CtWRKY41 Transcription Factor from Cynanchum thesioides Mediates Salt Stress Resistance and Controls Flowering Time
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Cultivation and Treatment of Cynanchum thesioides Seedlings
2.3. Quantitative Real-Time PCR (qRT-PCR) Analysis
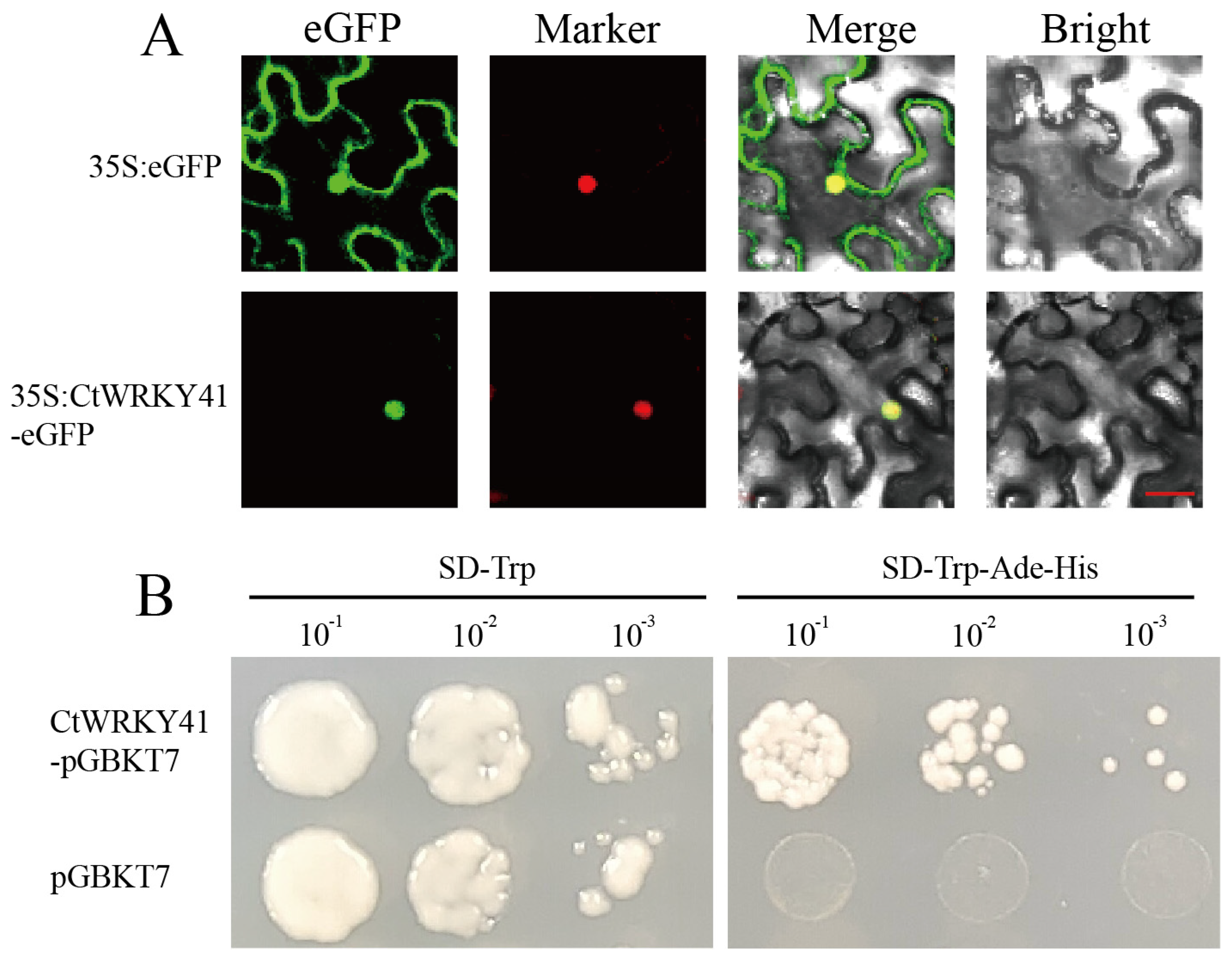
2.4. Detection of CtWRKY41 Subcellular Localization and Transcriptional Activation Assay
2.5. Phylogenetic and Motif Analysis of CtWRKY41
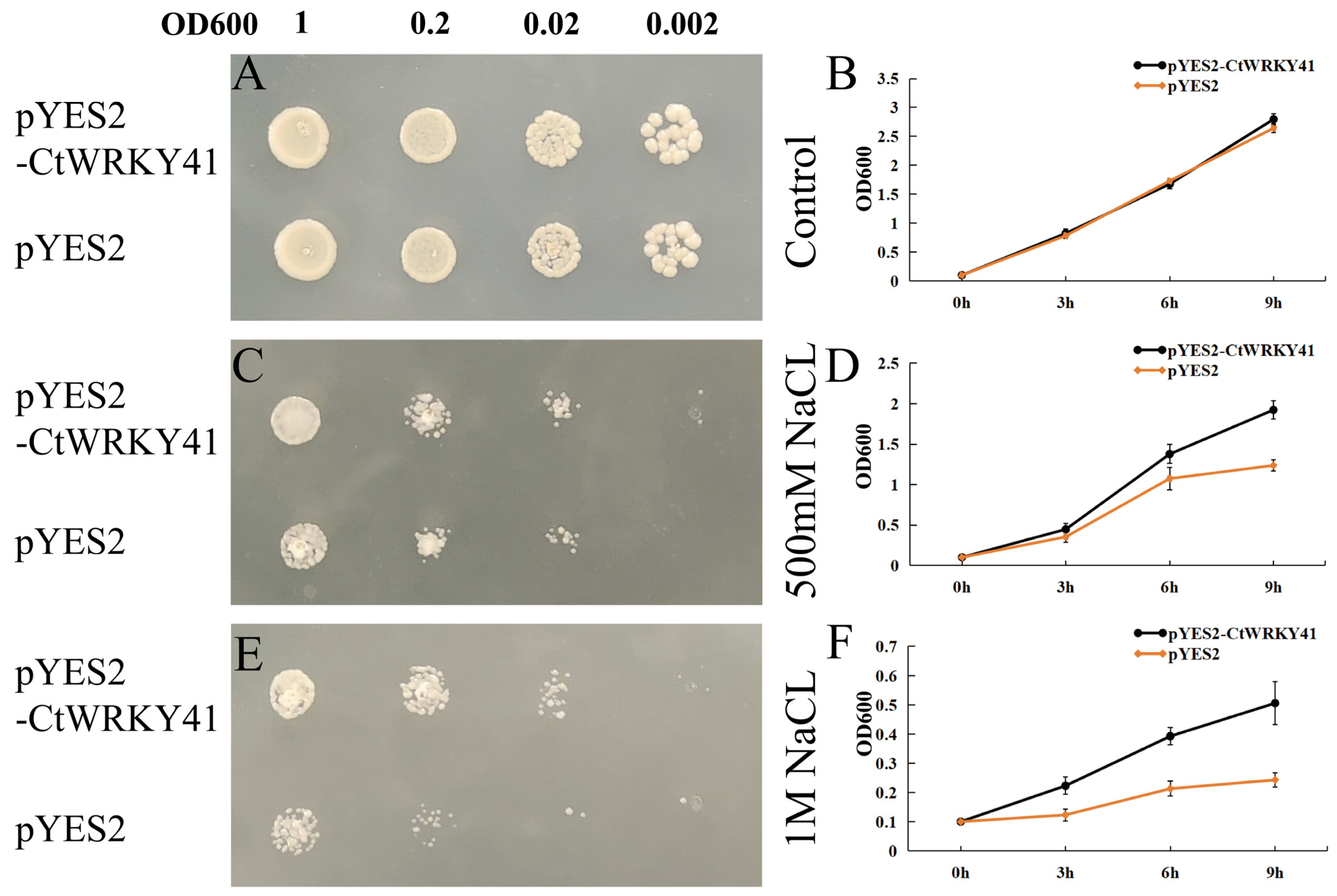
2.6. Evaluation of Tolerance of INVSc1 to Salt Stress
2.7. Stable Arabidopsis Lines Overexpressing CtWRKY41
2.8. Phenotypic Observation of Arabidopsis thaliana and Determination of Related Physiological and Biochemical Indexes
2.9. Statistical Analysis
3. Results
3.1. Sequence Characteristics and Expression Patterns of CtWRKY41
3.2. Subcellular Localization and Transcriptional Activation of CtWRKY41
3.3. Overexpression CtWRKY41 Enhanced Yeast Resistance to NaCl
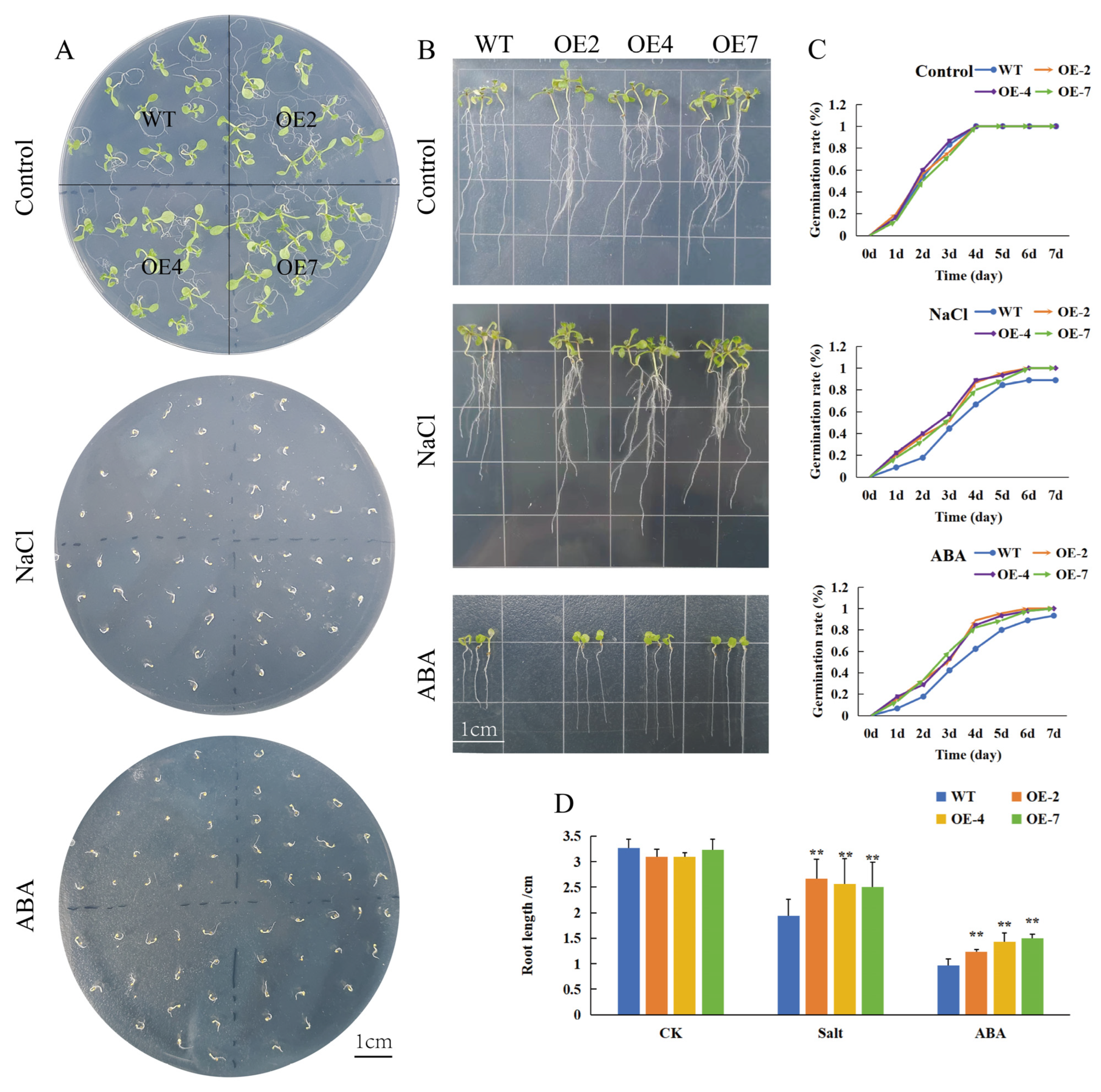
3.4. CtWRKY41 Participates in ABA Response and Enhances Salt Tolerance in Arabidopsis
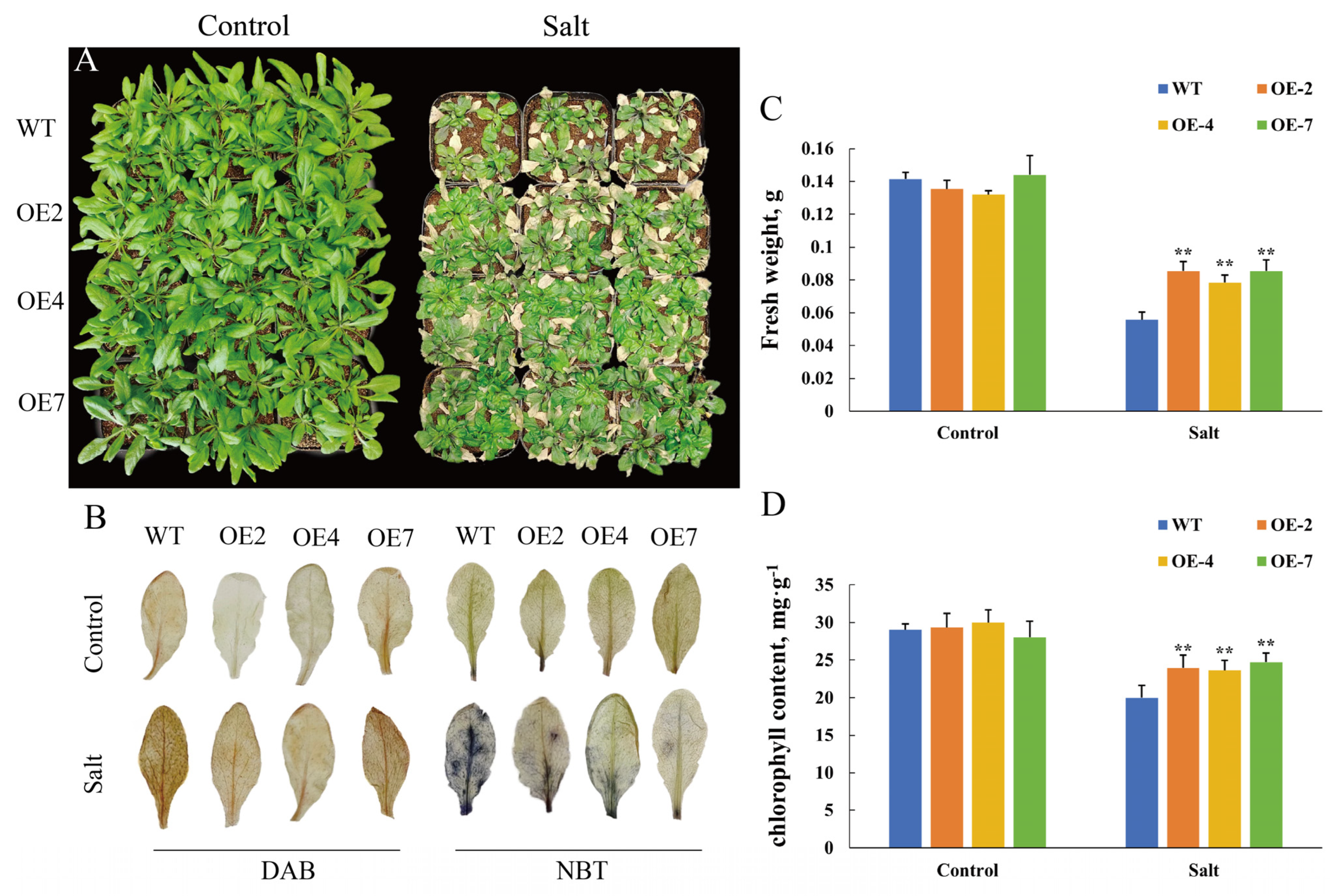
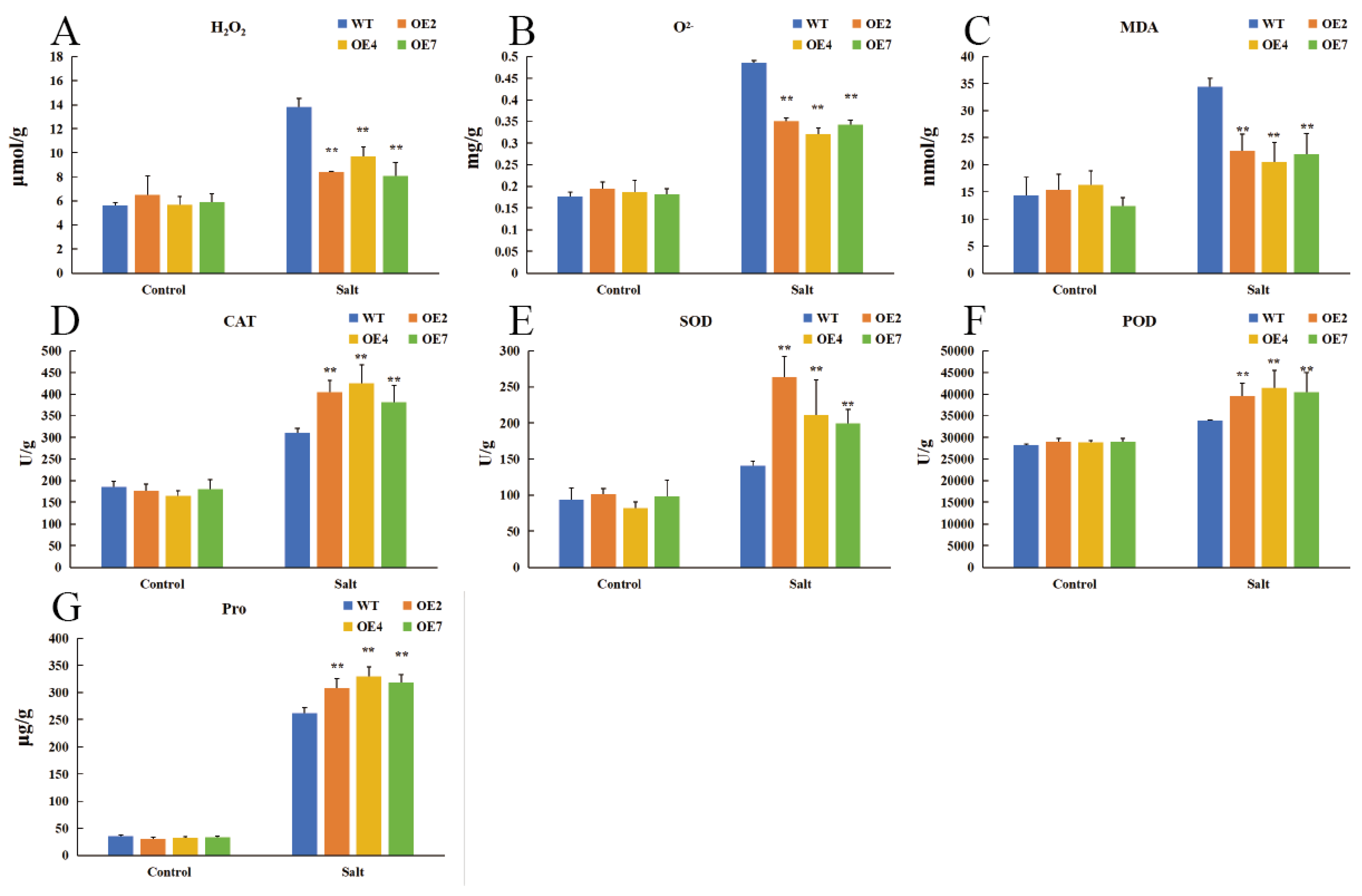
