Abstract
Basic helix–loop–helix (bHLH) transcription factors play significant roles in plant growth and organ development and diverse biochemical processes. However, the function of bHLH transcription factors in woody plants is not fully understood. In this study, the bHLH gene family in Rhododendron × pulchrum Sweet was identified and characterized using whole-genome data. A total of 109 bHLH family genes (RpbHLHs) were identified in R. pulchrum, and their expression levels were analyzed in flowers of different colors and developmental stages. The results showed that the RpbHLH family is divided into 24 subfamilies. Chromosomal localization and collinearity analyses revealed numerous duplication events during evolution, which is one of the main reasons for the diversification of gene functions. The bHLH domains showed relative conservation of RpbHLH proteins. In the promoter regions of the RpbHLHs, various cis-regulatory elements involved in light response, gibberellic acid (GA) response, and abscisic acid (ABA) response were identified. These elements may regulate flower development and pigment synthesis. A Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis of the target RpbHLHs revealed that 25 genes are enriched in the flavonoid biosynthetic pathway. Potential RpbHLHs related to flower development and pigment synthesis were identified through a transcriptome analysis and validated through quantitative reverse transcription PCR (qRT-PCR). This study will enhance our understanding of RpbHLH functions and provide a reference for the study of flower development and coloration in R. pulchrum.
1. Introduction
Rhododendron × pulchrum Sweet (R. pulchrum) belongs to the Rhododendron genus in the Ericaceae family. As an alpine flower, it has been extensively cultivated in China for a considerable period. Recognized as one of China’s top ten most famous flowers, R. pulchrum boasts lush foliage and vibrant blooms, making it a popular choice for landscaping [1]. In addition to its ornamental value, R. pulchrum possesses significant economic and medicinal utility. Its leaves are used for fragrance extraction, while the petals serve as a source of essential oils. The chemical compounds derived from this plant find applications as cardiovascular, anesthetic, and antimalarial medications [1].
The basic helix–loop–helix (bHLH) transcription factor is the largest family of transcription factors in plants [2]. The bHLH transcription factor has a highly conserved basic helix–loop–helix domain [3], which consists of 50–60 amino acids and is divided into two regions: the N-terminal basic region and the C-terminal α-helix 1-loop-α-helix 2 (HLH) region [2]. The basic region contains 15–20 conserved amino acids, which determine the recognition of the core E-box (5′-CANNTG-3′) and the binding of bHLH transcription factors to DNA [3]. The HLH region is composed of two hydrophobic residues linked by a more diverse loop region, allowing the HLH domain to facilitate protein–protein interactions and form homo- or heterodimer complexes, giving rise to two amphipathic α-helices separated by a loop of a variable length [2,4,5]. Previous studies have analyzed the whole-genome sequences of nine terrestrial plants and algae, classifying the bHLH gene family into 15–26 subfamilies [2,4]. Many bHLH gene families have been identified in Arabidopsis, Prunus mume (P. mume), Passiflora edulis (P.mume), Solanum lycopersicum (S. lycopersicum), and Malus domestica [6,7,8,9,10]. However, the identification of the bHLH (RpbHLH) gene family in R. pulchrum has not been reported.
In anthocyanin biosynthesis, bHLH transcription factors act as key regulators. Anthocyanins, one of the major pigments in plants, determine flower color diversity. bHLH transcription factors regulate key genes in the anthocyanin biosynthesis pathway, directly influencing flower color formation. Specifically, bHLH transcription factors form complexes with other transcription factors such as MYB and WD40 to regulate the expression of genes involved in anthocyanin biosynthesis, including CHS (chalcone synthase), F3H (flavanone 3-hydroxylase), and DFR (dihydroflavonol 4-reductase), thereby promoting anthocyanin accumulation and influencing flower color [11]. For instance, in Arabidopsis, bHLH transcription factors such as TT8 interact with MYB-type transcription factors like MYB75, forming a complex that activates the expression of key genes in the anthocyanin biosynthesis pathway, thereby promoting anthocyanin accumulation and influencing flower color formation [12].
In addition to flower color regulation, bHLH genes also play crucial roles in flower development. Studies have shown that bHLH transcription factors regulate the development of various plant organs, including anther and carpel development. In Arabidopsis, the bHLH transcription factor SPATULA, expressed in the anthers, regulates carpel development [13]. In rice, UDT1 regulates anther development and is essential for tapetum and microspore maturation [14]. By regulating downstream target genes, bHLH transcription factors promote the normal development of flowers, ensuring the correct formation and function of the floral organs.
