Phytochemical Characterization, Antioxidant Activity, and Anti-Melanoma Mechanism of Flower Buds of Magnolia biondii Pamp.
Abstract
1. Introduction
2. Results
2.1. Quantitative Analysis of Total Flavonoid and Polyphenol Contents
2.2. Qualitative Detection of Phytochemical Constituents
2.3. In Vitro Antioxidant Activity of MBP
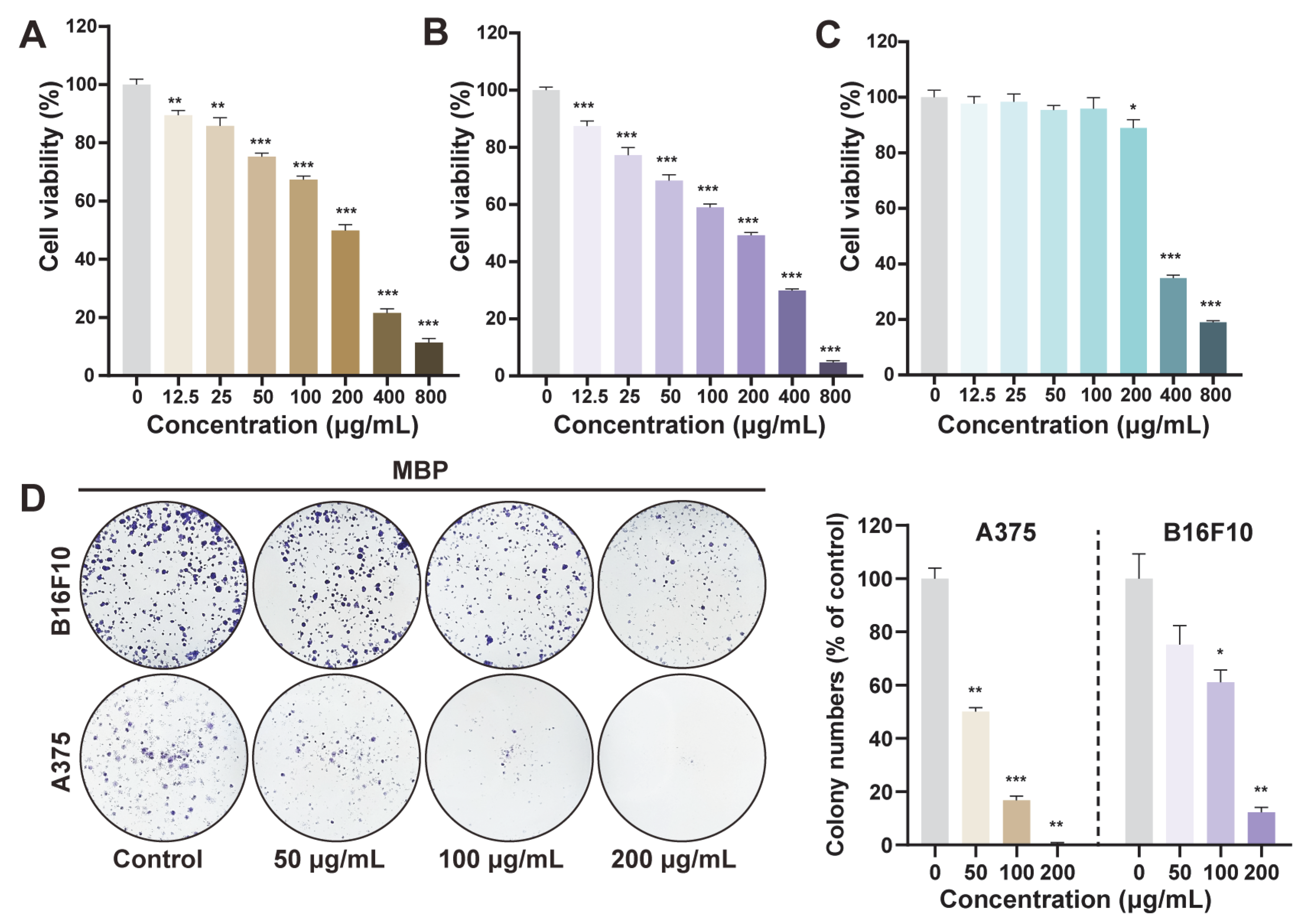
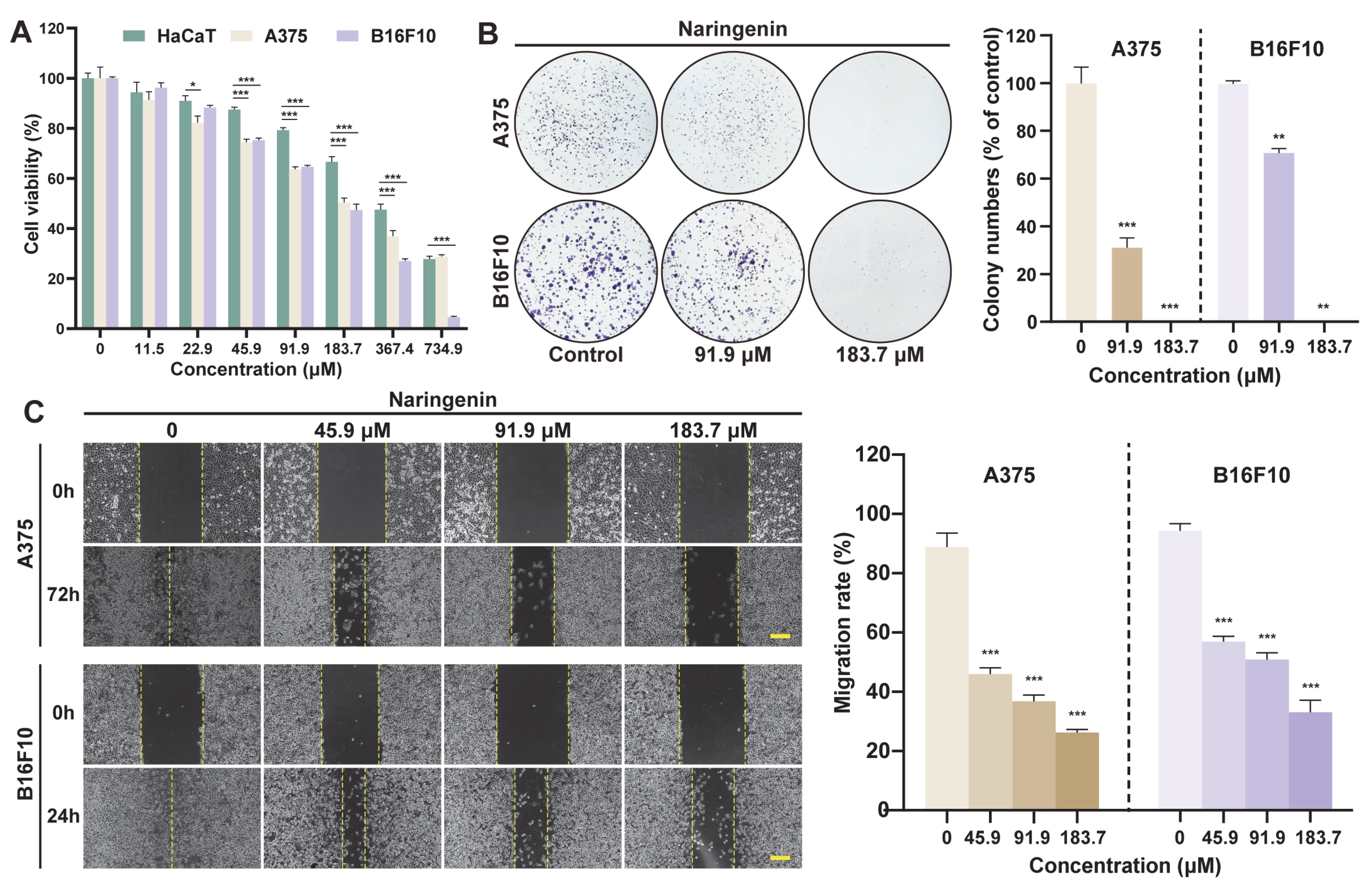
2.4. Anti-Proliferative Effect of MBP on Melanoma Cells
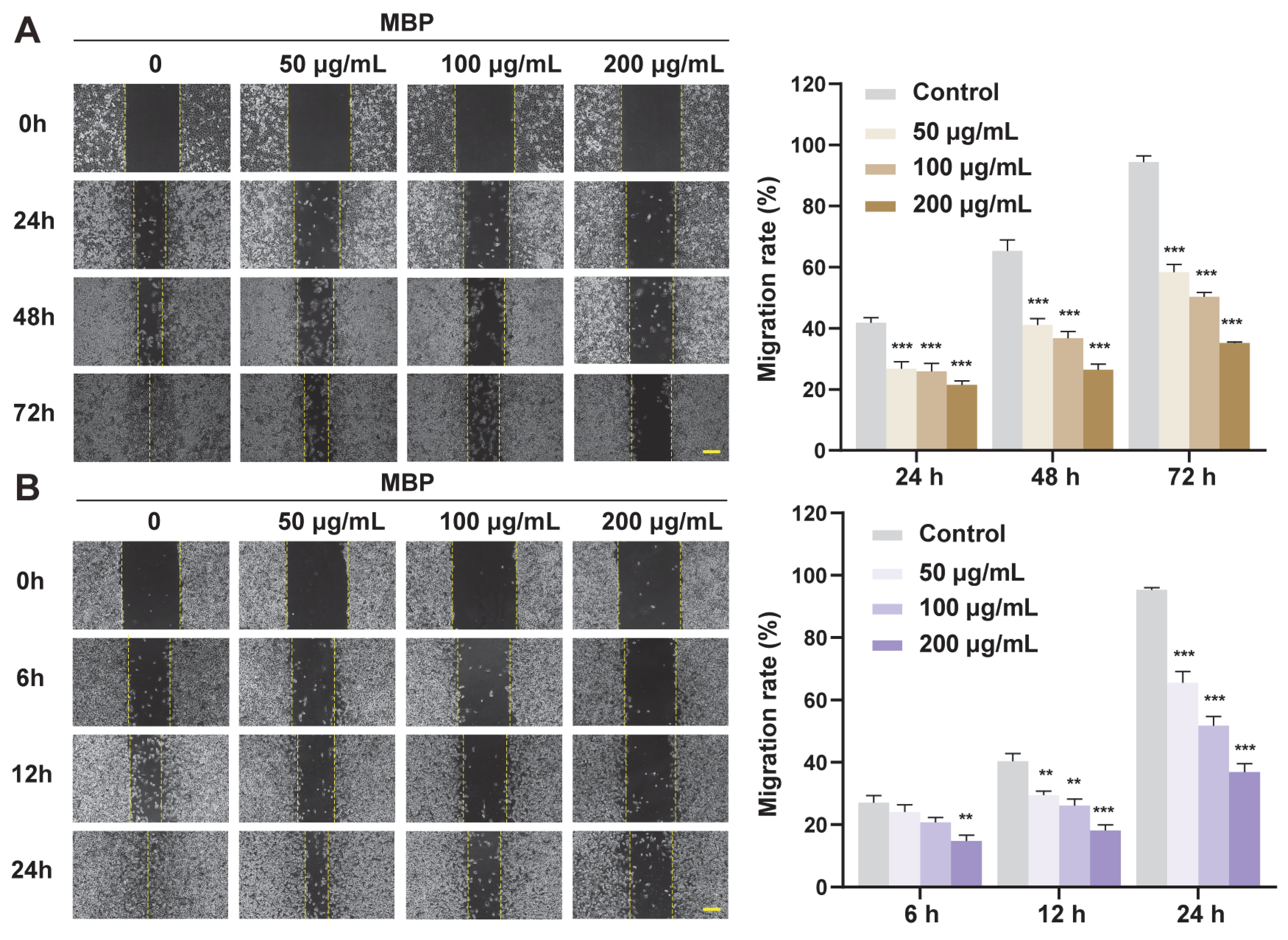
2.5. Anti-Migratory Effect of MBP on Melanoma Cells
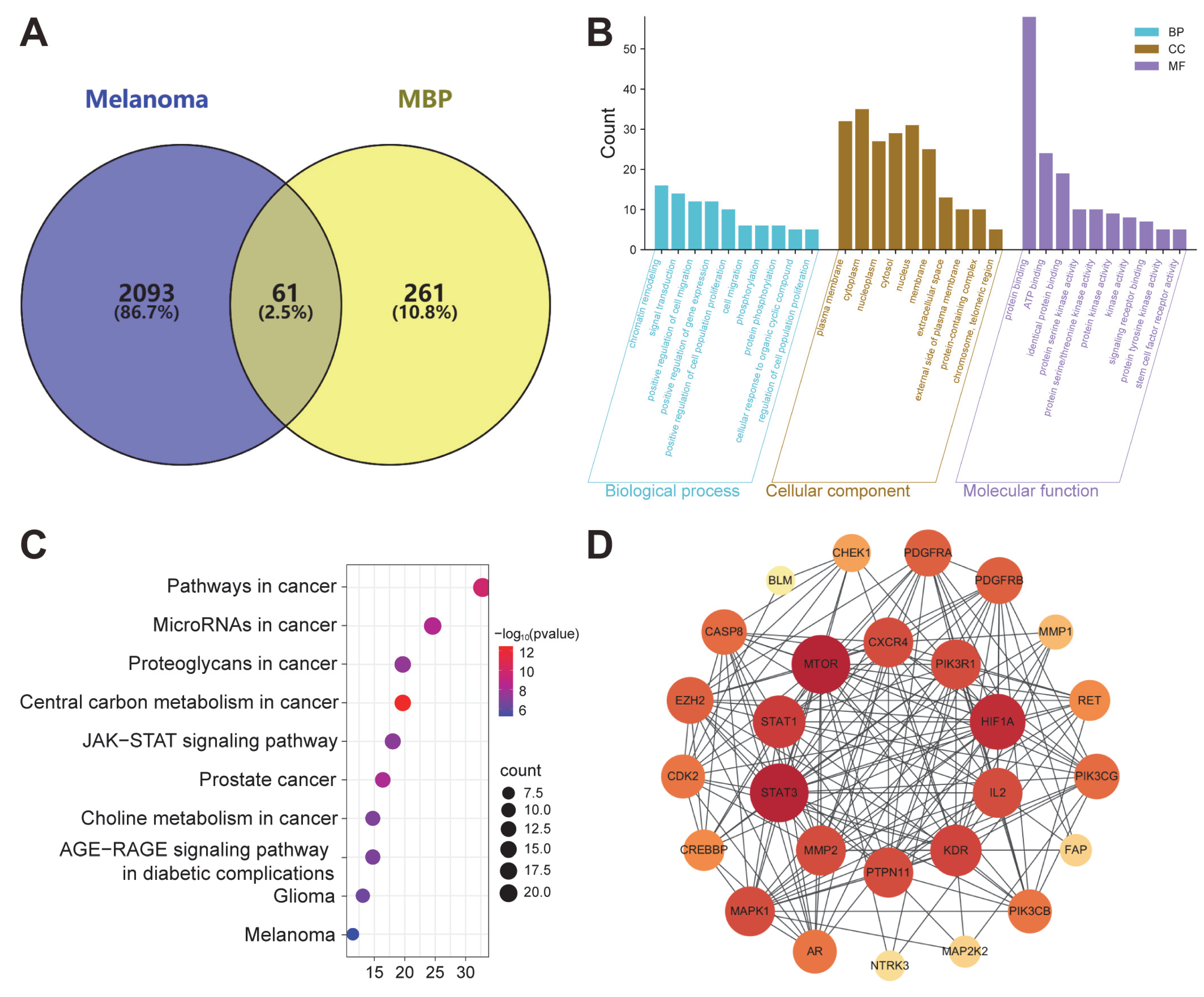
2.6. Network Analysis of MBP Targets in Melanoma Treatment
2.6.1. Predicted Targets of MBP
2.6.2. GO and KEGG Analysis
2.6.3. PPI Analysis
2.6.4. “Component-Disease-Target” Network Construction
2.6.5. Identification of Primary Anti-Melanoma Active Compounds Using CMap
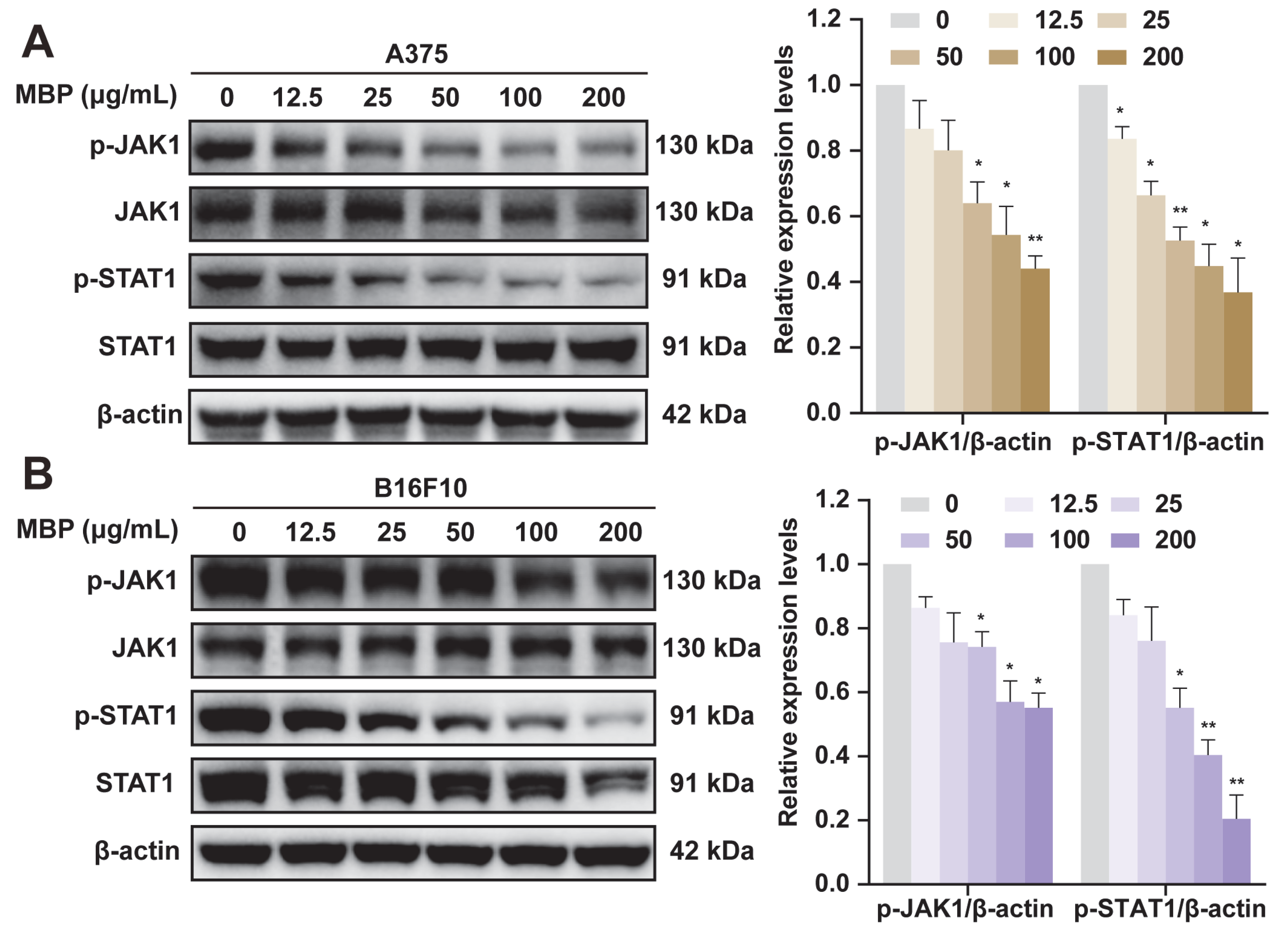
2.7. Validation of JAK1/STAT1 Pathway Involvement in MBP-Treated Melanoma Cells
2.8. Anti-Melanoma Activity of Naringenin
2.9. Molecular Docking Analysis
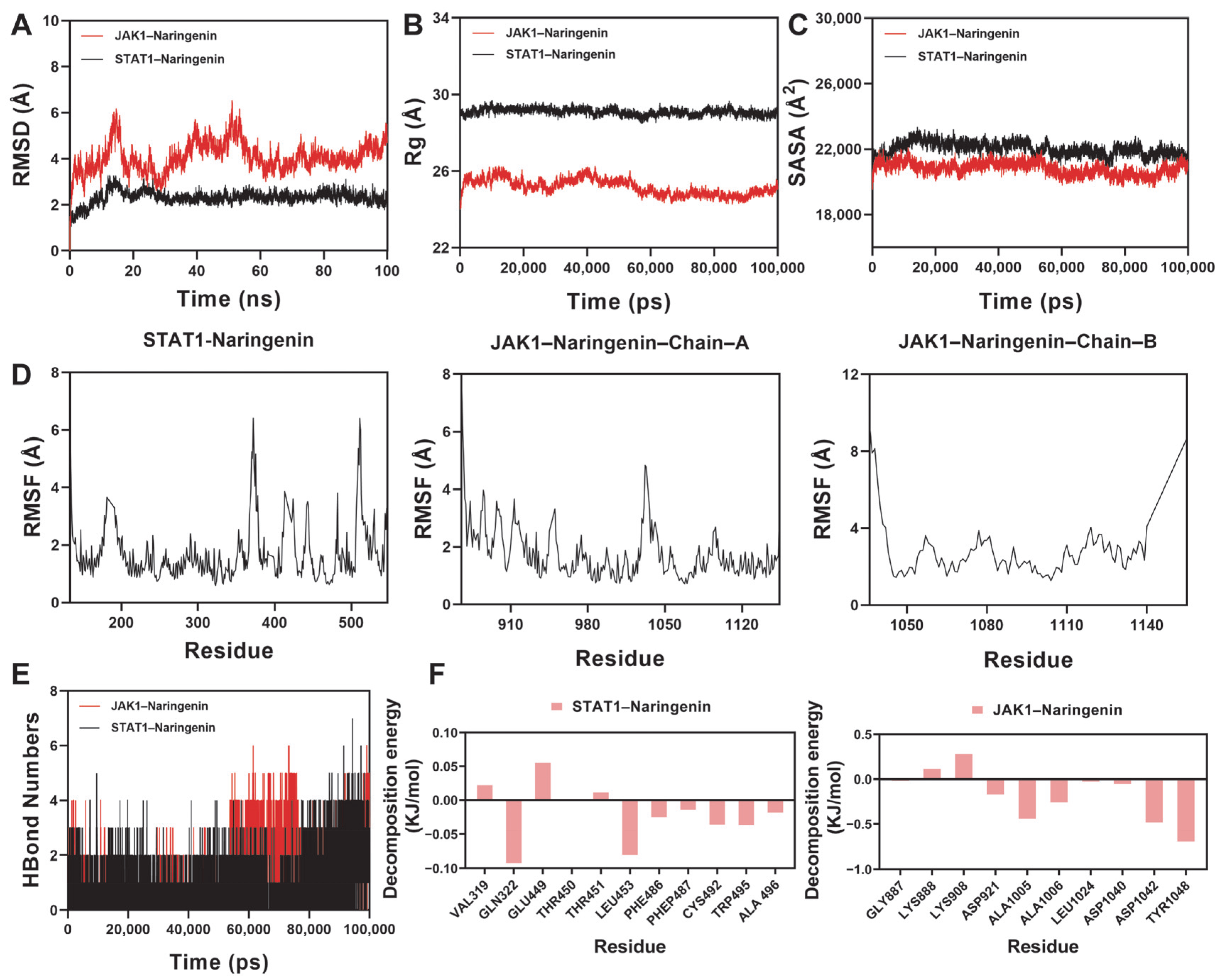
2.10. Molecular Dynamics Simulation Analysis
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Plant Materials
4.3. Quantitative Phytochemical Analysis
4.4. Qualitative Analysis Based on UHPLC-Q Exactive™ HFX-LC-MS/MS
4.5. Invitro Antioxidant Activity Evaluation
4.5.1. DPPH Radical Scavenging Assay
4.5.2. ABTS Radical Scavenging Assay
4.6. Cell Culture
4.7. Cell Viability Assay
4.8. Colony Formation Assay
4.9. Cell Migration Assay
4.10. Network Pharmacology Analysis
4.10.1. Targets Prediction of MBP
4.10.2. Screening of Potential Targets for Melanoma
4.10.3. GO Function and KEGG Pathway Enrichment Analysis of the Target
4.10.4. Construction and Analysis of Protein–Protein Interaction (PPI) Network
4.10.5. Construction of “Component-Disease-Target” Network
4.10.6. CMap Screening
4.11. Western Blot Assay
4.12. Molecular Docking
4.13. Molecular Dynamics Simulation
4.14. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MBP | The Flower Buds of Magnolia biondii Pamp. |
| CMap | Connectivity Map |
| TCM | Traditional Chinese Medicine |
| TFC | Total Flavonoid Content |
| TPC | Total Polyphenol Content |
| RE | Rutin Equivalents |
| GAE | Gallic Acid Equivalents |
| GNPS | Global Natural Products Social Molecular Networking |
| GO | Gene Ontology |
| BP | Biological Processes |
| CC | Cellular Components |
| MF | Molecular Functions |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| C-T-D | Component–Disease–Target |
| PPI | Protein–Protein Interaction |
| STAT | Signal Transducer and Activator of Transcription |
| JAK | Janus Kinase |
| raw-cs | raw connectivity score |
| norm-cs: | normalized connectivity score |
| MD | Molecular Dynamics |
| RMSD | Root-Mean-Square Deviation |
| SASA | Solvent-Accessible Surface Area |
| RMSF | Root-Mean-Square Fluctuation |
| MM/PBSA | Molecular Mechanics/Poisson–Boltzmann Surface Area |
References
- Gray-Schopfer, V.; Wellbrock, C.; Marais, R. Melanoma Biology and New Targeted Therapy. Nature 2007, 445, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.W.; Fisher, D.E. Treatment of Advanced Melanoma in 2020 and Beyond. J. Investig. Dermatol. 2021, 141, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, D.; Biswasroy, P.; Sahu, A.; Sahu, D.K.; Ghosh, G.; Rath, G. Recent Advances in Herbal Nanomedicines for Cancer Treatment. Curr. Mol. Pharmacol. 2021, 14, 292–305. [Google Scholar] [CrossRef]