3.5. CtWRKY41 Enhances Physiological Indicators and Antioxidant Capacity in Transgenic Arabidopsis
3.6. CtWRKY41 Regulates the Expression of Stress-Responsive Genes
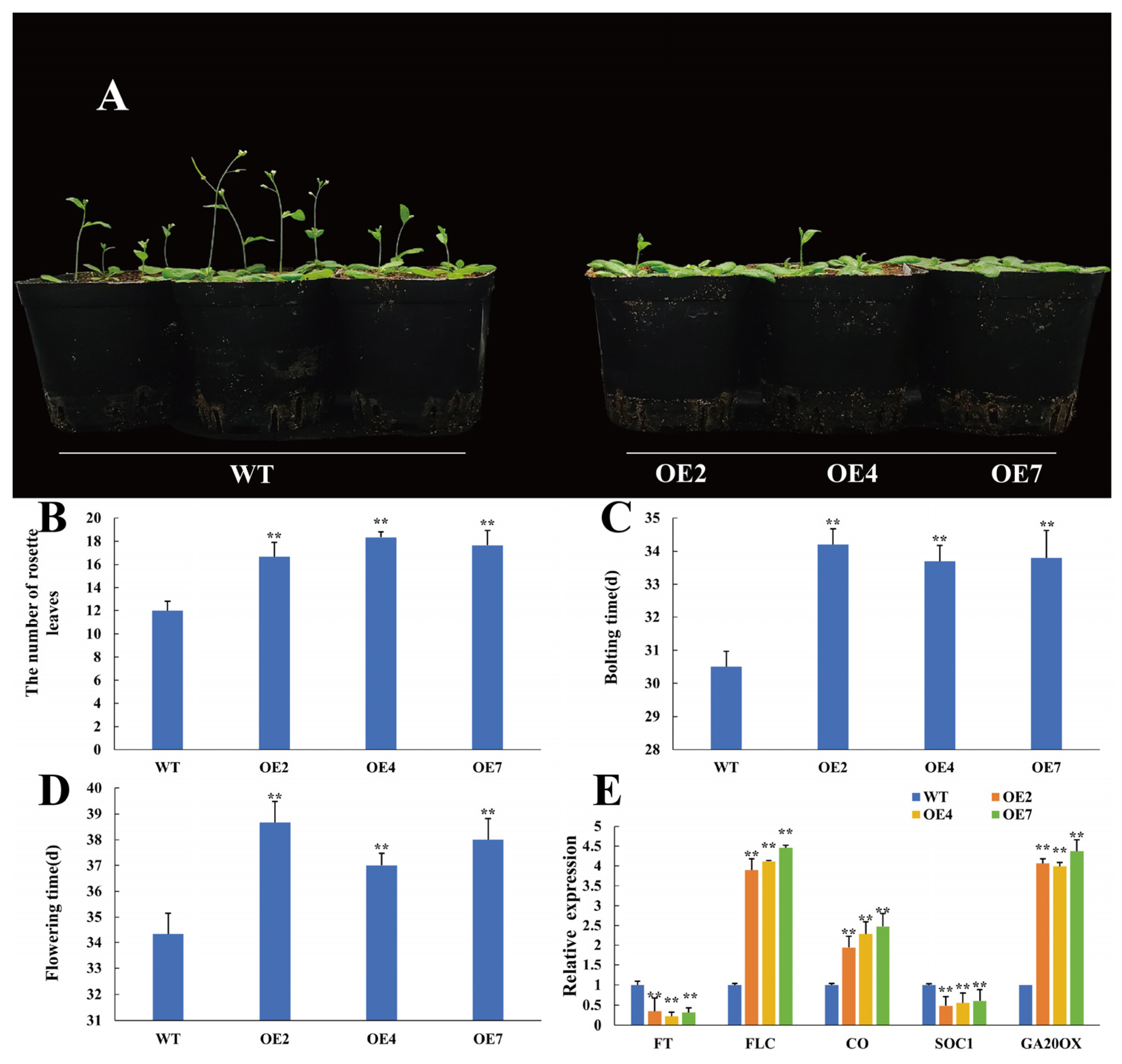
3.7. CtWRKY41 Delays Flowering Time in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, X.; Wang, Y.; Song, S.; Ma, L.; He, Q.; Lu, M.; Zhang, K.; Yang, Y.; Zhao, Q.; et al. Inhibition of the maize salt overly sensitive pathway by ZmSK3 and ZmSK4. J. Genet. Genom. 2023, 50, 960–970. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, L.; Chen, L.; Pan, X.; Lin, K.; Fang, Y.; Wang, X.E.; Zhang, W. Salt-responsive genes are differentially regulated at the chromatin levels between seedlings and roots in rice. Plant Cell Physiol. 2019, 60, 1790–1803. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Guo, X.; Ullah, A.; Siuta, D.; Kukfisz, B.; Iqbal, S. Role of WRKY transcription factors in regulation of abiotic stress responses in cotton. Life 2022, 12, 1410. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Qiao, L.Y.; Guo, H.Y.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-a is referred to drought and salt resistances. Front. Plant Sci. 2021, 12, 663118. [Google Scholar] [CrossRef]
