The Toxicity Effects of Metformin and the Bioremediation of Metformin in Aquatic Plant Duckweed
Abstract
1. Introduction
2. Results
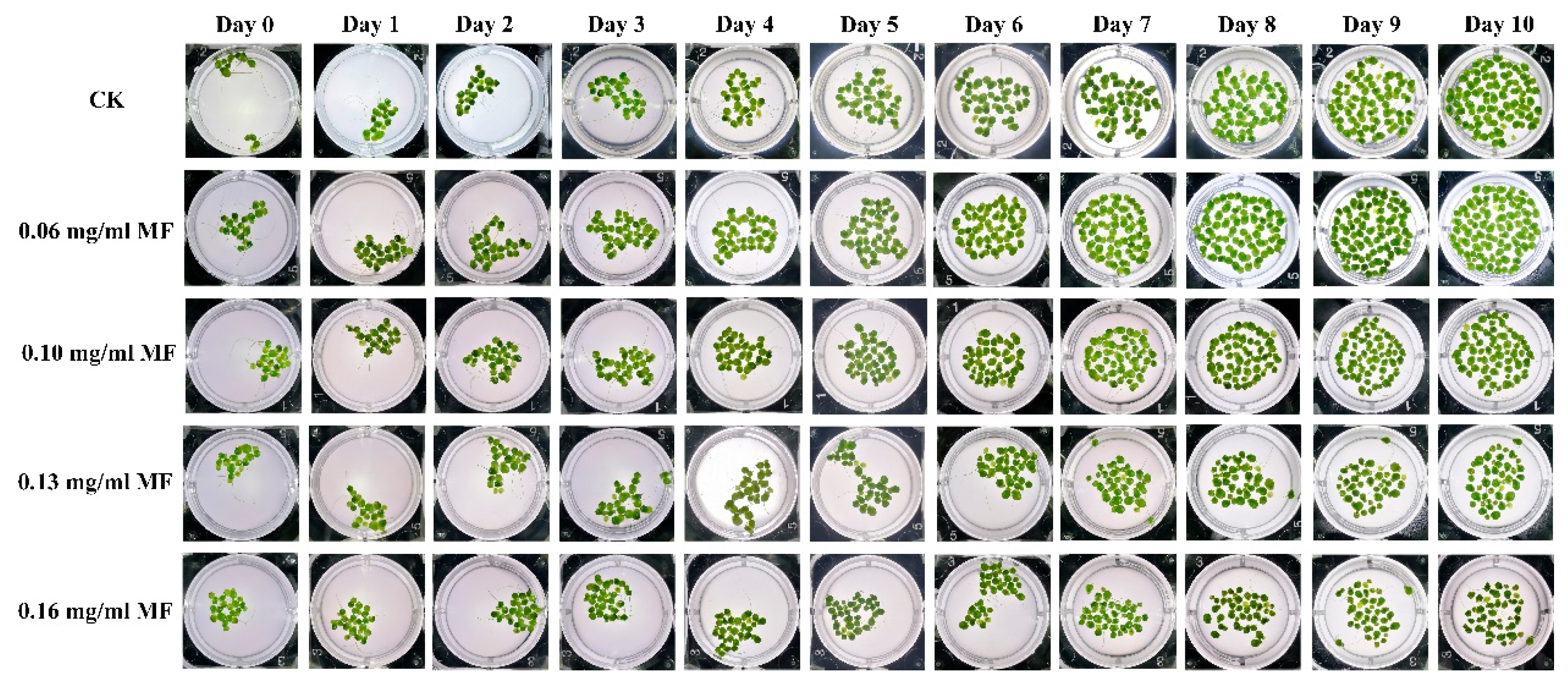
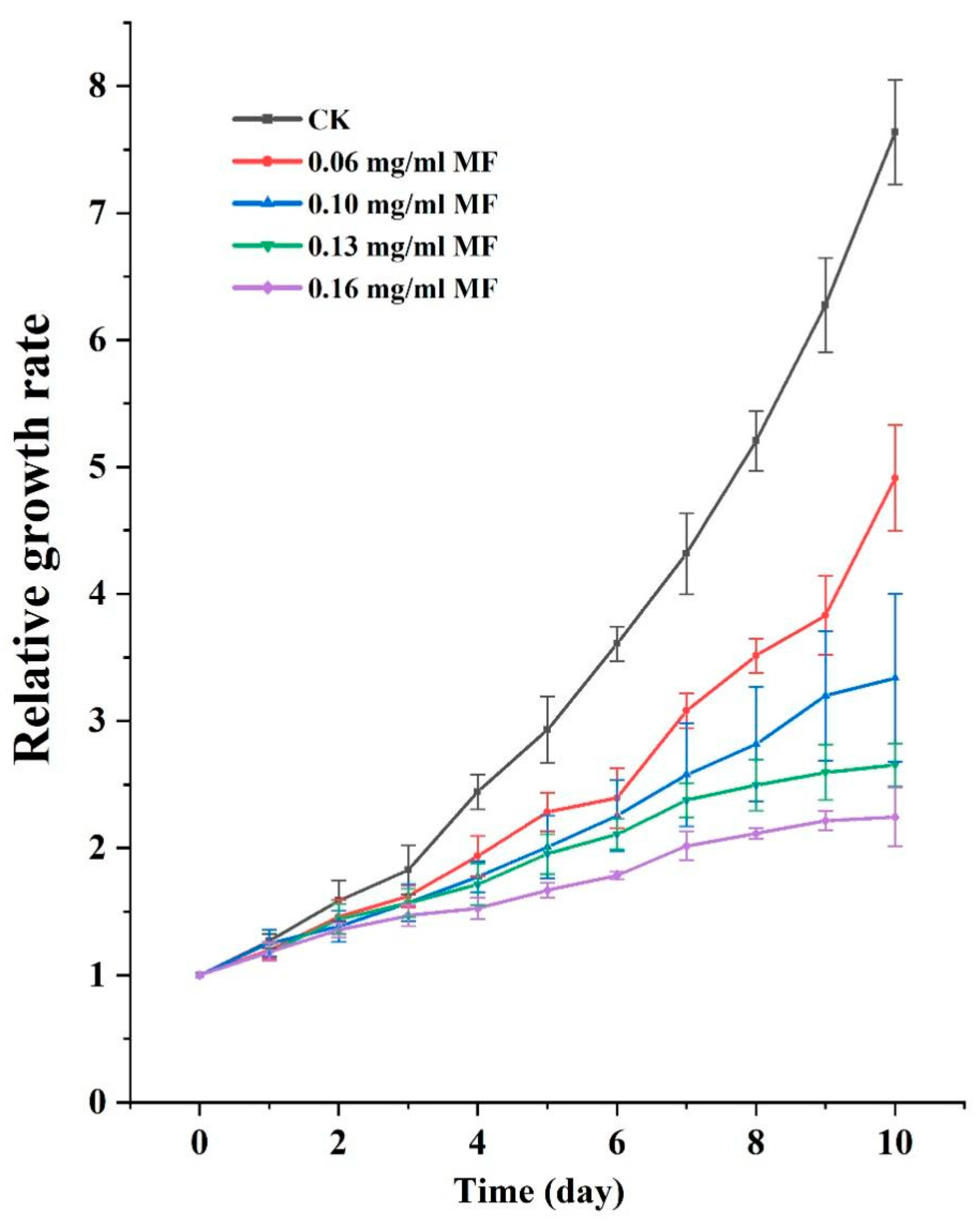
2.1. MF Exposure Inhibited Duckweed Growth
2.2. Photosynthetic Efficiency Was Affected by MF
2.3. The Expression of Critical Genes Involved in Photosynthesis and Photosynthesis—Antenna Pathways Were Downregulated Due to MF Exposure
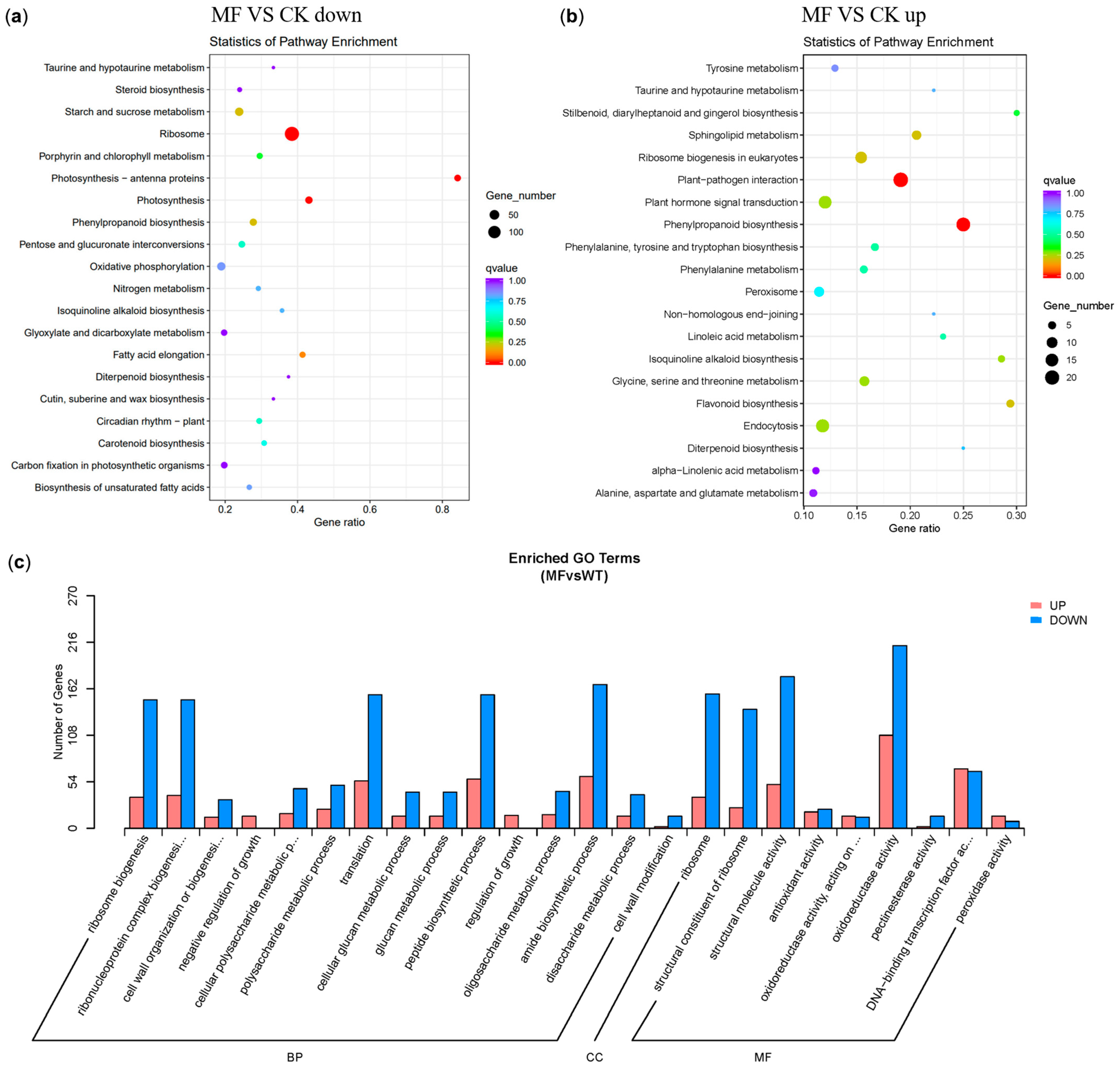
2.4. Differentially Expressed Genes (DEGs) After MF Treatment
2.5. Gene Ontology and KEGG Pathway Analyses of DEGs
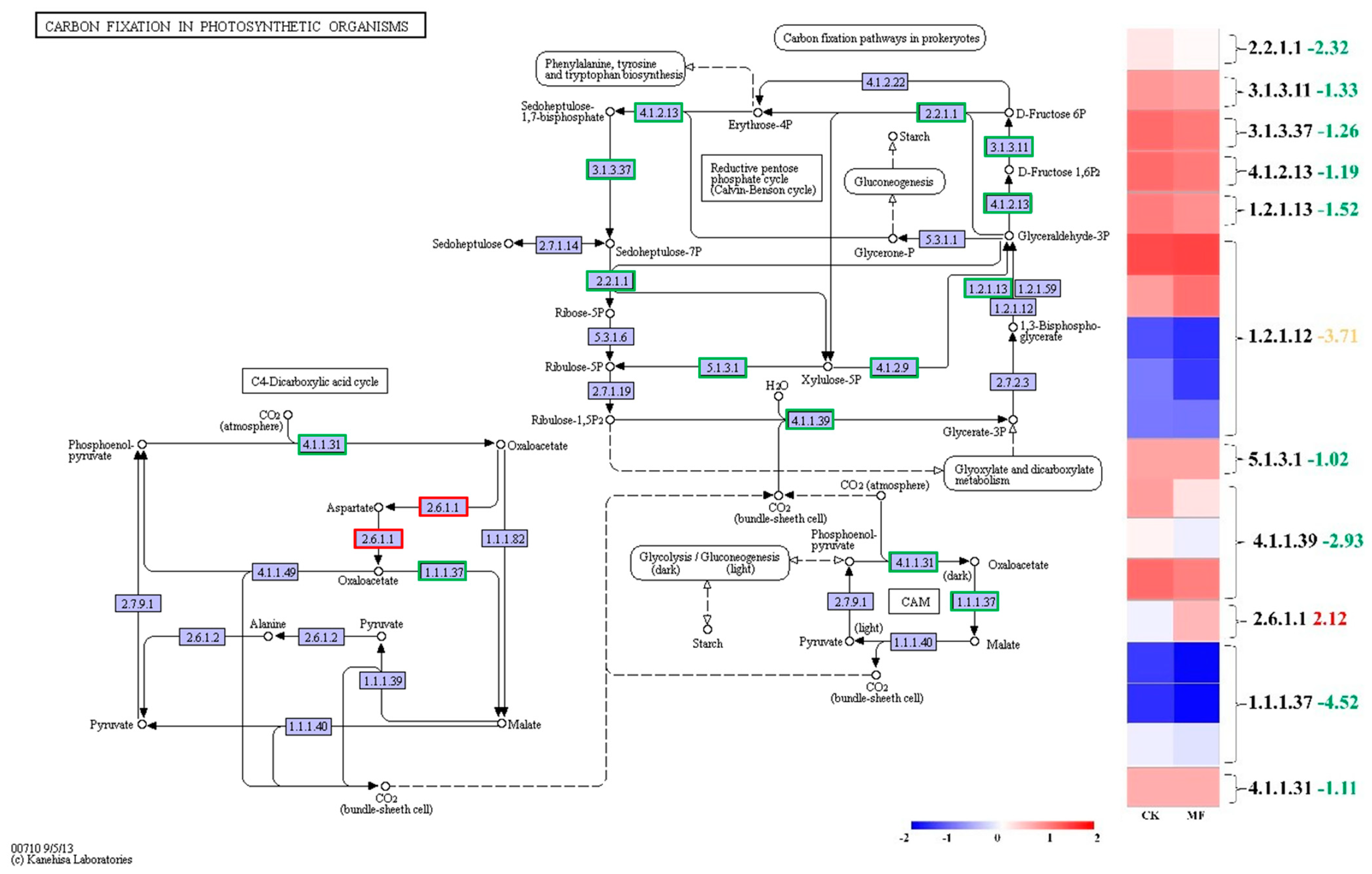
2.6. MF Suppressed Photosynthetic Carbon Fixation
2.7. MF Modified Signal Transduction of Auxin and Abscisic Acid
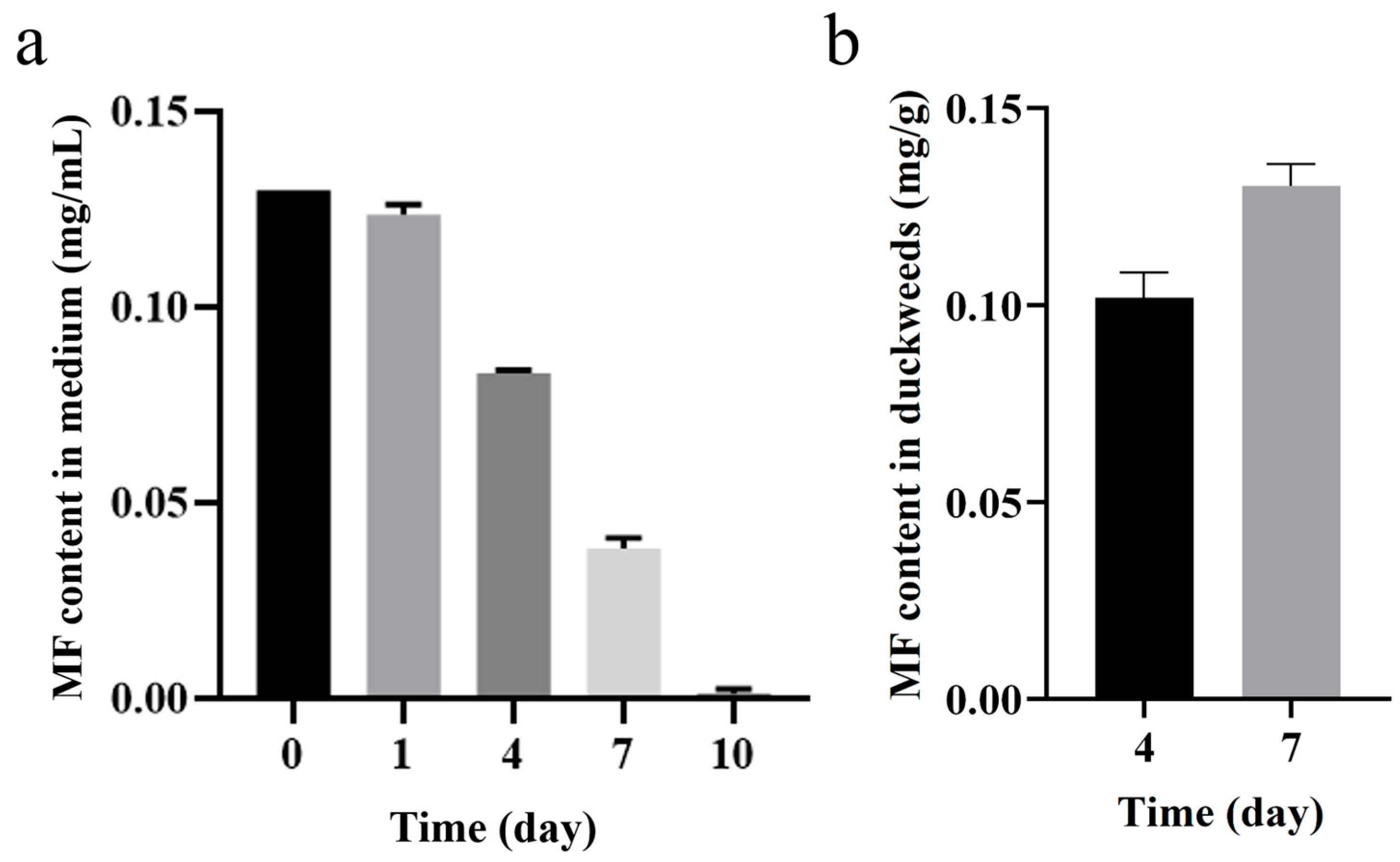
2.8. Analysis of MF Enrichment by Duckweed
3. Discussion and Conclusions
3.1. Effects of Metformin on Duckweed Growth and Photosynthesis
3.2. Effects of Metformin on Plant Hormone Signal Transduction in Duckweed
3.3. Potential of Duckweed in Remediation of Metformin-Contaminated Water
4. Materials and Methods
4.1. Culture of Duckweed and Metformin (MF) Exposure
4.2. Determination of the Growth Curves of Duckweed Under MF Treatment
4.3. Measurement of Photosynthetic Fluorescence Parameters
4.4. RNA Extraction and Sequencing
4.5. Determination of MF Enrichment by Duckweed
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiao, Y.; Shao, X.-T.; Tan, D.-Q.; Yan, J.-H.; Pei, W.; Wang, Z.; Yang, M.; Wang, D.-G. Assessing the trend of diabetes mellitus by analyzing metformin as a biomarker in wastewater. Sci. Total Environ. 2019, 688, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.G.; Evangelopoulos, A.; Kazazis, C. Metformin and cancer. Rev. Diabet. Stud. RDS 2013, 10, 228. [Google Scholar] [CrossRef]