- Wanderley, C.W.; Colón, D.F.; Luiz, J.P.M.; Oliveira, F.F.; Viacava, P.R.; Leite, C.A.; Pereira, J.A.; Silva, C.M.; Silva, C.R.; Silva, R.L.; et al. Paclitaxel Reduces Tumor Growth by Reprogramming Tumor-Associated Macrophages to an M1 Profile in a TLR4-Dependent Manner. Cancer Res. 2018, 78, 5891–5900. [Google Scholar] [CrossRef] [PubMed]
- Radha, R.; Paul, V.; Anjum, S.; Bouakaz, A.; Pitt, W.G.; Husseini, G.A. Enhancing Curcumin’s Therapeutic Potential in Cancer Treatment through Ultrasound Mediated Liposomal Delivery. Sci. Rep. 2024, 14, 10499. [Google Scholar] [CrossRef]
- Danciu, C.; Soica, C.; Antal, D.; Alexa, E.; Pavel, I.Z.; Ghiulai, R.; Ardelean, F.; Babuta, R.M.; Popescu, A.; Dehelean, C.A. Natural Compounds in the Chemoprevention of Malignant Melanoma. Anticancer Agents Med. Chem. 2018, 18, 631–644. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia Commission. Monographs of Traditional Chinese Medicines (TCMs). In Pharmacopoeia of the People’s Republic of China, 11th ed.; China Medical Science and Technology Press: Beijing, China, 2020; p. 292. [Google Scholar]
- Wu, X.; Yin, W.; Li, Y.; Wu, H.; Cheng, Q.; He, Q.; Wu, H.; Hu, M. Assessment of Quality in Volatile Oil from Three Basic Sources of Xinyi from Hubei by Anatomy, GC-MS, and Chemometric Methods. Sci. Rep. 2025, 15, 6857. [Google Scholar] [CrossRef]
- Shen, Y.; Pang, E.C.K.; Xue, C.C.L.; Zhao, Z.Z.; Lin, J.G.; Li, C.G. Inhibitions of Mast Cell-Derived Histamine Release by Different Flos Magnoliae Species in Rat Peritoneal Mast Cells. Phytomedicine 2008, 15, 808–814. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, Y.R.; Nam, J.H. Flos Magnoliae Inhibits Chloride Secretion via ANO1 Inhibition in Calu-3 Cells. Am. J. Chin. Med. 2018, 46, 1079–1092. [Google Scholar] [CrossRef]
- Chen, C.-H.; Chen, H.-C.; Chang, W.-T.; Lee, M.-S.; Liu, Y.-C.; Lin, M.-K. Magnoliae Flos Essential Oil as an Immunosuppressant in Dendritic Cell Activation and Contact Hypersensitivity Responses. Am. J. Chin. Med. 2020, 48, 597–613. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Tang, G.; Li, R.; Guo, H.; Weng, W.; Wang, D.; Ji, S. Development of a Comprehensive Quality Control Method for the Quantitative Analysis of Volatiles and Lignans in Magnolia Biondii Pamp. by near Infrared Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 230, 118080. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Z.; Zhao, H.; Liu, M.; Lin, C.; Li, L.; Ma, B. Pectin Polysaccharide from Flos Magnoliae (Xin Yi, Magnolia Biondii Pamp. Flower Buds): Hot-Compressed Water Extraction, Purification and Partial Structural Characterization. Food Hydrocolloids 2022, 122, 107061. [Google Scholar] [CrossRef]
- Gil, T.-Y.; Jin, B.-R.; Cha, Y.-Y.; An, H.-J. Magnoliae Flos Downregulated Lipopolysaccharide-Induced Inflammatory Responses via NF-κB/ERK-JNK MAPK/STAT3 Pathways. Mediators Inflamm. 2022, 2022, 6281892. [Google Scholar] [CrossRef]
- Ham, J.R.; Yun, K.W.; Lee, M.-K. Anti-Inflammatory and Antioxidant in Vitro Activities of Magnoliae Flos Ethanol Extract. Prev. Nutr. Food Sci. 2021, 26, 485–491. [Google Scholar] [CrossRef]