- Chang, X.; Yang, Z.; Zhang, X.; Zhang, F.; Huang, X.; Han, X. Transcriptome-wide identification of WRKY transcription factors and their expression profiles under different stress in Cynanchum thesioides. PeerJ 2022, 10, e14436. [Google Scholar] [CrossRef]
- Song, H.; Cao, Y.; Zhao, L.; Zhang, J.; Li, S. Review: WRKY transcription factors: Understanding the functional divergence. Plant Sci. 2023, 334, 111770. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Grzechowiak, M.; Ruszkowska, A.; Sliwiak, J.; Urbanowicz, A.; Jaskolski, M.; Ruszkowski, M. New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain. Int. J. Biol. Macromol. 2022, 213, 589–601. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, Y.; He, L. A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant. Plant Mol. Biol. 2023, 113, 171–191. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.K.; Yang, F.M.; Zhang, G.Y.; Wang, D.; Zhang, L.; Ou, Y.B.; Yao, Y.A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant. 2020, 168, 98–117. [Google Scholar] [CrossRef]
- Ping, X.; Ye, Q.; Yan, M.; Wang, J.; Zhang, T.; Chen, S.; Siddique, K.H.M.; Cowling, W.A.; Li, J.; Liu, L. Overexpression of BnaA10.WRKY75 decreases cadmium and salt tolerance via increasing ROS accumulation in Arabidopsis and Brassica napus L. Int. J. Mol. Sci. 2024, 25, 8002. [Google Scholar]
- Zhang, J.H.; Jia, W.S.; Yang, J.C.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Shen, Y.Y.; Wang, X.F.; Wu, F.Q.; Du, S.Y.; Cao, Z.; Shang, Y.; Wang, X.L.; Peng, C.C.; Yu, X.C.; Zhu, S.Y.; et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 2006, 443, 823–826. [Google Scholar] [CrossRef]
- Min, M.K.; Kim, R.; Hong, W.J.; Jung, K.H.; Lee, J.Y.; Kim, B.G. OsPP2C09 is a bifunctional regulator in both ABA-dependent and independent abiotic stress signaling pathways. Int. J. Mol. Sci. 2021, 22, 393. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Nelson, D.C.; Assmann, S.M. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 2009, 136, 136–148. [Google Scholar] [CrossRef]
- Qin, Y.; Tian, Y.; Han, L.; Yang, X. Constitutive expression of a salinity-induced wheat WRKY transcription factor enhances salinity and ionic stress tolerance in transgenic Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2013, 441, 476–481. [Google Scholar] [CrossRef]