- Ma, R.; Yi, B.; Riker, A.I.; Xi, Y. Metformin and cancer immunity. Acta Pharmacol. Sin. 2020, 41, 1403–1409. [Google Scholar] [CrossRef]
- Glossmann, H.H.; Lutz, O. Metformin and aging: A review. Gerontology 2019, 65, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Yerevanian, A.; Soukas, A.A. Metformin: Mechanisms in human obesity and weight loss. Curr. Obes. Rep. 2019, 8, 156–164. [Google Scholar] [CrossRef]
- Yang, D.; Zheng, Q.; Thai, P.K.; Ahmed, F.; O’Brien, J.W.; Mueller, J.F.; Thomas, K.V.; Tscharke, B. A nationwide wastewater-based assessment of metformin consumption across Australia. Environ. Int. 2022, 165, 107282. [Google Scholar] [CrossRef]
- Shao, X.-T.; Cong, Z.-X.; Liu, S.-Y.; Wang, Z.; Zheng, X.-Y.; Wang, D.-G. Spatial analysis of metformin use compared with nicotine and caffeine consumption through wastewater-based epidemiology in China. Ecotoxicol. Environ. Saf. 2021, 208, 111623. [Google Scholar] [CrossRef]
- Phillips, J.D.; Pooler, D.B.; Ness, D.B.; Fay, K.; Tau, S.; Demidenko, E.; Hampsch, R.A.; Lewis, L.D.; Miller, T.W. Tumour, whole-blood, plasma and tissue concentrations of metformin in lung cancer patients. Br. J. Clin. Pharmacol. 2023, 89, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Wilcock, C.; Bailey, C. Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica 1994, 24, 49–57. [Google Scholar] [CrossRef]