Although bHLH genes have been well studied in many plants and their roles in flower development and color regulation are well established, the functional roles of the bHLH gene family in woody plants, particularly in the genus Rhododendron, remain underexplored. This study aims to systematically identify the bHLH gene family in R. pulchrum through a genome-wide analysis and to explore their roles in flower development and color regulation through a gene expression analysis. We combined gene expression data to investigate further how bHLH genes influence flower color by regulating the anthocyanin biosynthesis pathway and explored their potential roles in flower development. In addition, this study lays the foundation for future functional validation experiments, including the identification of specific pigments involved in flower colors, and aims to provide valuable theoretical support for understanding the regulatory mechanisms of bHLH genes in flower development. Ultimately, this research not only contributes to the identification of bHLH genes related to flower colors in R. pulchrum but also offers a foundation for further functional studies and applications in horticultural practices and ornamental plant breeding.
2. Results
2.1. Flower-Color- and Stage-Specific Variations in Flavonoid and Anthocyanin Contents
Distinct pigmentation patterns were observed among flower colors during development (Figure 1). From the bud stage (S1) to the fully open flower stage (S3), both the flavonoid and anthocyanin contents increased continuously in purple flowers. Pink flowers also showed an increasing trend in their pigment content, though at significantly lower levels than those in purple flowers. In contrast, white flowers maintained low levels of flavonoids and had almost undetectable anthocyanin levels throughout development.
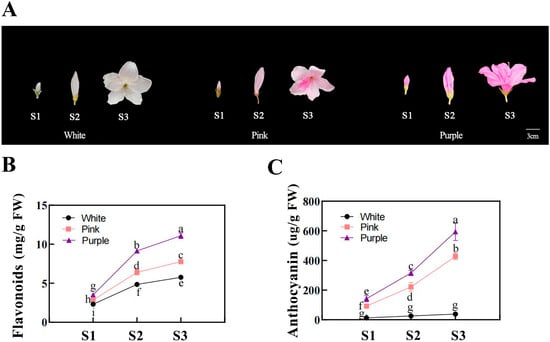
Figure 1.
Flower development and pigment analyses of different flower colors in Rhododendron simsii at various stages. (A) Photographs of Rhododendron simsii flowers at three developmental stages—the bud stage (S1), the flowering stage (S2), and the fully open flower stage (S3)—for white, pink, and purple flower colors. Scale bar: 3 cm. (B) The flavonoid content (mg/g) in flower petals at different developmental stages (S1, S2, S3) for white, pink, and purple flowers. Different letters indicate significant differences between stages within each flower color (p < 0.05). (C) Anthocyanin content (µg/g) in flower petals at different developmental stages (S1, S2, S3) for white, pink, and purple flowers. Different letters indicate significant differences between stages within each flower color (p < 0.05).
2.2. The Identification of the RpbHLH Family and an Analysis of the Physicochemical Properties of RpbHLH Proteins
To screen and identify the bHLH gene family in R. pulchrum, Hmmscan was employed to align with the PFAM database. A total of 109 candidate genes encoding basic helix–loop–helix (bHLH) proteins (RpbHLH) were selected from the genomic database of R. pulchrum and subsequently renamed RpbHLH01–RpbHLH109.
The basic features, including the amino acid count, protein molecular weight, isoelectric point, coding sequence length, grand average of hydropathicity, aliphatic index, and subcellular localization, were analyzed for the RpbHLH proteins identified (Table S1). Among the 109 RpbHLH proteins, the average amino acid count was 397. The molecular weight of the RpbHLH proteins ranged from 1.82 kDa (for RpbHLH105, which had the lowest amino acid count of 163) to 16.03 kDa (for RpbHLH99, which had the highest amino acid count of 1407). The isoelectric points were between 4.48 (RpbHLH80) and 10.37 (RpbHLH95), averaging 6.73. The longest coding sequence was found in RpbHLH99, with 4221 nucleotides, while the shortest was in RpbHLH105, with 489 nucleotides. The grand average of hydropathicity values were all negative, indicating that all of the RpbHLHs were predominantly hydrophilic proteins. The aliphatic index was between 46.87 (RpbHLH61) and 103.9 (RpbHLH72). In terms of their subcellular localization, a total of 98 of the RpbHLH proteins were found to be predominantly localized in the nucleus, with 4 proteins detected in the cytoplasm. Additionally, two proteins were associated with the chloroplasts, and one protein each was identified in both the nucleus and the cytoplasm, the Golgi apparatus, the endoplasmic reticulum, the vacuole, and the peroxisome.