- Kim, E.-K.; Song, M.-Y.; Kim, I.-S.; Moon, W.S.; Ryu, D.-G.; So, H.-S.; Park, R.; Park, J.-W.; Kwon, K.-B.; Park, B.-H. Beneficial Effect of Flos Magnoliae Extract on Multiple Low Dose Streptozotocin-Induced Type 1 Diabetes Development and Cytokine-Induced Beta-Cell Damage. Int. J. Mol. Med. 2008, 22, 481–488. [Google Scholar] [PubMed]
- Kim, M.R.; Kim, D.-I.; Park, S.Y.; Kang, H.J.; Park, S.-D.; Lee, J.-H. The Protective Role of Magnoliae Flos in Preventing Ovotoxicity and Managing Ovarian Function: An In Vitro and In Vivo Study. Int. J. Mol. Sci. 2024, 25, 6456. [Google Scholar] [CrossRef]
- Kim, H.M.; Yi, J.M.; Lim, K.S. Magnoliae Flos Inhibits Mast Cell-Dependent Immediate-Type Allergic Reactions. Pharmacol. Res. 1999, 39, 107–111. [Google Scholar] [CrossRef]
- Hu, S.; Li, S.; Xu, Y.; Huang, X.; Mai, Z.; Chen, Y.; Xiao, H.; Ning, W.; Gaus, S.; Savkovic, V.; et al. The Antitumor Effects of Herbal Medicine Triphala on Oral Cancer by Inactivating PI3K/Akt Signaling Pathway: Based on the Network Pharmacology, Molecular Docking, in Vitro and in Vivo Experimental Validation. Phytomedicine 2024, 128, 155488. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network Pharmacology: The next Paradigm in Drug Discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Zhao, X.; Ren, D.; Jin, R.; Chen, W.; Xu, L.; Guo, D.; Zhang, Q.; Luo, Z. Development of UHPLC-Q-Exactive Orbitrap/MS Technique for Determination of Proanthocyanidins (PAs) Monomer Composition Content in Persimmon. Plants 2024, 13, 1440. [Google Scholar] [CrossRef]
- Li, X.; Zeng, J.; Cai, R.; Li, C.; Chen, X.; Chen, B.; Zhao, X.; Khan, S. Putative Identification of 47 Compounds from Jieyu Anshen Granule and Proposal of Pharmacopeia Quality-Assessment Strategy Using TCM-Specific Library with UHPLC-Q-Exactive-Orbitrap-MS. ChemistryOpen 2025, 14, e202400046. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Fissistigma Oldhamii (Hemsl.) Merr.: Ethnomedicinal, Phytochemistry, and Pharmacological Aspects. Plants 2023, 12, 4094. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.; Chun, J.; Park, S.-M.; Yeo, H.; Na, S.W.; Ha, I.J.; Kim, B.; Jeong, M.-K. An Investigation of the Anticancer Mechanism of Caesalpinia sappan L. Extract Against Colorectal Cancer by Integrating a Network Pharmacological Analysis and Experimental Validation. Plants 2025, 14, 263. [Google Scholar] [CrossRef]
- Li, K.; Xiao, S.; Zhu, L.; Shu, L.; Zou, Y.; Wang, J.; Chen, Y.; Yan, F.; Cai, W. Integrated Strategy of Mass Spectrometry Imaging and LC/MS-Based GNPS for Spatial Characterization of Alkaloids from Menispermi Rhizoma and Study on Potential Anti-Inflammatory Mechanism by Network Pharmacology and Molecular Docking. Ind. Crops Prod. 2024, 222, 119952. [Google Scholar] [CrossRef]
- Zia-ur-Rehman; Gurgul, A.; Youn, I.; Maldonado, A.; Wahid, F.; Che, C.-T.; Khan, T. UHPLC-MS/MS-GNPS Based Phytochemical Investigation of Equisetum arvense L. And Evaluation of Cytotoxicity against Human Melanoma and Ovarian Cancer Cells. Saudi J. Biol. Sci. 2022, 29, 103271. [Google Scholar] [CrossRef]