- Naidu, R.; Espana, V.A.A.; Liu, Y.; Jit, J. Emerging contaminants in the environment: Risk-based analysis for better management. Chemosphere 2016, 154, 350–357. [Google Scholar] [CrossRef]
- Xin, X.; Huang, G.; Zhang, B. Review of aquatic toxicity of pharmaceuticals and personal care products to algae. J. Hazard. Mater. 2021, 410, 124619. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, J.; Winter, M.J.; Tyler, C.R. Pharmaceuticals in the aquatic environment: A critical review of the evidence for health effects in fish. Crit. Rev. Toxicol. 2010, 40, 287–304. [Google Scholar] [CrossRef]
- Lee, J.W.; Shin, Y.-J.; Kim, H.; Kim, H.; Kim, J.; Min, S.-A.; Kim, P.; Do Yu, S.; Park, K. Metformin-induced endocrine disruption and oxidative stress of Oryzias latipes on two-generational condition. J. Hazard. Mater. 2019, 367, 171–181. [Google Scholar] [CrossRef]
- Cummings, B.M.; Needoba, J.A.; Peterson, T.D. Effect of metformin exposure on growth and photosynthetic performance in the unicellular freshwater chlorophyte, Chlorella vulgaris. PLoS ONE 2018, 13, e0207041. [Google Scholar] [CrossRef]
- Viollet, B.; Guigas, B.; Garcia, N.S.; Leclerc, J.; Foretz, M.; Andreelli, F. Cellular and molecular mechanisms of metformin: An overview. Clin. Sci. 2012, 122, 253–270. [Google Scholar] [CrossRef]
- Nespor, B.; Andrianova, A.; Pollack, S.; Pfau, C.; Arifuzzaman, M.; Islam, N.; Kubátová, A.; Hossain, K. Metformin uptake and translocation in chickpeas: Determination using liquid chromatography–mass spectrometry. ACS Omega 2020, 5, 1789–1795. [Google Scholar] [CrossRef] [PubMed]
