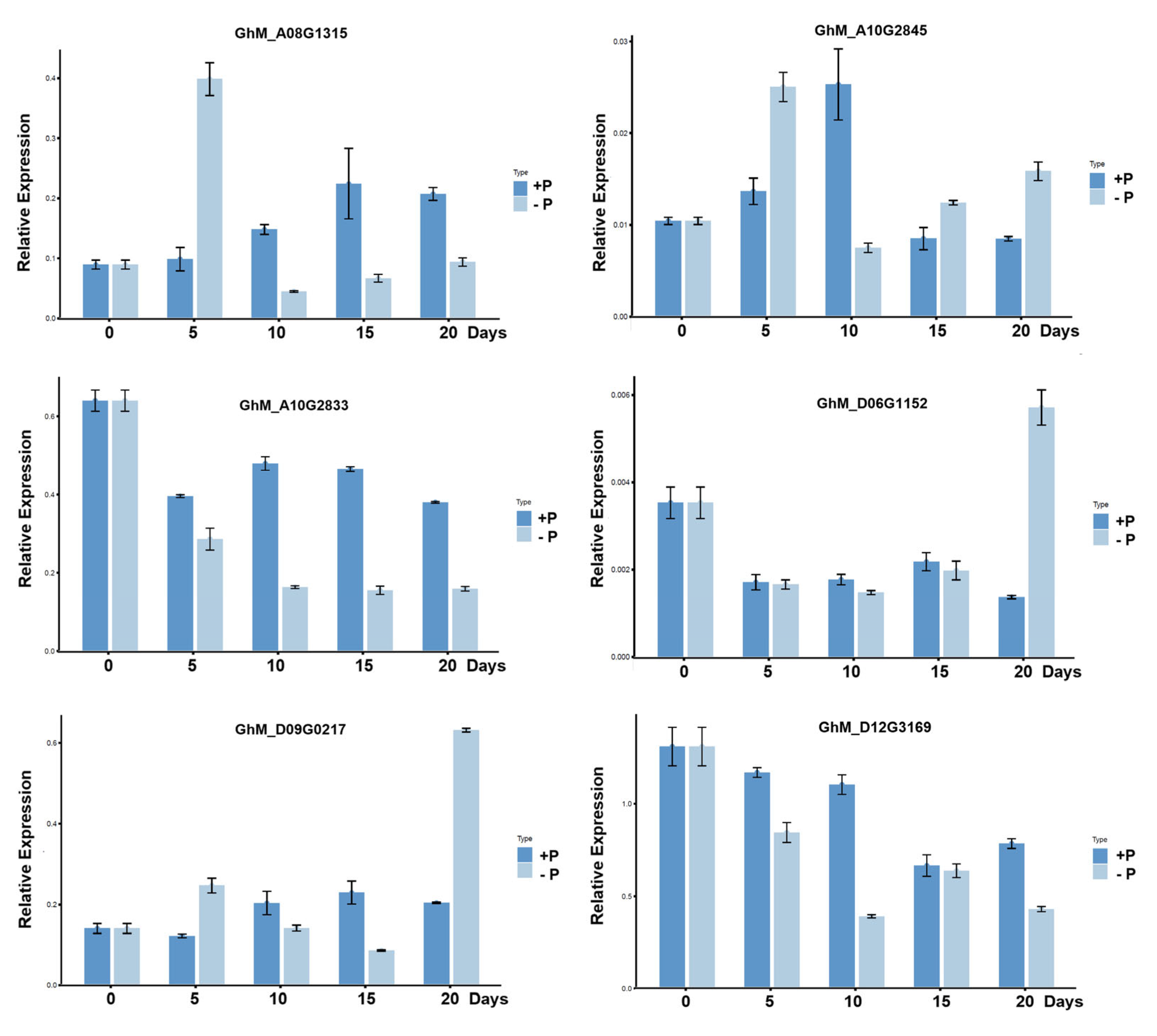
2.4. Validation of Candidate Gene Expression
3. Discussion
4. Materials and Methods
4.1. Materials and Experimental Processing
4.2. Phenotypic Identification
4.3. GWASs and Candidate Gene Identification
4.4. Candidate Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, A.; Brown, T. Global Cotton Production and Trade; Food and Agriculture Organization: Rome, Italy, 2020. [Google Scholar]
- Marschner, H. Mineral Nutrition of Higher Plants, 3rd ed.; Academic Press: London, UK, 2012. [Google Scholar]
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Alewell, C.; Ringeval, B.; Ballabio, C.; Robinson, D.A.; Panagos, P.; Borrelli, P. Global phosphorus shortage will be aggravated by soil erosion. Nat. Commun. 2020, 11, 4546. [Google Scholar] [CrossRef] [PubMed]
- Péret, B.; Desnos, T.; Jost, R.; Kanno, S.; Berkowitz, O.; Nussaume, L. Root architecture responses: In search of phosphate. Plant Physiol. 2014, 166, 1713–1723. [Google Scholar] [CrossRef]
- Wang, J.; Qin, Q.; Pan, J.; Sun, L.; Sun, Y.; Xue, Y.; Song, K. Transcriptome analysis in roots and leaves of wheat seedlings in response to low-phosphorus stress. Sci. Rep. 2019, 9, 19802. [Google Scholar] [CrossRef]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Yasir, M.; Kanwal, H.H.; Hussain, Q.; Riaz, M.W.; Sajjad, M.; Rong, J.; Jiang, Y. Status and prospects of genome-wide association studies in cotton. Front. Plant Sci. 2022, 13, 1019347. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, T.; Zhou, Y.; Yin, S.; Li, P.; Liu, J.; Xu, S.; Yang, Z.; Xu, C. Genome-wide association mapping of starch pasting properties in maize using single-locus and multi-locus models. Front. Plant Sci. 2018, 9, 1311. [Google Scholar] [CrossRef]
- Han, Z.; Ke, H.; Li, X.; Peng, R.; Zhai, D.; Xu, Y.; Wu, L.; Wang, W.; Cui, Y. Detection of epistasis interaction loci for fiber quality-related trait via 3VmrMLM in upland cotton. Front. Plant Sci. 2023, 14, 1250161. [Google Scholar] [CrossRef]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef]
- Marín, M.; Ott, T. Phosphorylation of intrinsically disordered regions in remorin proteins. Front. Plant Sci. 2012, 3, 86. [Google Scholar] [CrossRef] [PubMed]