- Aras, A.; Bursal, E.; Dogru, M. UHPLC-ESI-MS/MS Analyses for Quantification of Phenolic Compounds of Nepeta Nuda Subsp. Lydiae. J. Appl. Pharm. Sci. 2016, 6, 009–013. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, L.; Huang, H.; Yao, J.; Zhou, L.; Wang, D.; Qiu, X. Development of an Ultra-High Performance Liquid Chromatography Method for Simultaneous Determination of Six Active Compounds in Fructus Aurantii and Rat Plasma and Its Application to a Comparative Pharmacokinetic Study in Rats Administered with Different Doses. J. Anal. Methods Chem. 2018, 2018, 7579136. [Google Scholar] [CrossRef]
- Nguyen, C.; Baskaran, K.; Pupulin, A.; Ruvinov, I.; Zaitoon, O.; Grewal, S.; Scaria, B.; Mehaidli, A.; Vegh, C.; Pandey, S. Hibiscus Flower Extract Selectively Induces Apoptosis in Breast Cancer Cells and Positively Interacts with Common Chemotherapeutics. BMC Complement. Altern. Med. 2019, 19, 98. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, Q.; Shao, Y.; Yin, S.; Liu, C.; Liu, Y.; Wang, R.; Wang, T.; Qiu, Y.; Yu, H. Anticancer Activities of TCM and Their Active Components against Tumor Metastasis. Biomed. Pharmacother. 2021, 133, 111044. [Google Scholar] [CrossRef]
- Furumoto, Y.; Gadina, M. The Arrival of JAK Inhibitors: Advancing the Treatment of Immune and Hematologic Disorders. BioDrugs 2013, 27, 431–438. [Google Scholar] [CrossRef]
- Wang, R.; Liu, L.; Lai, L.; Tang, Y. SCORE: A New Empirical Method for Estimating the Binding Affinity of a Protein-Ligand Complex. J. Mol. Med. 1998, 4, 379–394. [Google Scholar] [CrossRef]
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as Anticancer Agents. Nutrients 2020, 12, 457. [Google Scholar] [CrossRef]
- Chahar, M.K.; Sharma, N.; Dobhal, M.P.; Joshi, Y.C. Flavonoids: A Versatile Source of Anticancer Drugs. Pharmacogn. Rev. 2011, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- García-Lafuente, A.; Guillamón, E.; Villares, A.; Rostagno, M.A.; Martínez, J.A. Flavonoids as Anti-Inflammatory Agents: Implications in Cancer and Cardiovascular Disease. Inflamm. Res. 2009, 58, 537–552. [Google Scholar] [CrossRef] [PubMed]
- You, O.H.; Shin, E.A.; Lee, H.; Kim, J.-H.; Sim, D.Y.; Kim, J.H.; Kim, Y.; Khil, J.-H.; Baek, N.-I.; Kim, S.-H. Apoptotic Effect of Astragalin in Melanoma Skin Cancers via Activation of Caspases and Inhibition of Sry-Related HMg-Box Gene 10. Phytother. Res. 2017, 31, 1614–1620. [Google Scholar] [CrossRef]
- Pinzaru, I.; Chioibas, R.; Marcovici, I.; Coricovac, D.; Susan, R.; Predut, D.; Georgescu, D.; Dehelean, C. Rutin Exerts Cytotoxic and Senescence-Inducing Properties in Human Melanoma Cells. Toxics 2021, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Benot-Dominguez, R.; Tupone, M.G.; Castelli, V.; d’Angelo, M.; Benedetti, E.; Quintiliani, M.; Cinque, B.; Forte, I.M.; Cifone, M.G.; Ippoliti, R.; et al. Olive Leaf Extract Impairs Mitochondria by Pro-Oxidant Activity in MDA-MB-231 and OVCAR-3 Cancer Cells. Biomed. Pharmacother. 2021, 134, 111139. [Google Scholar] [CrossRef]
- Owen, K.L.; Brockwell, N.K.; Parker, B.S. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers 2019, 11, 2002. [Google Scholar] [CrossRef]
- Orlova, A.; Wagner, C.; de Araujo, E.D.; Bajusz, D.; Neubauer, H.A.; Herling, M.; Gunning, P.T.; Keserű, G.M.; Moriggl, R. Direct Targeting Options for STAT3 and STAT5 in Cancer. Cancers 2019, 11, 1930. [Google Scholar] [CrossRef]
- Tabolacci, C.; De Vita, D.; Facchiano, A.; Bozzuto, G.; Beninati, S.; Failla, C.M.; Di Martile, M.; Lintas, C.; Mischiati, C.; Stringaro, A.; et al. Phytochemicals as Immunomodulatory Agents in Melanoma. Int. J. Mol. Sci. 2023, 24, 2657. [Google Scholar] [CrossRef]