- LaMoia, T.E.; Shulman, G.I. Cellular and molecular mechanisms of metformin action. Endocr. Rev. 2021, 42, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Zhang, X.; Yong, T.; Bie, N.; Zhan, G.; Li, X.; Liang, Q.; Li, J.; Yu, J.; Huang, G.; et al. Boosting anti-PD-1 therapy with metformin-loaded macrophage-derived microparticles. Nat. Commun. 2021, 12, 440. [Google Scholar] [CrossRef]
- Rezania, S.; Taib, S.M.; Md Din, M.F.; Dahalan, F.A.; Kamyab, H. Comprehensive review on phytotechnology: Heavy metals removal by diverse aquatic plants species from wastewater. J. Hazard. Mater. 2016, 318, 587–599. [Google Scholar] [CrossRef]
- Farid, M.; Sajjad, A.; Asam, Z.U.Z.; Zubair, M.; Rizwan, M.; Abbas, M.; Farid, S.; Ali, S.; Alharby, H.F.; Alzahrani, Y.M.; et al. Phytoremediation of contaminated industrial wastewater by duckweed (Lemna minor L.): Growth and physiological response under acetic acid application. Chemosphere 2022, 304, 135262. [Google Scholar] [CrossRef]
- Dinh, T.T.U.; Soda, S.; Nguyen, T.A.H.; Nakajima, J.; Cao, T.H. Nutrient removal by duckweed from anaerobically treated swine wastewater in lab-scale stabilization ponds in Vietnam. Sci. Total Environ. 2020, 722, 137854. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Li, C.; Zhou, Y.; Wu, Y.; Wang, W. Genomes and transcriptomes of duckweeds. Front. Chem. 2018, 6, 230. [Google Scholar] [CrossRef]
- Guo, L.; Jin, Y.; Xiao, Y.; Tan, L.; Tian, X.; Ding, Y.; He, K.; Du, A.; Li, J.; Yi, Z.; et al. Energy-efficient and environmentally friendly production of starch-rich duckweed biomass using nitrogen-limited cultivation. J. Clean. Prod. 2020, 251, 119726. [Google Scholar] [CrossRef]