- Lukowitz, W.; Nickle, T.C.; Meinke, D.W.; Last, R.L.; Conklin, P.L.; Somerville, C.R. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 2262–2267. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.J.; Chung, M.Y.; Lee, H.A.; Lee, S.B.; Grandillo, S.; Giovannoni, J.J.; Lee, J.M. Natural overexpression of CAROTENOID CLEAVAGE DIOXYGENASE 4 in tomato alters carotenoid flux. Plant Physiol. 2023, 192, 1289–1306. [Google Scholar] [CrossRef]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.Y. Regulation, Signaling, and Physiological Functions of G-Proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef]
- Iqbal, A.; Dong, Q.; Wang, X.; Gui, H.; Zhang, H.; Zhang, X.; Song, M. Phosphorus and carbohydrate metabolism contributes to low phosphorus tolerance in cotton. BMC Plant Biol. 2023, 23, 97. [Google Scholar] [CrossRef]
- Iqbal, B.; Kong, F.; Ullah, I.; Ali, S.; Li, H.; Wang, J.; Khattak, W.A.; Zhou, Z. Phosphorus application improves the cotton yield by enhancing reproductive organ biomass and nutrient accumulation in two cotton cultivars with different phosphorus sensitivity. Agronomy 2020, 10, 153. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Li, W.; Ku, L.; Wang, C.; Ye, J.; Li, K.; Yang, N.; Li, Y.; Zhong, T.; et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2013, 110, 16969–16974. [Google Scholar] [CrossRef]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Rouached, H.; Arpat, A.B.; Poirier, Y. Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 2010, 3, 288–299. [Google Scholar] [CrossRef]
- Nacry, P.; Canivenc, G.; Muller, B.; Azmi, A.; Van Onckelen, H.; Rossignol, M.; Doumas, P. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005, 138, 2061–2074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Jarsch, I.K.; Ott, T. Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant Microbe Interact. 2011, 24, 7–12. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, Y.; Wu, L.; Zhang, G.; Sun, Z.; Li, Z.; Jiang, Y.; Ke, H.; Chen, B.; Liu, Z.; et al. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement. Nat. Genet. 