2.3. The Phylogenetic Analysis of the RpbHLHs
To investigate the evolutionary relationships of the Rhododendron × pulchrum Sweet (RpbHLH) gene family further and classify its subfamilies, the protein sequences of the 109 identified RpbHLHs were aligned with 166 Arabidopsis bHLH proteins (AtbHLHs) using Mafft v7.313 and FastTree 2.1.11 to construct a phylogenetic tree. As shown in Figure 2, the 275 bHLH genes of R. pulchrum were grouped into 31 subfamilies according to the taxonomic classification proposed in previous studies [5,15], with the subfamily numbers ranging from 1 to 31. The distribution of the 109 RpbHLHs across the subfamilies was uneven. Subfamily 25 contained the highest number of genes (14), while subfamily 19 included only a single gene (RpbHLH). Notably, no RpbHLHs were found in subfamilies 6, 8, 18, 20, 21, 22, or 29, suggesting the potential loss of these subfamilies during the evolution of R. pulchrum. Subfamilies 15, 16, 17, and 23 were exclusively composed of AtbHLH genes, while the remaining subfamilies contained a mixture of both RpbHLH and AtbHLH genes, indicating species-specific differences in gene family evolution.
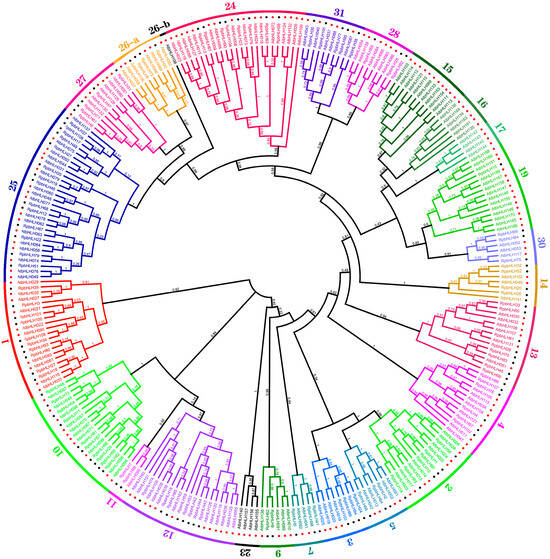
Figure 2.
The maximum likelihood phylogenetic tree of bHLH proteins from R. pulchrum and Arabidopsis thaliana. The tree was constructed using FastTree software. Branches of different colors represent distinct subfamilies. The numbers in the outer circles denote the subgroup names. The red dots represent Arabidopsis genes, and the black dots represent Rhododendron genes.
These findings reveal significant evolutionary divergence between Rhododendron × pulchrum Sweet and Arabidopsis thaliana, potentially linked to the unique flower color characteristics of R. pulchrum. The clustering of bHLH genes into specific subfamilies suggests that certain subfamilies may be involved in regulating flower color and developmental stages. The uneven distribution of the RpbHLHs across these subfamilies, with some subfamilies containing only R. pulchrum-specific genes, suggests that these genes may specialize in controlling flower pigmentation and developmental processes. This study underscores the crucial role of the bHLH gene family in regulating both flower color and developmental stages in R. pulchrum. Specific subfamilies may influence the formation of different flower colors, as illustrated in Figure 2. A further analysis of the expression patterns of these genes across various developmental stages and flower color morphs will provide deeper insights into the molecular mechanisms underlying flower color variations in R. pulchrum.
2.4. Analysis of RpbHLH Structures
First, a phylogenetic analysis of the 109 RpbHLH protein sequences was carried out, and the RpbHLH proteins were divided into 31 subfamilies (1–31) (Figure 3A).