- Ullah, A.; Sun, H.; Hakim; Yang, X.; Zhang, X. A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 2018, 162, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Z.; Wang, L.; Kim, S.G.; Seo, P.J.; Qiao, M.; Wang, N.; Li, S.; Cao, X.; Park, C.M.; et al. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016, 85, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Niu, Z.; Shi, G.; Song, Z.; Yang, Q.; Zhou, S.; Wang, L. WRKY22 Transcription Factor from Iris laevigata Regulates Flowering Time and Resistance to Salt and Drought. Plants 2024, 13, 1191. [Google Scholar] [CrossRef]
- Du, C.; Ma, B.; Wu, Z.; Li, N.; Zheng, L.; Wang, Y. Reaumuria trigyna transcription factor RtWRKY23 enhances salt stress tolerance and delays flowering in plants. J. Plant Physiol. 2019, 239, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, L.; Chen, J.; Liu, Z.; Park, C.M.; Xiang, F. WRKY71 Acts Antagonistically Against Salt-Delayed Flowering in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 414–422. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Z.; Li, Z.; Zhang, F.; Hao, L. De novo transcriptome assembly and co-expression network analysis of Cynanchum thesioides: Identification of genes involved in resistance to drought stress. Gene 2019, 710, 375–386. [Google Scholar] [CrossRef]