- Kamal, F.; Radziwon, A.; Davis, C.M.; Dennis, J.J. Duckweed (Lemna minor) and Alfalfa (Medicago sativa) as Bacterial Infection Model Systems. In Bacteriophages: Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1898, pp. 191–198. [Google Scholar]
- Markovic, M.; Neale, P.A.; Nidumolu, B.; Kumar, A. Combined toxicity of therapeutic pharmaceuticals to duckweed, Lemna minor. Ecotoxicol. Environ. Saf. 2021, 208, 111428. [Google Scholar] [CrossRef] [PubMed]
- Briones, R.M.; Sarmah, A.K. Modelling degradation kinetics of metformin and guanylurea in soil microcosms to derive degradation end-points. Environ. Pollut. 2019, 245, 735–745. [Google Scholar] [CrossRef]
- Zheng, Y.; Shao, Y.; Zhang, Y.; Liu, Z.; Zhao, Z.; Xu, R.; Ding, J.; Li, W.; Wang, B.; Zhang, H. Metformin as an emerging pollutant in the aquatic environment: Occurrence, analysis, and toxicity. Toxics 2024, 12, 483. [Google Scholar] [CrossRef]
- Straub, J.O.; Caldwell, D.J.; Davidson, T.; D’Aco, V.; Kappler, K.; Robinson, P.F.; Simon-Hettich, B.; Tell, J. Environmental risk assessment of metformin and its transformation product guanylurea. I. Environmental fate. Chemosphere 2019, 216, 844–854. [Google Scholar] [CrossRef]
- Thingujam, D.; Pajerowska-Mukhtar, K.M.; Mukhtar, M.S. Duckweed: Beyond an Efficient Plant Model System. Biomolecules 2024, 14, 628. [Google Scholar] [CrossRef]
- Yang, L.; Ma, X.; Guo, Y.; He, Y.; Yang, Y.; Wang, W.; Xu, Z.; Zuo, Z.; Xue, Y.; Yang, R.; et al. Acetylcholine (ACh) enhances Cd tolerance through transporting ACh in vesicles and modifying Cd absorption in duckweed (Lemna turionifera 5511). Environ. Pollut. 2023, 335, 122305. [Google Scholar] [CrossRef]