- Yu, X.; Lv, J.; Wu, J.; Chen, Y.; Chen, F.; Wang, L. The Autoimmune Encephalitis-Related Cytokine TSLP in the Brain Primes Neuroinflammation by Activating the JAK2-NLRP3 Axis. Clin. Exp. Immunol. 2022, 207, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Laudisi, F.; Cherubini, F.; Monteleone, G.; Stolfi, C. STAT3 Interactors as Potential Therapeutic Targets for Cancer Treatment. Int. J. Mol. Sci. 2018, 19, 1787. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Wang, Q.; Li, S.; Huang, R.; Wang, X.; Liao, X.; Mo, H.; Zhang, L.; Zhou, X. Prognostic Value of Key Genes of the JAK-STAT Signaling Pathway in Patients with Cutaneous Melanoma. Oncol. Lett. 2020, 19, 1928–1946. [Google Scholar] [CrossRef] [PubMed]
- Khodarev, N.N.; Roach, P.; Pitroda, S.P.; Golden, D.W.; Bhayani, M.; Shao, M.Y.; Darga, T.E.; Beveridge, M.G.; Sood, R.F.; Sutton, H.G.; et al. STAT1 Pathway Mediates Amplification of Metastatic Potential and Resistance to Therapy. PLoS ONE 2009, 4, e5821. [Google Scholar] [CrossRef]
- Aydoğmuş-Öztürk, F.; Günaydin, K.; Öztürk, M.; Jahan, H.; Duru, M.E.; Choudhary, M.I. Effect of Sideritis Leptoclada against HT-144 Human Malignant Melanoma. Melanoma Res. 2018, 28, 502–509. [Google Scholar] [CrossRef]
- Choi, J.; Lee, D.-H.; Jang, H.; Park, S.-Y.; Seol, J.-W. Naringenin Exerts Anticancer Effects by Inducing Tumor Cell Death and Inhibiting Angiogenesis in Malignant Melanoma. Int. J. Med. Sci. 2020, 17, 3049–3057. [Google Scholar] [CrossRef]
- Martínez Conesa, C.; Vicente Ortega, V.; Yáñez Gascón, M.J.; Alcaraz Baños, M.; Canteras Jordana, M.; Benavente-García, O.; Castillo, J. Treatment of Metastatic Melanoma B16F10 by the Flavonoids Tangeretin, Rutin, and Diosmin. J. Agric. Food Chem. 2005, 53, 6791–6797. [Google Scholar] [CrossRef]
- Memariani, Z.; Abbas, S.Q.; ul Hassan, S.S.; Ahmadi, A.; Chabra, A. Naringin and Naringenin as Anticancer Agents and Adjuvants in Cancer Combination Therapy: Efficacy and Molecular Mechanisms of Action, a Comprehensive Narrative Review. Pharmacol. Res. 2021, 171, 105264. [Google Scholar] [CrossRef]
- Li, T.; Zhang, X.; Wang, H.; Li, J.; Wang, H.; Zhang, X. Development, Physical–Chemical Characterization, and Molecular Docking Simulations of Ursolic Acid–Sodium Alginate Complexes. J. Agric. Food Chem. 2021, 69, 14311–14319. [Google Scholar] [CrossRef]
- Al-Khafaji, K.; Taskin Tok, T. Molecular Dynamics Simulation, Free Energy Landscape and Binding Free Energy Computations in Exploration the Anti-Invasive Activity of Amygdalin against Metastasis. Comput. Methods Programs Biomed. 2020, 195, 105660. [Google Scholar] [CrossRef]
- Jain, N.K.; Chandrasekaran, B.; Khazaleh, N.; Jain, H.K.; Lal, M.; Joshi, G.; Jha, V. Computational Network Pharmacology, Molecular Docking, and Molecular Dynamics to Decipher Natural Compounds of Alchornea Laxiflora for Liver Cancer Chemotherapy. Pharmaceuticals 2025, 18, 508. [Google Scholar] [CrossRef] [PubMed]
- Nohutçu, L.; Tunçtürk, M.; Tunçtürk, R.; Şelem, E.; Eroğlu, H. Determination of Morphological and Quality Characteristics of Naturally Growing Thymus Kotschyanus Boiss. & Hohen. Var. Kotschyanus Populations Around of Van/Türkiye. Plants 2025, 14, 729. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Pan, X.; Tian, L.; Xu, Y.; Yang, M.; Deng, R. Application of Salicylic Acid Improves the Production of Medicinal Components in Mucuna Macrocarpa Wall by Regulating Endogenous Hormone and Nutrient Balance. Plants 2025, 14, 1023. [Google Scholar] [CrossRef]