- Chang, X.; Zhang, X.; Huang, X.; Yang, Z.; Zhang, F. Transcriptome and metabolome analysis of the developmental changes in Cynanchum thesioides anther. Genomics 2024, 116, 110884. [Google Scholar] [CrossRef]
- Acosta-Motos, J.; Ortuño, M.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.; Hernandez, J. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Jaspers, P.; Kangasjarvi, J. Reactive oxygen species in abiotic stress signaling. Physiol. Plant. 2010, 138, 405–413. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Ben Rejeb, K.; Abdelly, C.; Savouré, A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014, 80, 278–284. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Liu, J.; Wang, W.; Wang, L.; Sun, Y. Exogenous melatonin improves seedling health index and drought tolerance in tomato. Plant Growth Regul. 2015, 77, 317–326. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, L. WRKY transcription factor responses and tolerance to abiotic stresses in plants. Int. J. Mol. Sci. 2024, 25, 6845. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, C.; He, X.; Zhang, X.; Zhu, L. ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 2020, 39, 181–194. [Google Scholar] [CrossRef]
- Jiang, Y.; Tong, S.; Chen, N.; Liu, B.; Bai, Q.; Chen, Y.; Bi, H.; Zhang, Z.; Lou, S.; Tang, H.; et al. The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus. Plant J. 2021, 105, 1258–1273. [Google Scholar] [CrossRef]
- Zhang, A.; Shang, J.; Xiao, K.; Zhang, M.; Wang, S.; Zhu, W.; Wu, X.; Zha, D. WRKY transcription factor 40 from eggplant (Solanum melongena L.) regulates ABA and salt stress responses. Sci. Rep. 2024, 14, 19289. [Google Scholar] [CrossRef]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Seo, Y.J.; Park, J.B.; Cho, Y.J.; Jung, C.; Seo, H.S.; Park, S.K.; Nahm, B.H.; Song, J.T. Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol. Cells 2010, 30, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hu, X.; Song, J.; Zong, X.; Li, D. Over-expression of a Zea mays L. protein phosphatase 2C gene (ZmPP2C) in Arabidopsis thaliana decreases tolerance to salt and drought. J. Plant Physiol. 2009, 166, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.; Dean, C. The FLC Locus: A Platform for Discoveries in Epigenetics and Adaptation. Annu. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar] [CrossRef]