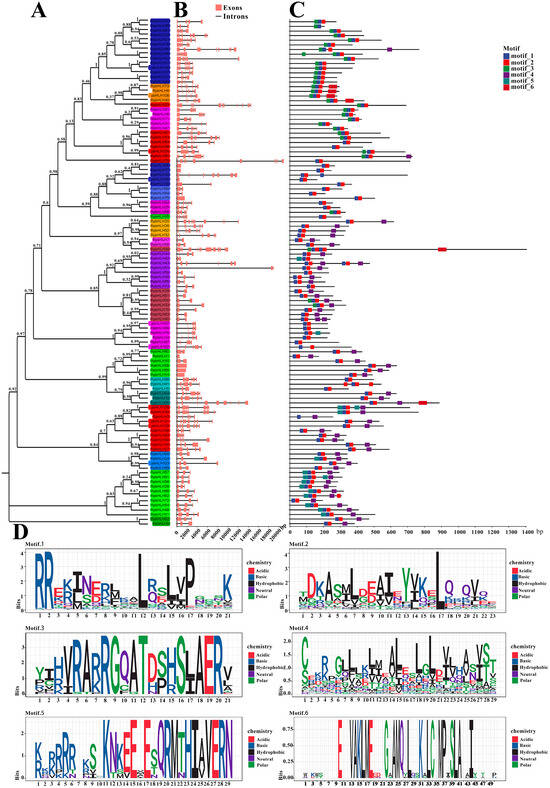
Figure 3.
The phylogenetic relationship, conserved motifs, and gene structure of RpbHLHs. (A) A neighbor-joining phylogenetic tree of the RpbHLHs. (B) The gene structures of RpbHLHs, including exons (pink boxes) and introns (black lines), with the gene lengths displayed at the bottom. (C) Identification of conserved motifs in RpbHLHs using MEME software; six patterns were identified. (D) Display of sequences of conserved Motifs 1–6: motif 1 consists of an α-helix 1 region and half a loop region; motif 2 contains an α-helix 2 region and a loop region; motif 3 represents an acidic region; and motifs 4–6 denote non-conserved regions.
The analysis of the RpbHLH structures revealed that the numbers of introns and exons of the 109 identified RpbHLHs within the same subfamily were also similar, especially among homologous branches. As indicated in Figure 3B, the RpbHLH family contained between 1 and 15 exons, with 20 RpbHLHs having 3 exons and 42 RpbHLHs possessing 2 to 7 exons.
The conservation motifs and gene structures of the 109 identified RpbHLHs were analyzed using MEME 5.4.1. As illustrated in Figure 3D, the analysis revealed six conserved motifs (motifs 1–6). Motif 1 contained an α-helix 1 region and a half-loop region, while motif 2 included an α-helix 2 region and a loop region. Motif 3 was primarily composed of an acidic region. The majority of the RpbHLH family contained motifs 1 and 2, with the exception of RpbHLH35/3, which solely contained motif 1 (Figure 3C). Furthermore, the conserved motifs of RpbHLHs within the same subfamily were highly similar, suggesting functional conservation within subfamilies. These findings provide valuable insights into the evolutionary and structural diversity of the RpbHLH family. The conserved motifs and gene structures are crucial for understanding the functional roles of these genes in regulating key processes, such as flower color and development, in R. pulchrum.
2.5. Chromosomal Localization and Collinearity Analysis of RpbHLHs
To examine the chromosomal distributions of the RpbHLH families, each identified RpbHLH was mapped to 13 physical positions on the R. pulchrum chromosomes according to the gene annotation information (Table S2). As shown in Figure 4, on average, each chromosome contained 5–7 RpbHLHs. Chromosome 3 and chromosome 7 contained 15 and 11 RpbHLHs, respectively, while only 1 RpbHLH was found in chromosomes 143, 216, 228, and 235. A homology analysis of the Rhododendron genes was performed using OrthoFinder2 2.5.4, identifying 59 pairs of homologous genes. Among them, 56 pairs exhibited homology between gene chromosomes, while 3 pairs of genes, namely RpbHLH42 and RpbHLH43, RpbHLH42 and RpbHLH44, and RpbHLH100 and RpbHLH101, exhibited homology within the same chromosome. The ratio of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks), or Ka/Ks, was calculated for the 59 pairs of homologous genes. The Ka/Ks ratio reflects a gene’s evolutionary processes and selection pressures. A Ka/Ks ratio of less than 1 for all pairs indicates that the RpbHLHs underwent purifying selection during evolution, which plays a critical role in maintaining the stability of the bHLH conserved domain (Table S3). These findings underscore the importance of the RpbHLH family in maintaining the genetic integrity of R. pulchrum while also suggesting potential roles in flower color regulation and development. The distribution and evolutionary patterns of these genes could help elucidate their involvement in the molecular mechanisms that govern flower pigmentation and developmental processes in R. pulchrum.
