Genetic Characterization and Symbiotic Performance of Soybean Rhizobia Under Cold and Water-Deficient Conditions in Poland
Abstract
1. Introduction
2. Results
2.1. Isolation and Phenotypical Characterization of Rhizobia from Polish Soils
2.2. Environmental Stress Potential of Polish Rhizobia
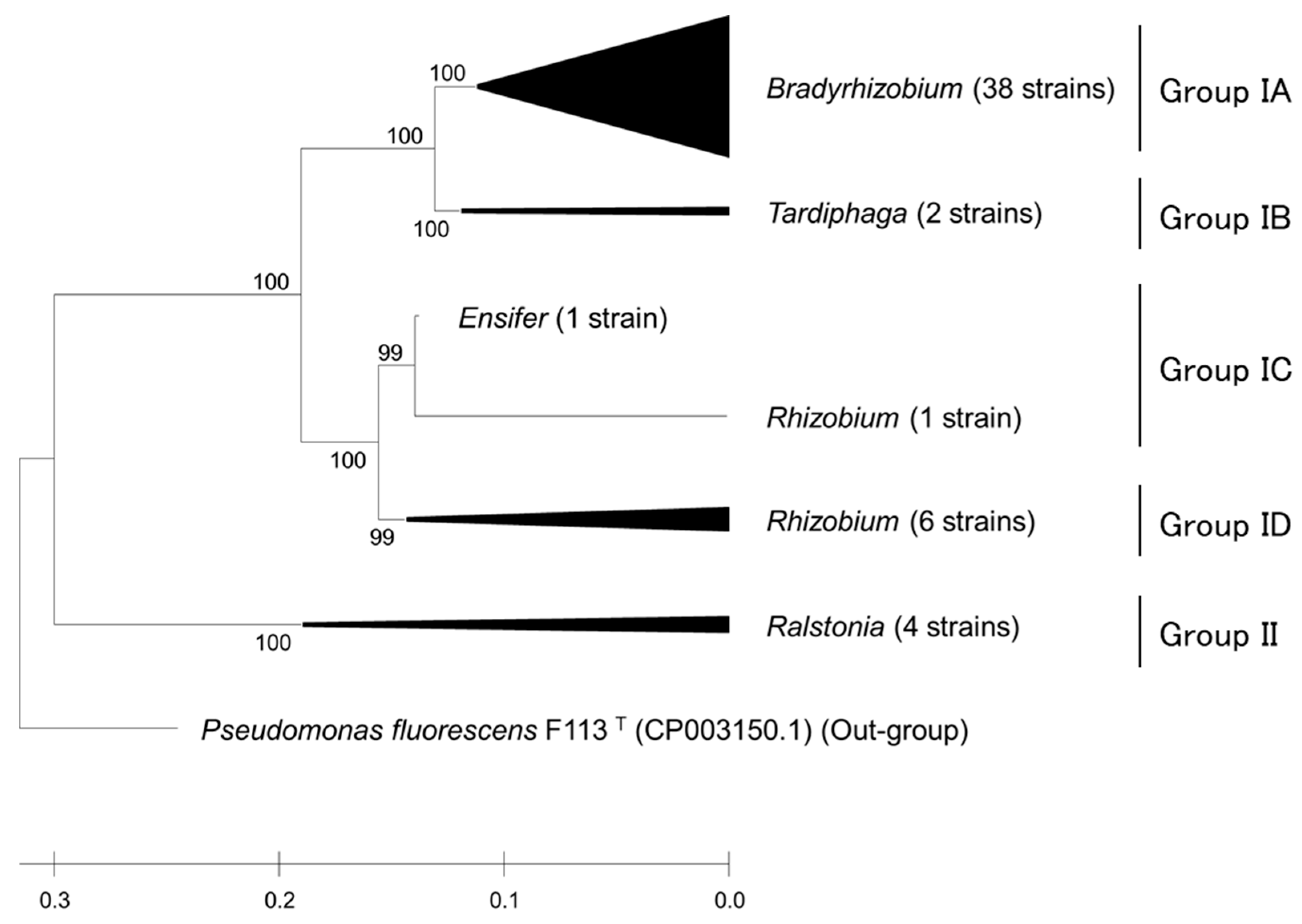
2.3. Phylogenetic Characterization of Rhizobia
2.4. Nitrogen Fixation-Associated Gene Sequencing and MSLT in Rhizobia
2.5. Plant Inoculation Assays
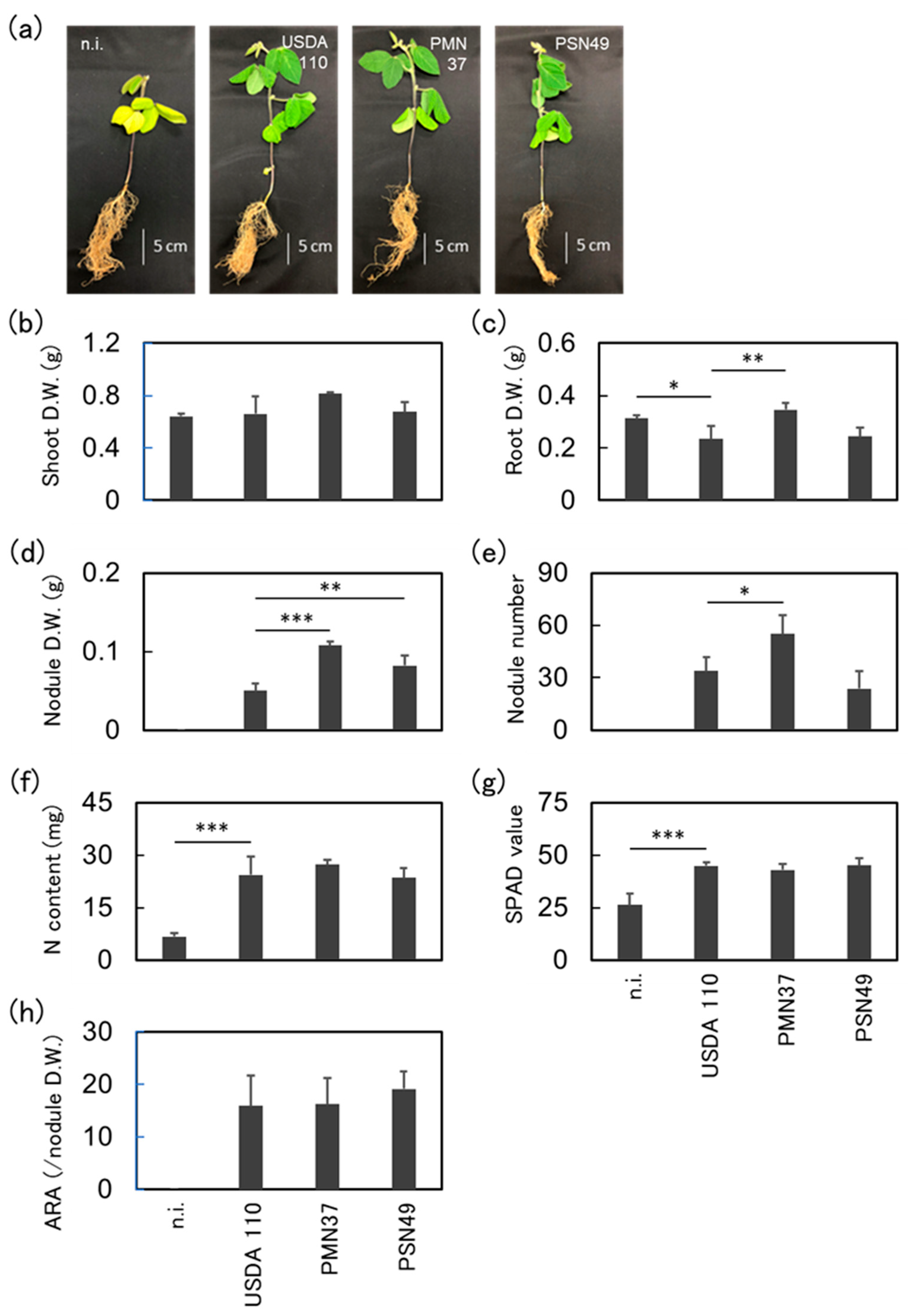
2.6. Plant Inoculation Assays Under Cold Conditions
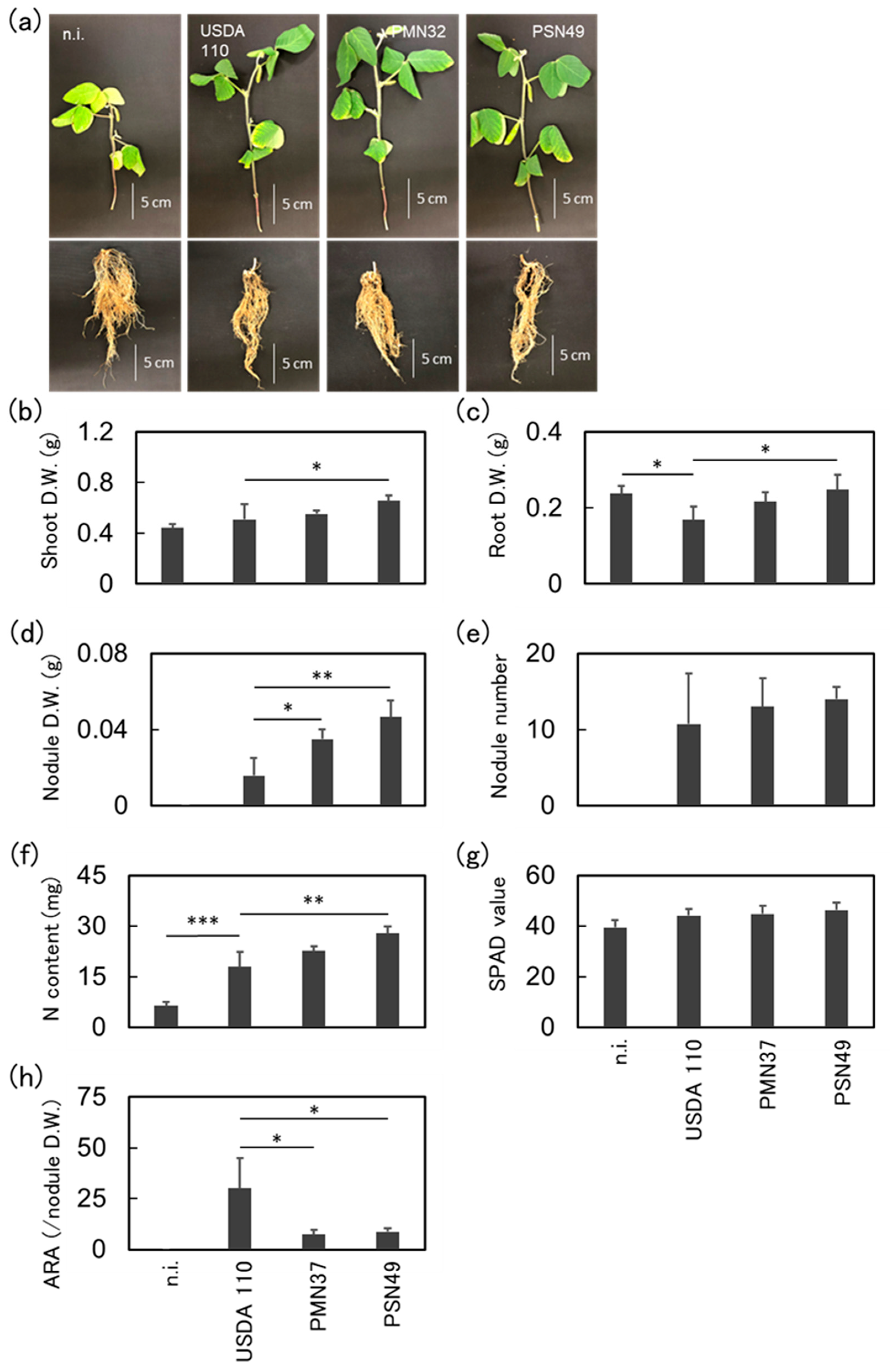
2.7. Plant Inoculation Assays Under Water-Deficient Conditions
2.8. Whole Genome Sequencing of PSN49
3. Discussion
3.1. Diversity of the Soybean Rhizobia Isolated from Polish Soil with Inoculant History
3.2. High Compatibility Between Abelina and Highly Osmotic-Tolerant Rhizobia
3.3. Phylogenetic Characteristics of Locally Adapted Bradyrhizobium Strains Contribute to Soybean Growth Under Abiotic Stress Conditions
3.4. Genomic Approach for Assessment of the Abiotic Stress Adaptation of PSN49
4. Materials and Methods