2021, 53, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Zhang, G.; Yu, T.; Zhang, C.; Yang, G.; Luo, X.; Zhang, S.; Guo, J.; Zhang, H.; Zheng, H.; et al. Genome-wide association studies dissect low-phosphorus stress response genes underlying seedling traits in maize. Theor. Appl. Genet. 2024, 137, 172. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]


| Trait | Max. | Min. | Mean | SD | CV | H2b |
|---|---|---|---|---|---|---|
| LP-PH (cm) | 8.52 | 2.73 | 4.75 | 0.93 | 19.65 | 0.90 |
| NP-PH (cm) | 13.84 | 3.54 | 7.57 | 1.65 | 21.80 | |
| LP-SDW (g) | 0.60 | 0.08 | 0.21 | 0.07 | 33.69 | 0.90 |
| NP-SDW (g) | 0.68 | 0.18 | 0.43 | 0.09 | 20.71 | |
| LP-RDW (g) | 0.53 | 0.16 | 0.34 | 0.07 | 19.75 | 0.76 |
| NP-RDW(g) | 0.77 | 0.22 | 0.44 | 0.09 | 19.86 | |
| LP-RSR | 4.03 | 0.78 | 1.73 | 0.50 | 28.76 | 0.75 |
| NP-RSR | 1.99 | 0.52 | 1.08 | 0.23 | 21.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Yao, S.; Di, J.; Guan, J.; Wang, A.; Yang, J.; Zhang, L.; Liu, Y.; Liang, M.; Niu, Z.; et al. Single- and Multi-Locus GWAS Unravels Novel Genomic Regions Related to Low-Phosphate Stress in Cotton Seedlings. Plants 2025, 14, 1803. https://doi.org/10.3390/plants14121803
Wei X, Yao S, Di J, Guan J, Wang A, Yang J, Zhang L, Liu Y, Liang M, Niu Z, et al. Single- and Multi-Locus GWAS Unravels Novel Genomic Regions Related to Low-Phosphate Stress in Cotton Seedlings. Plants. 2025; 14(12):1803. https://doi.org/10.3390/plants14121803
Chicago/Turabian StyleWei, Xianxu, Siyu Yao, Jiangnuo Di, Jiaxin Guan, Aohan Wang, Jie Yang, Luyao Zhang, Yang Liu, Mengyao Liang, Zhihao Niu, and et al. 2025. "Single- and Multi-Locus GWAS Unravels Novel Genomic Regions Related to Low-Phosphate Stress in Cotton Seedlings" Plants 14, no. 12: 1803. https://doi.org/10.3390/plants14121803
APA StyleWei, X., Yao, S., Di, J., Guan, J., Wang, A., Yang, J., Zhang, L., Liu, Y., Liang, M., Niu, Z., Zhang, X., Xue, J., Shen, M., Li, L., Su, Y., & Sun, Z. (2025). Single- and Multi-Locus GWAS Unravels Novel Genomic Regions Related to Low-Phosphate Stress in Cotton Seedlings. Plants, 14(12), 1803. https://doi.org/10.3390/plants14121803