- Yu, G.-R.; Kim, D.-H.; Kim, H.; Lim, D.-W. Evaluation of Cannabis sativa L. Callus Extract as a Novel Cosmetic Ingredient with Dual Anti-Inflammatory and Antioxidant Effects. Plants 2025, 14, 1148. [Google Scholar] [CrossRef]
- Conta, A.; Simirgiotis, M.J.; Martínez Chamás, J.; Isla, M.I.; Zampini, I.C. Extraction of Bioactive Compounds from Larrea Cuneifolia Cav. Using Natural Deep Eutectic Solvents: A Contribution to the Plant Green Extract Validation of Its Pharmacological Potential. Plants 2025, 14, 1016. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Kan, H.; Wu, Q.; Li, C.; Zheng, Y.; Peng, G. Inhibitory Effect of Salvia Miltiorrhiza Extract and Its Active Components on Cervical Intraepithelial Neoplastic Cells. Molecules 2022, 27, 1582. [Google Scholar] [CrossRef]
- Liberato, T.; Pessotti, D.S.; Fukushima, I.; Kitano, E.S.; Serrano, S.M.T.; Zelanis, A. Signatures of Protein Expression Revealed by Secretome Analyses of Cancer Associated Fibroblasts and Melanoma Cell Lines. J. Proteom. 2018, 174, 1–8. [Google Scholar] [CrossRef]
- Pavez Lorie, E.; Stricker, N.; Plitta-Michalak, B.; Chen, I.-P.; Volkmer, B.; Greinert, R.; Jauch, A.; Boukamp, P.; Rapp, A. Characterisation of the Novel Spontaneously Immortalized and Invasively Growing Human Skin Keratinocyte Line HaSKpw. Sci. Rep. 2020, 10, 15196. [Google Scholar] [CrossRef]
- Danciu, C.; Falamas, A.; Dehelean, C.; Soica, C.; Radeke, H.; Barbu-Tudoran, L.; Bojin, F.; Pînzaru, S.C.; Munteanu, M.F. A Characterization of Four B16 Murine Melanoma Cell Sublines Molecular Fingerprint and Proliferation Behavior. Cancer Cell Int. 2013, 13, 75. [Google Scholar] [CrossRef]
- Chittasupho, C.; Samee, W.; Na Takuathung, M.; Okonogi, S.; Nimkulrat, S.; Athikomkulchai, S. Clerodendrum Chinense Stem Extract and Nanoparticles: Effects on Proliferation, Colony Formation, Apoptosis Induction, Cell Cycle Arrest, and Mitochondrial Membrane Potential in Human Breast Adenocarcinoma Breast Cancer Cells. Int. J. Mol. Sci. 2024, 25, 978. [Google Scholar] [CrossRef]
- Lim, W.-C.; Choi, H.-K.; Kim, K.-T.; Lim, T.-G. Rose (Rosa Gallica) Petal Extract Suppress Proliferation, Migration, and Invasion of Human Lung Adenocarcinoma A549 Cells through via the EGFR Signaling Pathway. Molecules 2020, 25, 5119. [Google Scholar] [CrossRef]
- Nickel, J.; Gohlke, B.-O.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on Drug Classification and Target Prediction. Nucleic Acids Res. 2014, 42, W26–W31. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]



| Solvent | TFC (mg RE/g Extract) | TPC (mg GAE/g Extract) |
|---|---|---|
| Water | 67.55 ± 1.09 | 6.53 ± 0.12 |
| 80% Ethanol | 123.81 ± 1.33 | 10.79 ± 0.33 |
| No. | Name | Formula | Rt (min) | Measured m/z | Adducts | MS/MS Fragment Ions (m/z) |
|---|---|---|---|---|---|---|
| 1 | Quinic acid [27] | C7H12O6 | 0.9461 | 383.1199 | 2M−H | 75.0088; 85.0294; 99.0088; 113.036; 135.0448; 155.0465; 164.072; 191.0564; 208.0613; 226.0647 |
| 2 | Hinokitiol | C10H12O2 | 2.6254 | 165.0909 | M+H | 101.0087; 107.0491; 120.0808; 121.0651; 124.0243; 137.0597; 142.0347; 150.0669; 164.0718; 165.0916 |