4.1. Isolation of Soybean Rhizobia from Polish Soils
4.2. Abiotic Stress Tolerance Assays
4.3. Genomic DNA Extraction
4.4. DNA Amplification and Sequencing of 16S rRNA and Symbiosis-Associated Genes
4.5. Multilocus Sequence Typing (MLST)
4.6. Plant Inoculation Assay
4.7. Whole Genome Sequencing of PSN49
4.8. Nucleotide Sequence Accession Numbers
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, F.J.; Chen, J.Q. Consumption of soybean, soy foods, soy isoflavones and breast cancer incidence: Differences between Chinese women and women in Western countries and possible mechanisms. Food Sci. Hum. Wellness 2013, 2, 146–161. [Google Scholar] [CrossRef]
- De Visser, L.C.M.; Schreuder, R.; Stoddard, F. Protein sources in animal feed. Les sources de protéines dans l’alimentation du bétail The EU’s dependency on soya bean import for the animal feed industry and potential for EU produced alternatives. OCL 2014, 21, D407. [Google Scholar] [CrossRef]
- Biermann, U.; Friedt, W.; Lang, S.; Lühs, W.; Machmüller, G.; Metzger, J.O.; Mark, M.R.; Schäfer, H.J.; Schneider, M.P. New Syntheses with Oils and Fats as Renewable Raw Materials for the Chemical Industry. Biorefineries-Ind. Process. Prod. Status Quo Future Dir. 2008, 2, 253–289. [Google Scholar] [CrossRef]
- Carrera, C.S.; Dardanelli, J.L.; Soldini, D.O. Chemical compounds related to nutraceutical and industrial qualities of non-transgenic soybean genotypes. J. Sci. Food Agric. 2014, 94, 1463–1469. [Google Scholar] [CrossRef]
- Zaheer, K.; Humayoun Akhtar, M. An updated review of dietary isoflavones: Nutrition, processing, bioavailability and impacts on human health. Crit. Rev. Food Sci. Nutr. 2017, 57, 1280–1293. [Google Scholar] [CrossRef]
- Messina, M. Soy and health update: Evaluation of the clinical and epidemiologic literature. Nutrients 2016, 8, 754. [Google Scholar] [CrossRef]
- Lewandowska, S.; Michalak, I. Effect of algal extracts on germination ability of different varieties of soybean. Seed and Seedlings. In Proceedings of the XIV Scientific and Technical Seminar, Prague, Czech Republic, 10–14 June 2019; pp. 84–90. [Google Scholar]
- Lewandowska, S. Perspectives of Soybean Cultivation in Poland. 2016. Available online: https://www.researchgate.net/publication/304898171_Perspectives_of_soybean_cultivation_in_Poland (accessed on 1 April 2025).