- Moon, J.; Lee, H.; Kim, M.; Lee, I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005, 46, 292–299. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, X.; Zhang, X.; Huang, X.; Zhang, F.; Yang, Z. CtWRKY41 Transcription Factor from Cynanchum thesioides Mediates Salt Stress Resistance and Controls Flowering Time. Plants 2025, 14, 1716. https://doi.org/10.3390/plants14111716
Chang X, Zhang X, Huang X, Zhang F, Yang Z. CtWRKY41 Transcription Factor from Cynanchum thesioides Mediates Salt Stress Resistance and Controls Flowering Time. Plants. 2025; 14(11):1716. https://doi.org/10.3390/plants14111716
Chicago/Turabian StyleChang, Xiaoyao, Xiaoyan Zhang, Xiumei Huang, Fenglan Zhang, and Zhongren Yang. 2025. "CtWRKY41 Transcription Factor from Cynanchum thesioides Mediates Salt Stress Resistance and Controls Flowering Time" Plants 14, no. 11: 1716. https://doi.org/10.3390/plants14111716
APA StyleChang, X., Zhang, X., Huang, X., Zhang, F., & Yang, Z. (2025). CtWRKY41 Transcription Factor from Cynanchum thesioides Mediates Salt Stress Resistance and Controls Flowering Time. Plants, 14(11), 1716. https://doi.org/10.3390/plants14111716