| 3 | Aucubin | C15H22O9 | 3.6226 | 345.1196 | M−H | 59.0138; 71.0139; 89.0245; 101.0244; 103.0553; 106.0422; 119.0344; 121.066; 165.0561; 345.1239 |
| 4 | Puerarin | C21H20O9 | 5.1640 | 415.1294 | M−H | 268.0349; 282.0161; 295.0245; 296.0323; 297.0392; 311.0559; 338.1033; 340.0841; 355.1074; 370.1335 |
| 5 | Rutin | C27H30O16 | 5.6846 | 611.1604 | M+H | 57.0342; 71.0497; 85.0288; 129.0551; 137.0226; 153.018; 165.0172; 229.0502; 257.041; 303.0494 |
| 6 | Verbenalin | C17H24O10 | 5.9029 | 411.1284 | M+Na | 68.9977; 85.0291; 99.0442; 125.0232; 151.0388; 161.0607; 207.0648; 217.0505; 231.0655; 249.0754 |
| 7 | Hyperoside | C21H20O12 | 5.9329 | 463.0884 | M−H | 71.0139; 101.0245; 107.0138; 151.0038; 243.0297; 255.0299; 271.0247; 300.0279; 301.0296; 463.0873 |
| 8 | Aloenin | C19H22O10 | 5.9329 | 409.1146 | M−H | 59.0137; 127.0556; 163.0397; 171.0456; 188.0485; 203.0716; 204.041; 215.0351; 232.0373; 247.0614 |
| 9 | Astragalin | C21H20O11 | 5.9514 | 449.1074 | M+H | 85.0289; 103.0545; 107.0493; 131.0491; 163.0751; 175.0758; 269.1171; 286.1434; 287.0547; 448.2018 |
| 10 | Isorhamnetin | C16H12O7 | 6.0029 | 317.0653 | M+H | 153.0180; 228.0412; 229.0487; 245.0438; 246.0508; 257.0428; 273.0378; 274.0468; 285.0389; 317.0649 |
| 11 | Morusin | C25H24O6 | 6.0447 | 438.1905 | M+NH4 | 143.0490; 157.0647; 175.0752; 178.0854; 227.0699; 237.0909; 252.1154; 259.0962; 421.1636; 438.1900 |
| 12 | Cynaroside | C21H20O11 | 6.1693 | 449.1075 | M+H | 57.0345; 59.0139; 71.0137; 99.0452; 101.0243; 125.0247; 143.0349; 331.1775; 373.1867; 475.2188 |
| 13 | Eriocitrin | C27H32O15 | 6.2042 | 595.1685 | M−H | 59.0139; 65.0032; 107.014; 135.0452; 151.0039; 191.0696; 287.0565; 359.1519; 360.1542; 595.1638 |
| 14 | Eriodictyol | C15H12O6 | 6.2751 | 289.0705 | M+H | 67.0547; 69.0704; 109.1013; 153.0181; 161.0599; 173.0596; 187.0754; 271.1695; 288.1979; 289.0695 |
| 15 | Luteolin | C15H10O6 | 6.2884 | 285.0405 | M−H | 107.0135; 133.0295; 149.0249; 151.0038; 175.0404; 199.0394; 217.051; 257.1544; 284.127; 285.0404 |
| 16 | Ononin | C22H22O9 | 6.4809 | 472.1599 | M+ACN+H | 85.0288; 207.0675; 236.069; 254.0806; 263.0576; 264.0648; 280.0585; 281.0677; 310.1066; 472.1595 |
| 17 | Prunin | C21H22O10 | 6.6258 | 433.1149 | M−H | 59.0138; 65.0032; 83.0142; 107.0141; 119.0505; 151.0039; 271.0610; 313.0572; 432.2338; 433.1253 |
| 18 | Diosmin | C28H32O15 | 6.6730 | 607.1100 | M−H | 59.0138; 89.0246; 227.0361; 255.0288; 271.0236; 284.0325; 285.0388; 299.0193; 373.1678; 607.1041 |
| 19 | Isovitexin | C21H20O10 | 6.7012 | 487.1228 | M+CH3OH+K | 308.0604; 327.0789; 334.0365; 336.0555; 349.058; 351.0791; 377.0579; 395.0687; 452.0895; 467.1799 |
| 20 | Tribuloside | C30H26O13 | 6.9509 | 595.1440 | M+H | 52.8999; 69.0341; 81.0338; 91.0547; 119.0492; 147.0438; 165.0546; 287.0544; 291.086; 309.0978 |
| 21 | Kaempferide | C16H12O6 | 7.2449 | 299.0201 | M−H | 133.03; 171.0452; 175.041; 199.0403; 201.0199; 215.0357; 227.0345; 243.0294; 271.0246; 299.0197 |
| 22 | Rhoifolin | C27H30O14 | 7.5514 | 577.1371 | M−H | 119.0505; 145.0297; 211.0404; 213.0555; 239.0352; 241.0503; 268.038; 269.0452; 414.1346; 577.1367 |
| 23 | Procyanidin B2 | C30H26O12 | 7.5530 | 579.1493 | M+H | 58.0658; 119.0493; 147.0439; 271.0595; 294.075; 373.1294; 401.1247; 416.1483; 418.1593; 578.1992 |