- Ifuku, K. The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol. Biochem. 2014, 81, 108–114. [Google Scholar] [CrossRef]
- Redekop, P.; Rothhausen, N.; Rothhausen, N.; Melzer, M.; Mosebach, L.; Dülger, E.; Bovdilova, A.; Caffarri, S.; Hippler, M.; Jahns, P. PsbS contributes to photoprotection in Chlamydomonas reinhardtii independently of energy dissipation. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148183. [Google Scholar] [CrossRef] [PubMed]
- García-Cerdán, J.G.; Kovács, L.; Tóth, T.; Kereïche, S.; Aseeva, E.; Boekema, E.J.; Mamedov, F.; Funk, C.; Schröder, W.P. The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 2011, 65, 368–381. [Google Scholar] [CrossRef]
- von Sydow, L.; Schwenkert, S.; Meurer, J.; Funk, C.; Mamedov, F.; Schröder, W.P. The PsbY protein of Arabidopsis Photosystem II is important for the redox control of cytochrome b559. Biochim. Biophys. Acta 2016, 1857, 1524–1533. [Google Scholar] [CrossRef] [PubMed]
- Grasse, N.; Mamedov, F.; Becker, K.; Styring, S.; Rögner, M.; Nowaczyk, M.M. Role of Novel Dimeric Photosystem II (PSII)-Psb27 Protein Complex in PSII Repair. J. Biol. Chem. 2011, 286, 29548–29555. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Wu, D.; Yong, W.; Liu, M.; Wang, S.; Liu, W.; Lu, M.; Wei, Y.; Sun, J. Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 2017, 192, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kandeler, R. Promotion of Flowering by a Tumor Promoter. J. Plant Physiol. 1994, 144, 710–713. [Google Scholar] [CrossRef]