- Michalak, I.; Lewandowska, S.; Detyna, J.; Olsztyńska-Janus, S.; Bujak, H.; Pacholska, P. The Effect of Macroalgal Extracts and Near Infrared Radiation on Germination of Soybean Seedlings: Preliminary Research Results. Open Chem. 2018, 16, 1066–1076. [Google Scholar] [CrossRef]
- Niwińska, B.; Witaszek, K.; Niedbała, G.; Pilarski, K. Seeds of n-GM soybean varieties cultivated in Poland and their processing products as high-protein feeds in cattle nutrition. Agriculture 2020, 10, 174. [Google Scholar] [CrossRef]
- Serafin-Andrzejewska, M.; Jama-Rodzeńska, A.; Helios, W.; Kozak, M.; Lewandowska, S.; Zalewski, D.; Kotecki, A. Influence of nitrogen fertilization, seed inoculation and the synergistic effect of these treatments on soybean yields under conditions in south-western Poland. Sci. Rep. 2024, 14, 6672. [Google Scholar] [CrossRef]
- Miladinovic, J.; Kurosaki, H.; Burton, J.W.; Hrustic, M.; Miladinovic, D. The adaptability of shortseason soybean genotypes to varying longitudinal regions. Eur. J. Agron. 2006, 25, 243–249. [Google Scholar] [CrossRef]
- Risal, C.P.; Yokoyama, T.; Ohkama-Ohtsu, N.; Djedidi, S.; Sekimoto, H. Genetic diversity of native soybean bradyrhizobia from different topographical regions along the southern slopes of the Himalayan Mountains in Nepal. Syst. Appl. Microbiol. 2010, 33, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.; Andrews, M.E. Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef]
- De Meyer, S.E.; Briscoe, L.; Martínez-Hidalgo, P.; Agapakis, C.M.; De-Los Santos, P.E.; Seshadri, R.; Reeve, W.; Weinstock, G.; O’Hara, G.; Howieson, J.G.; et al. Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical High-Affinity cytochrome cbb3 Oxidase genes. Mol. Plant-Microbe Interact. 2016, 29, 609–619. [Google Scholar] [CrossRef]
- Ramírez, M.D.A.; España, M.; Aguirre, C.; Kojima, K.; Ohkama-Ohtsu, N.; Sekimoto, H.; Yokoyama, T. Burkholderia and Paraburkholderia are predominant soybean rhizobial genera in venezuelan soils in different climatic and topographical regions. Microbes Environ. 2019, 34, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.; Maillet, F.; Plazanet, C.; Debellé, F.; Ferro, M.; Truchet, G.; Promé, J.C.; Dénarié, J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc. Natl. Acad. Sci. USA 1996, 93, 15305–15310. [Google Scholar] [CrossRef]
- Buendía-Clavería, A.M.; Rodriguez-Navarro, D.N.; Santamaría-Linaza, C.; Ruiz-Saínza, J.E.; Temprano-Vera, F. Evaluation of the Symbiotic Properties of Rhizobium fredii in European Soils. Syst. Appl. Microbiol. 1994, 17, 155–160. [Google Scholar] [CrossRef]
- Vinuesa, P.; Rojas-Jiménez, K.; Contreras-Moreira, B.; Mahna, S.K.; Prasad, B.N.; Moe, H.; Selvaraju, S.B.; Thierfelder, H.; Werner, D. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the asiatic continent. Appl. Environ. Microbiol. 2008, 74, 6987–6996. [Google Scholar] [CrossRef]
- Rumjanek, N.G.; Dobert, R.C.; Van Berkum, P.; Triplett, E.W. Common soybean inoculant strains in Brazil are members of Bradyrhizobium elkanii. Appl. Environ. Microbiol. 1993, 59, 4371–4373. [Google Scholar] [CrossRef]
- Fatima, Z.; Zia, M.; Chaudhary, M.F. Effect of Rhizobium strains and phosphorus on growth of soybean (Glycine max) and survival of Rhizobium and P solubilizing bacteria. Pak. J. Bot. 2006, 38, 459–464. [Google Scholar]
- Liu, H.; Cui, Y.; Zhou, J.; Penttinen, P.; Liu, J.; Zeng, L.; Chen, Q.; Gu, Y.; Zou, L.; Zhao, K.; et al. Nickel mine soil is a potential source for soybean plant growth promoting and heavy metal tolerant rhizobia. PeerJ 2022, 10, e13215. [Google Scholar] [CrossRef]
- Soulemanov, A.; Prithiviraj, B.; Carlson, R.W.; Jeyaretnam, B.; Smith, D.L. Isolation and characterization of the major nod factor of Bradyrhizobium japonicum strain 532C. Microbiol. Res. 2002, 157, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Melchiorre, M.; de Luca, M.J.; Anta, G.G.; Suarez, P.; Lopez, C.; Lascano, R.; Racca, R.W. Evaluation of Bradyrhizobia strains isolated from field-grown soybean plants in Argentina as improved inoculants. Biol. Fertil. Soils 2011, 47, 81–89. [Google Scholar] [CrossRef]
- Omari, R.A.; Yuan, K.; Anh, K.T.; Reckling, M.; Halwani, M.; Egamberdieva, D.; Ohkama-Ohtsu, N.; Bellingrath-Kimura, S.D. Enhanced Soybean Productivity by Inoculation with Indigenous Bradyrhizobium Strains in Agroecological Conditions of Northeast Germany. Front. Plant Sci. 2022, 12, 707080. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Reckling, M.; Ramirez, M.D.A.; Djedidi, S.; Fukuhara, I.; Ohyama, T.; Yokoyama, T.; Bellingrath-Kimura, S.D.; Halwani, M.; Egamberdieva, D.; et al. Characterization of rhizobia for the improvement of soybean cultivation at cold conditions in central Europe. Microbes Environ. 2020, 35, ME19124. [Google Scholar] [CrossRef]
- Haegele, J.W.; Below, F.E. Transgenic corn rootworm protection increases grain yield and nitrogen use of maize. Crop Sci. 2013, 53, 585–594. [Google Scholar] [CrossRef]
- EAG volume 19 issue 4 Cover and Back matter. Exp. Agric. 1983, 19, b1–b8. [CrossRef]
- Szczerba, A.; Płażek, A.; Pastuszak, J.; Kopeć, P.; Hornyák, M.; Dubert, F. Effect of low temperature on germination, growth, and seed yield of four soybean (Glycine max L.) cultivars. Agronomy 2021, 11, 800. [Google Scholar] [CrossRef]
- Kabała, C.; Charzyński, P.; Chodorowski, J.; Drewnik, M.; Glina, B.; Greinert, A.; Hulisz, P.; Jankowski, M.; Jonczak, J.; Łabaz, B.; et al. Polish soil classification, 6th edition—Principles, classification scheme and correlations. Soil Sci. Annu. 2019, 70, 71–97. [Google Scholar] [CrossRef]
- Soil Survey Division Staff. Soil Survey Manual—Chapter 3 Examination and description of soils. Soil Surv. Man. 1993, 18, 46–155. Available online: http://soils.usda.gov/technical/manual/ (accessed on 1 April 2025).