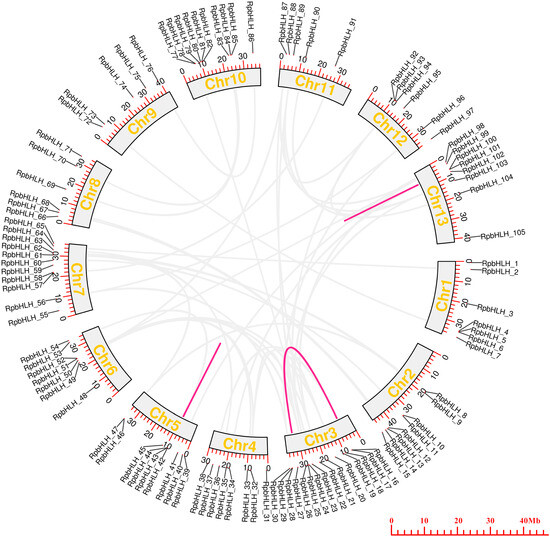
Figure 4.
The collinearity analysis of RpbHLHs in the R. pulchrum genome. The colored outer circles represent chromosomes, with duplicated RpbHLHs mapped to different chromosomes, as shown in the middle section. RpbHLHs exhibiting collinearity relationships are linked by lines. The light gray lines represent homology between genes on different chromosomes, while the magenta lines indicate homology within the same chromosome.
2.6. Prediction of the Regulatory Elements in RpbHLH Promoters
The analysis of cis-acting elements in the promoter regions of the RpbHLHs significantly enhanced our understanding of their regulation, function, and evolutionary dynamics (Table S4). As illustrated in Figure 5, all RpbHLHs contain cis-acting elements associated with light response, indicating that light signaling plays a crucial role in modulating the expression of these genes. Additionally, many RpbHLHs feature cis-acting elements related to various environmental factors and hormonal responses: 102 RpbHLHs contain GA_Response elements, 96 have Wound_Response elements, 92 include Ethylene_Response elements, 77 carry MeJA_Response elements, 60 feature Drought_Response elements, 57 contain Auxin_Response elements, and 5 include Heat_Response elements. The distribution of these cis-acting elements suggests that RpbHLHs play a pivotal role in regulating flower growth and development, particularly in response to environmental stresses such as light, drought, and mechanical damage. The presence of ethylene and MeJA response elements highlights the involvement of these genes in controlling flower senescence, color changes, and flowering processes. This is of particular significance for flower color formation and maintenance, as both the ethylene and jasmonate pathways are key regulators of flower opening and color transitions.
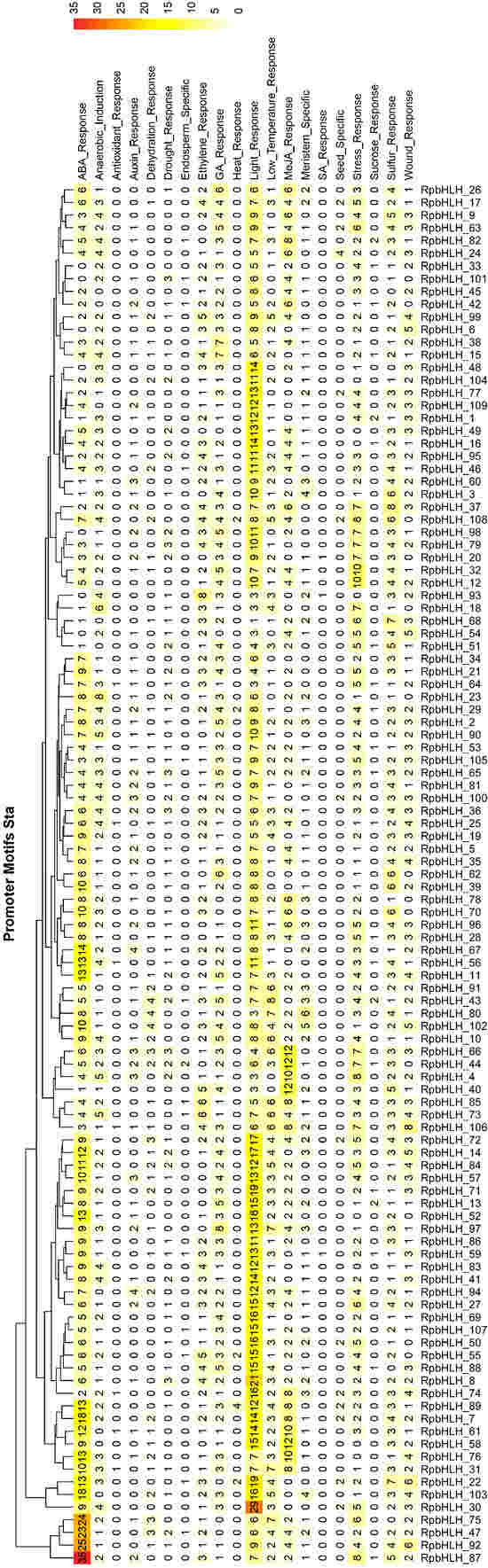
Figure 5.