- Yang, L.; Han, H.; Liu, M.; Zuo, Z.; Zhou, K.; Lü, J.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of the Arabidopsis photorespiratory pathway gene, serine: Glyoxylate aminotransferase (AtAGT1), leads to salt stress tolerance in transgenic duckweed (Lemna minor). Plant Cell Tissue Organ Cult. (PCTOC) 2013, 113, 407–416. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410–424. [Google Scholar] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]



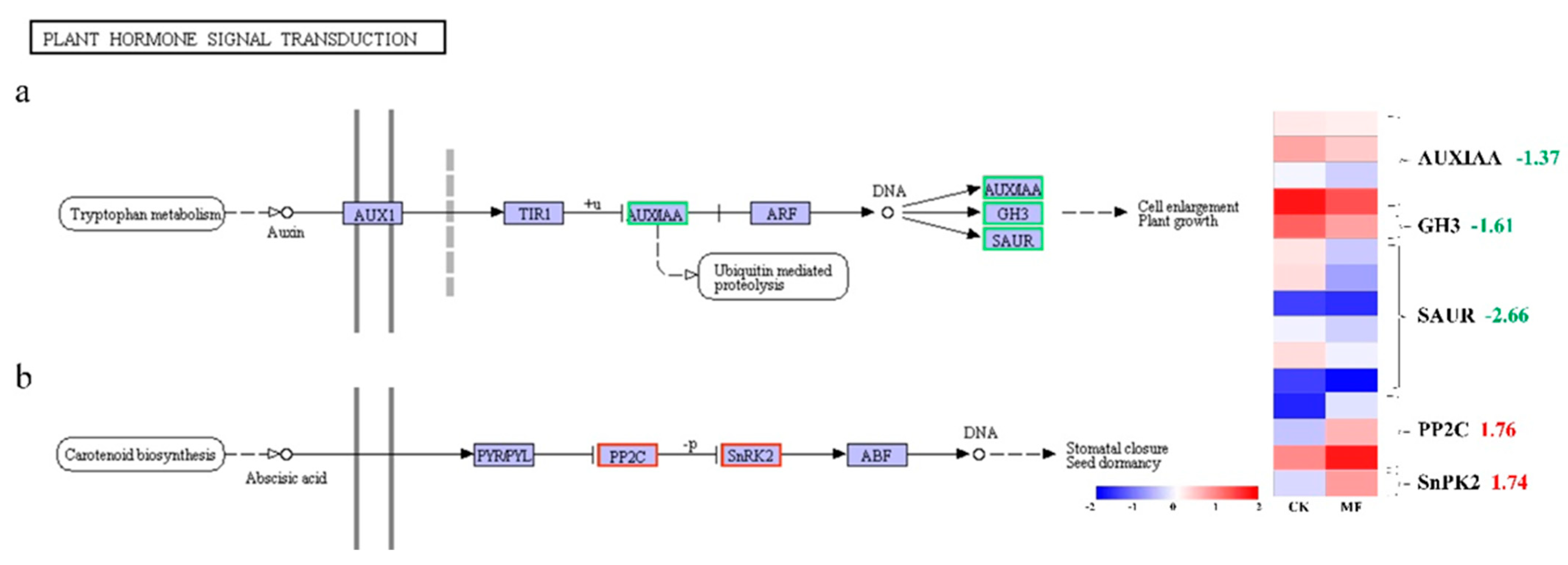
| Description | Gene_Id | MF_ Readcount | CK_ Readcount | log2Fold Change | Pval | Padj |
|---|---|---|---|---|---|---|
| auxin-responsive protein IAA | Cluster-3667.11150 | 110.05 | 226.59 | −1.04 | 3.21 × 10−6 | 3.07 × 10−5 |
| auxin-responsive protein IAA | Cluster-3667.3607 | 189.65 | 499.70 | −1.40 | 4.14 × 10−11 | 8.50 × 10−10 |
| auxin-responsive protein IAA | Cluster-3667.2138 | 41.97 | 155.55 | −1.89 | 5.77 × 10−6 | 5.27 × 10−5 |
| auxin-responsive protein IAA | Cluster-3667.8895 | 1082.36 | 2400.65 | −1.15 | 7.25 × 10−14 | 2.03 × 10−12 |
| auxin responsive GH3 gene family | Cluster-3667.9729 | 349.46 | 1069.31 | −1.61 | 8.22 × 10−13 | 2.07 × 10−11 |
| SAUR family protein | Cluster-6625.0 | 38.64 | 240.96 | −2.64 | 3.05 × 10−19 | 1.36 × 10−17 |
| SAUR family protein | Cluster-3667.1437 | 21.11 | 260.06 | −3.64 | 9.24 × 10−36 | 1.15 × 10−33 |
| SAUR family protein | Cluster-6287.0 | 3.36 | 18.92 | −2.45 | 0.003364 | 0.016163 |
| SAUR family protein | Cluster-3667.19491 | 42.41 | 148.76 | −1.81 | 2.08 × 10−10 | 3.89 × 10−9 |
| SAUR family protein | Cluster-3667.14101 | 69.64 | 263.02 | −1.92 | 2.16 × 10−11 | 4.59 × 10−10 |