- van Reeuwijk, L.P. Procedures for Soil Analysis; International Soil Reference and Information Centre: Wageningen, The Netherlands, 2002. [Google Scholar]
- Griebsch, A.; Matschiavelli, N.; Lewandowska, S.; Schmidtke, K. Presence of Bradyrhizobium sp. Under continental conditions in central europe. Agriculture 2020, 10, 446. [Google Scholar] [CrossRef]
- Narozna, D.; Pudełko, K.; Króliczak, J.; Golińska, B.; Sugawara, M.; Madrzak, C.J.; Sadowsky, M.J. Survival and competitiveness of Bradyrhizobium japonicum strains 20 years after introduction into field locations in Poland. Appl. Environ. Microbiol. 2015, 81, 5552–5559. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Chen, H.; Yang, Z.; Yuan, S.; Zhou, X. Research status of soybean symbiosis nitrogen fixation. Oil Crop Sci. 2020, 5, 6–10. [Google Scholar] [CrossRef]
- Rymuza, K.; Radzka, E.; Wysokiński, A. Nitrogen uptake from different sources by Non-GMO soybean varieties. Agronomy 2020, 10, 1219. [Google Scholar] [CrossRef]
- Thilakarathna, M.S.; Raizada, M.N. A meta-analysis of the effectiveness of diverse rhizobia inoculants on soybean traits under field conditions. Soil Biol. Biochem. 2017, 105, 177–196. [Google Scholar] [CrossRef]
- Caldwell, B.E.; Vest, G. Effects of Rhizobium japonicum Strains on Soybean Yields. Crop Sci. 1970, 10, 19–21. [Google Scholar] [CrossRef]
- Kvien, C.S.; Ham, G.E.; Lambert, J.W. Recovery of Introduced Rhizobium japonicum Strains by Soybean Genotypes 1. Agron. J. 1981, 73, 900–905. [Google Scholar] [CrossRef]
- Moawad, M.; Schmidt, E.L. Occurrence and nature of mixed infections in nodules of field-grown soybeans (Glycine max). Biol. Fertil. Soils 1987, 5, 112–114. [Google Scholar] [CrossRef]
- Madrzak, C.J.; Golinska, B.; Kroliczak, J.; Pudelko, K.; Lazewska, D.; Lampka, B.; Sadowsky, M.J. Diversity among field populations of Bradyrhizobium japonicum in Poland. Appl. Environ. Microbiol. 1995, 61, 1194–1200. [Google Scholar] [CrossRef]
- Streeter, J.G.; Salminen, S.O.; Whitmoyer, R.E.; Carlson, R.W. Formation of novel polysaccharides by Bradyrhizobium japonicum bacteroids in soybean nodules. Appl. Environ. Microbiol. 1992, 58, 607–613. [Google Scholar] [CrossRef]
- Shiro, S.; Kuranaga, C.; Yamamoto, A.; Sameshima-Saito, R.; Saeki, Y. Temperature-dependent expression of NODC and community structure of soybean-nodulating Bradyrhizobia. Microbes Environ. 2016, 31, 27–32. [Google Scholar] [CrossRef]
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial adaptation to cold. Microbiology 2013, 159 Pt 12, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gross, C.A. Cold Shock Response in Bacteria. Annu. Rev. Genet. 2021, 55, 377–400. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, H.; Jiang, W.; Inouye, M.; Heinemann, U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 5119–5123. [Google Scholar] [CrossRef]
- Jiang, W.; Hou, Y.; Inouye, M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997, 272, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Severinov, K. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 2005, 33, 5583–5590. [Google Scholar] [CrossRef]
- Gualerzi, C.O.; Giuliodori, A.M.; Pon, C.L. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003, 331, 527–539. [Google Scholar] [CrossRef]
- Teana, A.L.A.; Brandi, A.; Falconi, M.; Spurio, R.; Pon, C.L.; Gualerzi, C.O. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. USA 1991, 88, 10907–10911. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.G.; Krah, R.; Tafuri, S.R.; Wolffe, A.P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 1992, 174, 5798–5802. [Google Scholar] [CrossRef]
- Zhang, H.; Charles, T.C.; Driscoll, B.T.; Prithiviraj, B.; Smith, D.L. Low temperature-tolerant Bradyrhizobium japonicum strains allowing improved soybean yield in short-season areas. Agron. J. 2002, 94, 870–875. [Google Scholar] [CrossRef]