Prediction of cis-acting elements in RpbHLH promoter regions. The distribution of cis-acting elements in the upstream 2000 bp promoter regions of the RpbHLHs.
2.7. A KEGG Enrichment Analysis of Target RpbHLHs
A KEGG pathway analysis was conducted to perform functional enrichment of the predicted target RpbHLHs (Table S5). The resulting KEGG enrichment pathway map revealed the enrichment of potential target RpbHLHs in 20 distinct pathways (Figure 6). Among these, the pathways exhibiting the highest number of enriched genes were plant hormone signal transduction (109 genes), glycolysis/gluconeogenesis (65 genes), and the MAPK signaling pathway—plant (62 genes). Additionally, the flavonoid biosynthesis pathway, which plays a key role in plant pigmentation, showed the enrichment of 25 genes. These metabolic and signaling pathways provide the necessary energy and regulatory signals to support floral organ development and color formation and the plant’s response to environmental stressors.
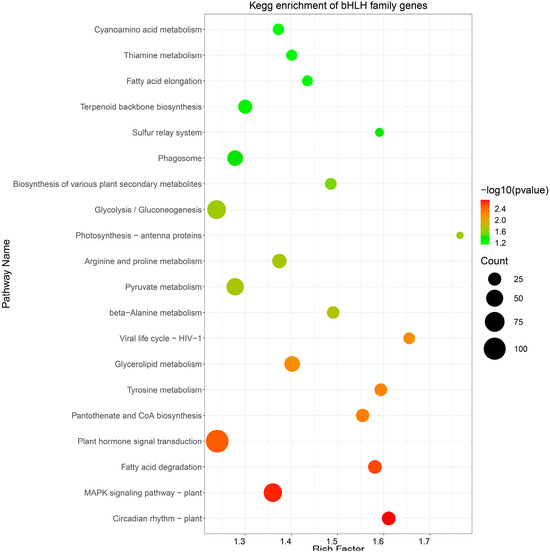
Figure 6.
KEGG enrichment analysis. The pathway enrichment of bHLH genes in R. pulchrum. The size of each circle indicates the number of genes enriched in each pathway, while the color gradient represents the significance of the enrichment, with darker green indicating higher significance (lower p-values).
2.8. The Expression of RpbHLHs at Different Developmental Stages and in Flowers of Different Colors
To explore the potential role of the identified RpbHLHs in different developmental stages, the expression of the RpbHLHs was analyzed. As shown in Figure 7A, the expression levels of the RpbHLHs were higher during the bud stage compared to those during the early flowering, blooming, and fading stages. Seven genes, including RpbHLH14, RpbHLH58, RpbHLH87, RpbHLH65, RpbHLH19, RpbHLH35, and RpbHLH27, exhibited high expression during the bud stage. In the early flowering stage, four genes (RpbHLH10, RpbHLH14, RpbHLH58, and RpbHLH65) were highly expressed. During the blooming stage, seven genes (RpbHLH10, RpbHLH14, RpbHLH58, RpbHLH65, RpbHLH19, RpbHLH35, and RpbHLH70) demonstrated high expression. In the fading stage, six genes (RpbHLH10, RpbHLH14, RpbHLH58, RpbHLH87, RpbHLH65, and RpbHLH19) were highly expressed. Moreover, the expression of RpbHLH14, RpbHLH58, and RpbHLH65 increased across all four stages (Figure 7A).