| SAUR family protein | Cluster-3667.17453 | 1.70 | 18.90 | −3.49 | 0.000268 | 0.001697 |
| protein phosphatase 2C | Cluster-3667.5265 | 55.00 | 13.81 | 2.00 | 1.06 × 10−5 | 9.17 × 10−5 |
| protein phosphatase 2C | Cluster-3667.11703 | 257.77 | 87.59 | 1.56 | 2.89 × 10−6 | 2.78 × 10−5 |
| protein phosphatase 2C | Cluster-3667.9131 | 2255.85 | 685.34 | 1.72 | 4.38 × 10−12 | 1.01 × 10−10 |
| serine/threonine-protein kinase SRK2 | Cluster-3667.6959 | 371.62 | 110.84 | 1.74 | 9.69 × 10−12 | 2.16 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, B.; Jiang, Y.; Wang, W.; Guo, Y.; Yang, Y.; He, Y.; Di, Q.; Qu, Z.; Xing, Y.; Yang, L. The Toxicity Effects of Metformin and the Bioremediation of Metformin in Aquatic Plant Duckweed. Plants 2025, 14, 1761. https://doi.org/10.3390/plants14121761
Han B, Jiang Y, Wang W, Guo Y, Yang Y, He Y, Di Q, Qu Z, Xing Y, Yang L. The Toxicity Effects of Metformin and the Bioremediation of Metformin in Aquatic Plant Duckweed. Plants. 2025; 14(12):1761. https://doi.org/10.3390/plants14121761
Chicago/Turabian StyleHan, Bing, Yumeng Jiang, Wenqiao Wang, Yuhan Guo, Yunwen Yang, Yuman He, Qiqi Di, Ziyang Qu, Yun Xing, and Lin Yang. 2025. "The Toxicity Effects of Metformin and the Bioremediation of Metformin in Aquatic Plant Duckweed" Plants 14, no. 12: 1761. https://doi.org/10.3390/plants14121761
APA StyleHan, B., Jiang, Y., Wang, W., Guo, Y., Yang, Y., He, Y., Di, Q., Qu, Z., Xing, Y., & Yang, L. (2025). The Toxicity Effects of Metformin and the Bioremediation of Metformin in Aquatic Plant Duckweed. Plants, 14(12), 1761. https://doi.org/10.3390/plants14121761