- Zhang, H.; Prithiviraj, B.; Charles, T.C.; Driscoll, B.T.; Smith, D.L. Low temperature tolerant Bradyrhizobium japonicum strains allowing improved nodulation and nitrogen fixation of soybean in a short season (cool spring) area. Eur. J. Agron. 2003, 19, 205–213. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Z.; Liu, H.; Yue, L.; Wang, F.; Liu, S.; Su, B.; Liu, B.; Kong, F.; Fang, C. Genome-Wide Identification and Characterization of the Soybean Snf2 Gene Family and Expression Response to Rhizobia. Int. J. Mol. Sci. 2023, 24, 7250. [Google Scholar] [CrossRef]
- Jiang, X.; Keto-Timonen, R.; Skurnik, M.; Korkeala, H. Role of DEAD-box RNA helicase genes in the growth of Yersinia pseudotuberculosis IP32953 under cold, pH, osmotic, ethanol and oxidative stresses. PLoS ONE 2019, 14, e0219422. [Google Scholar] [CrossRef]
- Cytryn, E.J.; Sangurdekar, D.P.; Streeter, J.G.; Franck, W.L.; Chang, W.S.; Stacey, G.; Emerich, D.W.; Joshi, T.; Xu, D.; Sadowsky, M.J. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 2007, 189, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.A.; Tuteja, N. Stress responsive DEAD-box helicases: A new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. B Biol. 2006, 84, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Ha, J.; Lee, H.; Lee, S.; Lee, J.; Choi, Y.; Oh, H.; Yoon, Y.; Choi, K.H. Role of Pseudomonas aeruginosa DesB in adaptation to osmotic stress. J. Food Prot. 2019, 82, 1278–1282. [Google Scholar] [CrossRef]
- Aguilar, P.S.; Hernandez-Arriaga, A.M.; Cybulski, L.E.; Erazo, A.C.; De Mendoza, D. Molecular basis of thermosensing: A two-component signal transduction thermometer in bacillus subtilis. EMBO J. 2001, 20, 1681–1691. [Google Scholar] [CrossRef]
- Sugawara, M.; Cytryn, E.J.; Sadowsky, M.J. Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation. Appl. Environ. Microbiol. 2010, 76, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Streeter, J.G. Effect of trehalose on survival of Bradyrhizobium japonicum during desiccation. J. Appl. Microbiol. 2003, 95, 484–491. [Google Scholar] [CrossRef]
- Streeter, J.G. Factors affecting the survival of Bradyrhizobium applied in liquid cultures to soya bean [Glycine max (L.) Merr.] seeds. J. Appl. Microbiol. 2007, 103, 1282–1290. [Google Scholar] [CrossRef]
- Streeter, J.G.; Gomez, M.L. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 2006, 72, 4250–4255. [Google Scholar] [CrossRef]
- Zézé, A.; Mutch, L.A.; Young, J.P.W. Direct amplification of nodD from community DNA reveals the genetic diversity of Rhizobium leguminosarum in soil. Environ. Microbiol. 2001, 3, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, G.; Nour, S.M.; Macheret, V.; Sanjuan, J.; Drouin, P.; Amarger, N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 2001, 147, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Widmer, F.; Shaffer, B.T.; Porteous, L.A.; Seidler, R.J. Analysis of nifH gene pool complexity in soil and litter at a douglas fir forest site in the Oregon cascade mountain range. Appl. Environ. Microbiol. 1999, 65, 374–380. [Google Scholar] [CrossRef]
- Del Cerro, P.; Megías, M.; López-Baena, F.J.; Gil-Serrano, A.; Pérez-Montaño, F.; Ollero, F.J. Osmotic stress activates nif and fix genes and induces the Rhizobium tropici CIAT 899 Nod factor production via NodD2 by up-regulation of the nodA2 operon and the nodA3 gene. PLoS ONE 2019, 14, e0213298. [Google Scholar] [CrossRef]
- Gaunt, M.W.; Turner, S.L.; Rigottier-Gois, L.; Lloyd-Macgilp, S.A.; Young, J.P.W. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 2001, 51, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.W.; Vandamme, P.; Morgan, S.H.; LiPuma, J.J.; Coenye, T.; Weightman, A.J.; Jones, T.H.; Mahenthiralingam, E. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 2005, 71, 3917–3927. [Google Scholar] [CrossRef]
- Martens, M.; Delaere, M.; Coopman, R.; De Vos, P.; Gillis, M.; Willems, A. Multilocus sequence analysis of Ensifer and related taxa. Int. J. Syst. Evol. Microbiol. 2007, 57, 489–503. [Google Scholar] [CrossRef]