| 24 | Kaempferitrin | C27H30O14 | 7.7164 | 633.1810 | M−H | 151.0049; 211.0408; 227.0352; 239.0315; 255.0299; 257.0429; 269.0457; 284.0329; 414.1344; 577.1367 |
| 25 | Naringenin [28] | C15H12O5 | 7.7348 | 271.0610 | M−H | 63.024; 65.0033; 83.0139; 93.0346; 107.0138; 119.0502; 151.0038; 169.0149; 177.0197; 271.0611 |
| 26 | Amygdalin | C20H27NO11 | 8.9810 | 475.1958 | M+NH4 | 151.0755; 165.0546; 181.0858; 186.0673; 217.0858; 229.0856; 366.1457; 397.1637; 415.1758; 457.1857 |
| Solvent | DPPH (IC50, μg/mL) | ABTS (IC50, μg/mL) |
|---|---|---|
| Water | 115.77 ± 4.45 | 458.27 ± 76.50 |
| 80% Ethanol | 88.14 ± 5.34 | 100.22 ± 6.84 |
| No. | Target Name | Common Name | Uniprot ID | Degree | Betweenness Centrality | Closeness Centrality |
|---|---|---|---|---|---|---|
| 1 | Signal transducer and activator of transcription 3 | STAT3 | P40763 | 28 | 0.0657624 | 0.66666667 |
| 2 | Mammalian target of rapamycin | MTOR | P42345 | 25 | 0.08375584 | 0.63291139 |
| 3 | Hypoxia Inducible Factor 1 Subunit Alpha | HIF1A | Q16665 | 24 | 0.06052699 | 0.63291139 |
| 4 | C-X-C Motif Chemokine Receptor 4 | CXCR4 | P61073 | 22 | 0.07601931 | 0.61728395 |
| 5 | Kinase Insert Domain Receptor | KDR | P35968 | 21 | 0.04383475 | 0.6097561 |
| 6 | Toll-Like Receptor 4 | TLR4 | O00206 | 21 | 0.13030473 | 0.58139535 |
| 7 | Interleukin 2 | IL2 | P60568 | 21 | 0.06060622 | 0.57471264 |
| 8 | Signal transducer and activator of transcription 1 | STAT1 | P42224 | 20 | 0.02031513 | 0.60240964 |
| 9 | Mitogen-Activated Protein Kinase 1 | MAPK1 | P28482 | 20 | 0.05702655 | 0.57471264 |
| 10 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit | EZH2 | Q15910 | 19 | 0.10237625 | 0.55555556 |
| No. | Compound | raw-cs | q-Value | norm-cs |
|---|---|---|---|---|
| 1 | Naringenin | −0.47 | 4.467 × 10−16 | −1.51 |
| 2 | Glycitein | −0.45 | 0.004 | −1.45 |
| 3 | Eugenol | −0.44 | 0.009 | −1.41 |
| 4 | Isoquercetin | −0.44 | 0.009 | −1.42 |
| 5 | Quercetagetin | −0.44 | 0.008 | −1.42 |
| 6 | Quercetin | −0.41 | 0.021 | −1.31 |
| 7 | Rosmarinic acid | −0.41 | 0.021 | −1.31 |
| 8 | Chaetocin | −0.41 | 0.020 | −1.32 |
| 9 | Butein | −0.40 | 0.029 | −1.29 |
| 10 | Arctigenin | −0.40 | 0.029 | −1.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Jiao, G.; Ou, P.; Zhang, X.; Yu, Y.; Wang, Y.; Yao, Q.; Wang, W. Phytochemical Characterization, Antioxidant Activity, and Anti-Melanoma Mechanism of Flower Buds of Magnolia biondii Pamp. Plants 2025, 14, 1725. https://doi.org/10.3390/plants14111725
Li S, Jiao G, Ou P, Zhang X, Yu Y, Wang Y, Yao Q, Wang W. Phytochemical Characterization, Antioxidant Activity, and Anti-Melanoma Mechanism of Flower Buds of Magnolia biondii Pamp. Plants. 2025; 14(11):1725. https://doi.org/10.3390/plants14111725
Chicago/Turabian StyleLi, Shanshan, Gege Jiao, Penghui Ou, Xiaona Zhang, Yang Yu, Yihui Wang, Qingping Yao, and Wei Wang. 2025. "Phytochemical Characterization, Antioxidant Activity, and Anti-Melanoma Mechanism of Flower Buds of Magnolia biondii Pamp." Plants 14, no. 11: 1725. https://doi.org/10.3390/plants14111725
APA StyleLi, S., Jiao, G., Ou, P., Zhang, X., Yu, Y., Wang, Y., Yao, Q., & Wang, W. (2025). Phytochemical Characterization, Antioxidant Activity, and Anti-Melanoma Mechanism of Flower Buds of Magnolia biondii Pamp. Plants, 14(11), 1725. https://doi.org/10.3390/plants14111725