- Mohkam, M.; Nezafat, N.; Berenjian, A.; Mobasher, M.A.; Ghasemi, Y. Identification of Bacillus Probiotics Isolated from Soil Rhizosphere Using 16S rRNA, recA, rpoB Gene Sequencing and RAPD-PCR. Probiotics Antimicrob. Proteins 2016, 8, 8–18. [Google Scholar] [CrossRef]
- Nzoué, A.; Miché, L.; Klonowska, A.; Laguerre, G.; de Lajudie, P.; Moulin, L. Multilocus sequence analysis of Bradyrhizobia isolated from Aeschynomene species in Senegal. Syst. Appl. Microbiol. 2009, 32, 400–412. [Google Scholar] [CrossRef]
- Khamis, A.; Colson, P.; Raoult, D.; La Scola, B. Usefulness of rpoB Gene Sequencing for Identification of Afipia and Bosea Species, Including a Strategy for Choosing Discriminative Partial Sequences. Appl. Environ. Microbiol. 2003, 69, 6740–6749. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Kim, J.; Na, S.I.; Kim, D.; Chun, J. UBCG2: Up-to-date bacterial core genes and pipeline for phylogenomic analysis. J. Microbiol. 2021, 59, 609–615. [Google Scholar] [CrossRef]
- Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [CrossRef]



| Soil Sample No. | Site Name | Soil Sample Collection Date | Soil Gravity Categories | Clay (%) (<0.02 mm) | Silt (%) (0.1–0.02 mm) | Sand (%) (1–0.1 mm) | Crop Rotation History (Winter–Spring–Autumn–Summer) | Soybean Harvest Date | pH |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NOWY ROŻNÓW | 15 November 2019 | Light | 39 | 45 | 16 | Spring wheat–oilseed rape– winter wheat–maize | - | 6.3 |
| 2 | PAWŁOWICE | 12 November 2019 | Light | 20 | 15 | 65 | Winter oilseed rape–winter wheat– winter oilseed rape–winter wheat | - | 7.9 |
| 3 | WROCŁAW | 14 November 2019 | Medium | 25 | 18 | 57 | Oat–winter oilseed rape–winter wheat–winter oilseed rape–winter wheat | - | 6.2 |
| 4 | PAWŁOWICE | 14 November 2019 | Medium | 23 | 15 | 62 | Winter oilseed rape–winter wheat– winter oilseed rape–winter wheat | - | 6.5 |
| 5 | PAWŁOWICE | 21 November 2019 | Light | 16 | 14 | 70 | Oat–winter triticale–flax– winter oilseed rape–winter triticale | - | 7.6 |
| 6 | TARNÓW | 26 November 2019 | Heavy | 40 | 45 | 15 | Winter wheat–sugar beet– spring barley–winter oilseed rape | - | 6.2 |
| 7 | TARNÓW | 27 November 2019 | Heavy | 41 | 48 | 11 | Spring barley–winter oilseed rape– winter wheat–sugar beet | - | 6.3 |
| 8 | TARNÓW | 28 November 2019 | Heavy | 43 | 42 | 15 | Sugar beet–spring barley– winter oilseed rape–winter wheat | - | 6.1 |
| 9 | TARNÓW | 26 November 2019 | Heavy | 45 | 41 | 14 | Spring barley–vegetable– spring barley–vegetable | - | 6.5 |
| 10 | TARNÓW | 20 November 2019 | Heavy | 47 | 43 | 10 | Soybean–winter wheat– potatoes–soybean | 2019 | 6.3 |
| 11 | TARNÓW | 17 November 2019 | Heavy | 42 | 47 | 11 | Winter wheat–winter oilseed rape– winter wheat–soybean | 2019 | 6.2 |
| 12 | TARNÓW | 13 November 2019 | Heavy | 44 | 42 | 14 | Soybean–spring wheat– vegetable–winter wheat | 2016 | 6.1 |
| 13 | TARNÓW | 8 November 2019 | Heavy | 39 | 46 | 15 | Maize–winter wheat–vegetable– spring wheat–soybean–vegetable– winter wheat–vegetable | 2015 | 6.6 |
| 14 | NOWY ROŻNÓW | 8 November 2019 | Heavy | 38 | 47 | 15 | Sugar beet–winter wheat– oilseed rape–winter wheat– soybean–maize–spring wheat | 2017 | 6.5 |
| 15 | NOWY ROŻNÓW | 5 November 2019 | Heavy | 36 | 51 | 13 | Sugar beet–winter wheat– oilseed rape–winter wheat– soybean–maize–spring wheat | 2016 | 6.4 |
| 16 | NOWY ROŻNÓW | 6 November 2019 | Heavy | 37 | 49 | 14 | Sugar beet–winter wheat–oilseed rape– winter wheat–soybean | 2019 | 6.7 |
| 17 | NOWY ROŻNÓW | 6 November 2019 | Heavy | 38 | 48 | 14 | Sugar beet–winter wheat–oilseed rape– winter wheat–soybean–maize | 2018 | 6.5 |
| 18 | NOWE GOLUSZOWICE | 7 November 2019 | Heavy | 36 | 41 | 23 | Winter wheat–oilseed rape– winter wheat–soybean | 2019 | 6.7 |
| 19 | NOWY ROŻNÓW | 4 November 2019 | Heavy | 38 | 42 | 20 | Winter wheat–oilseed rape– winter wheat–soybean | 2019 | 6.5 |
| Strain | Classification Based on MLST | Whole Plant Dry Weight (g) | Shoot Dry Weight (g) | Root Dry Weight (g) | Number of Nodules | Nodule Dry Weight (g) | ARA (/Nodule DW) | ARA (/Plant DW) |
|---|---|---|---|---|---|---|---|---|
| Non-inoculation (negative control) | 0.966 ± 0.147 * | 0.628 ± 0.064 ** | 0.338 ± 0.022 | 0.00 | - | 0.00 | 0.00 | |
| USDA 110 (positive control) | 1.643 ± 0.156 | 1.352 ± 0.077 | 0.291 ± 0.019 | 33.00 ± 4.73 | 0.056 ± 0.003 | 16.40 ± 2.26 | 0.57 ± 0.12 | |
| PAN18 | Bradyrhizobium japonicum | 1.684 ± 0.172 | 1.326 ± 0.092 | 0.358 ± 0.027 | 39.67 ± 8.41 | 0.094 ± 0.019 | 8.04 ± 1.61 | 0.47 ± 0.15 |
| PMN30 | Bradyrhizobium japonicum | 1.379 ± 0.272 | 1.045 ± 0.121 | 0.334 ± 0.040 | 49.67 ± 9.40 | 0.088 ± 0.011 | 7.88 ± 0.15 | 0.50 ± 0.02 |
| PMN35 | Bradyrhizobium japonicum | 1.604 ± 0.217 | 1.238 ± 0.113 | 0.366 ± 0.015 | 47.00 ± 3.46 | 0.116 ± 0.004 * | 7.85 ± 1.76 | 0.58 ± 0.16 |
| PMN37 | Bradyrhizobium japonicum | 1.749 ± 0.226 | 1.297 ± 0.113 | 0.453 ± 0.018 * | 51.67 ± 3.18 | 0.123 ± 0.015 ** | 9.11 ± 2.06 | 0.66 ± 0.20 |
| PSN49 | Bradyrhizobium japonicum | 1.921 ± 0.529 | 1.433 ± 0.219 | 0.489 ± 0.090 ** | 37.67 ± 3.33 | 0.108 ± 0.016 | 11.16 ± 3.32 | 0.58 ± 0.08 |
| PAT4 | Rhizobium sp. | 1.583 ± 0.078 | 1.163 ± 0.060 | 0.420 ± 0.020 | 45.67 ± 7.51 | 0.087 ± 0.027 | 21.14 ± 9.83 | 0.85 ± 0.19 |
| PAN13 | Rhizobium sp. | 1.620 ± 0.351 | 1.221 ± 0.163 | 0.399 ± 0.040 | 46.67 ± 1.67 | 0.103 ± 0.011 | 7.35 ± 1.30 | 0.47 ± 0.06 |
| PMN31 | Rhizobium sp. | 1.122 ± 0.250 | 0.830 ± 0.118 * | 0.292 ± 0.027 | 59.00 ± 6.03 | 0.089 ± 0.011 | 14.14 ± 3.70 | 1.09 ± 0.24 |
| PAN21 | Ensifer sp. | 1.013 ± 0.145 * | 0.748 ± 0.040 ** | 0.265 ± 0.054 | 75.33 ± 10.17 ** | 0.049 ± 0.003 | 12.22 ± 2.64 | 0.58 ± 0.09 |
| Strain | Bradyrhizobium japonicum PSN49 | Bradyrhizobium japonicum USDA 123 | Bradyrhizobium japonicum USDA 6T | Bradyrhizobium diazoefficiens USDA 110T |
|---|---|---|---|---|
| Total length | 11,082,922 | 10,457,665 | 9,207,384 | 9,106,064 |
| GC (%) | 63.23 | 63.27 | 63.67 | 64.06 |
| CDS | 10,931 | 10,253 | 8421 | 8586 |
| rRNA | 6 | 3 | 6 | 3 |
| tRNA | 61 | 59 | 58 | 55 |
| Accession number | CP187249–CP187256 | GCA_000482525.1 | GCA_000284375.1 | GCA_001642675.1 |
| OrthoANI value (%) to USDA 123 | 99.77 | - | - | - |
| OrthoANI value (%) to USDA 6 | 95.46 | 95.40 | - | - |
| OrthoANI value (%) to USDA 110 | 89.53 | 89.40 | 90.15 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, R.; Artigas Ramirez, M.D.; Agake, S.-i.; Bellingrath-Kimura, S.D.; Lewandowska, S.; Onishi, Y.; Nishikawa, Y.; Takeyama, H.; Yasuda, M.; Ohkama-Ohtsu, N. Genetic Characterization and Symbiotic Performance of Soybean Rhizobia Under Cold and Water-Deficient Conditions in Poland. Plants 2025, 14, 1786. https://doi.org/10.3390/plants14121786
Watanabe R, Artigas Ramirez MD, Agake S-i, Bellingrath-Kimura SD, Lewandowska S, Onishi Y, Nishikawa Y, Takeyama H, Yasuda M, Ohkama-Ohtsu N. Genetic Characterization and Symbiotic Performance of Soybean Rhizobia Under Cold and Water-Deficient Conditions in Poland. Plants. 2025; 14(12):1786. https://doi.org/10.3390/plants14121786
Chicago/Turabian StyleWatanabe, Riku, Maria Daniela Artigas Ramirez, Shin-ichiro Agake, Sonoko Dorothea Bellingrath-Kimura, Sylwia Lewandowska, Yuki Onishi, Yohei Nishikawa, Haruko Takeyama, Michiko Yasuda, and Naoko Ohkama-Ohtsu. 2025. "Genetic Characterization and Symbiotic Performance of Soybean Rhizobia Under Cold and Water-Deficient Conditions in Poland" Plants 14, no. 12: 1786. https://doi.org/10.3390/plants14121786
APA StyleWatanabe, R., Artigas Ramirez, M. D., Agake, S.-i., Bellingrath-Kimura, S. D., Lewandowska, S., Onishi, Y., Nishikawa, Y., Takeyama, H., Yasuda, M., & Ohkama-Ohtsu, N. (2025). Genetic Characterization and Symbiotic Performance of Soybean Rhizobia Under Cold and Water-Deficient Conditions in Poland. Plants, 14(12), 1786. https://doi.org/10.3390/plants14121786