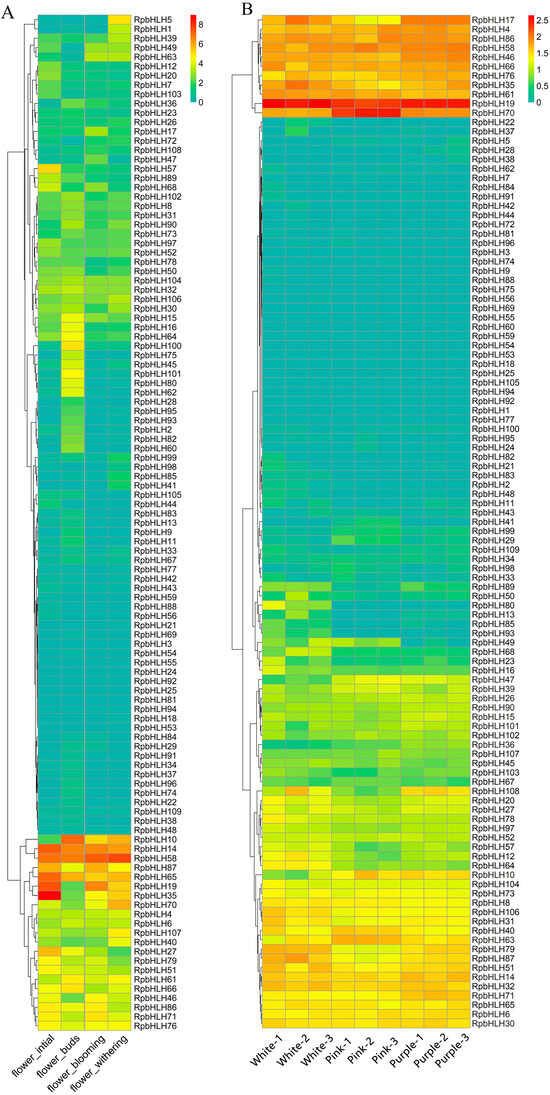
Figure 7.
The expression of RpbHLHs in different growth stages and flowers of different colors. (A) RpbHLH expression in R. pulchrum flower initiation, flower budding, flower withering, and flower blooming stages. The data were sourced from the NCBI bioprojects (PRJNA485857). (B) RpbHLH expression in white, pink, and purple flowers. The data were measured in this study. The color intensity represents the expression levels, with higher expression indicated by warmer colors (yellow to red) and lower expression by cooler colors (green to blue).
The expression of the RpbHLHs in flowers of different colors (white, pink, and purple) was also evaluated (Figure 7B). The overall RPKM (Reads Per Kilobase of transcript per Million mapped reads) values indicate that white flowers had the highest number of expressed genes, followed by purple and pink flowers (Table S6). RpbHLH4, RpbHLH19, RpbHLH86, RpbHLH58, RpbHLH46, RpbHLH66, RpbHLH76, RpbHLH 35, and RpbHLH61 exhibited high expression levels across all three flower colors. RpbHLH70 showed high expression in pink flowers and moderate expression in white and purple flowers, while RpbHLH17 was exclusively expressed in the white and purple flowers. Among the 11 genes (RpbHLH17, RpbHLH4, RpbHLH86, RpbHLH58, RpbHLH46, RpbHLH66, RpbHLH76, RpbHLH108, RpbHLH14, RpbHLH32, and RpbHLH71) highly expressed in the purple flowers, RpbHLH17 demonstrated the highest expression levels. In white flowers, the genes RpbHLH35, RpbHLH61, RpbHLH79, RpbHLH87, and RpbHLH51 were highly expressed. Conversely, in pink flowers, only three genes (RpbHLH22, RpbHLH40, and RpbHLH63) were highly expressed.
Overall, the expression profiles of the RpbHLHs across developmental stages and flower colors highlight their complex and dynamic role in flower development, color formation, and adaptation. The genes that show consistent expression across different stages and flower colors are likely involved in fundamental processes such as flower initiation, maturation, and pigmentation. These data underscore the potential of RpbHLHs in regulating the intricate network of pathways that govern flower color diversity and development in R. pulchrum.
2.9. Quantification of the RpbHLH Expression Using RT-qPCR
To elucidate the influence of RpbHLHs on varied developmental stages and floral pigmentation in R. pulchrum further, highly expressed RpbHLHs from the transcriptome data were assessed using RT-qPCR (Table S7). The expression of 14 genes from different subfamilies in flowers of different colors (white, pink, and purple) and at different developmental stages (the bud stage (S1), the early flowering stage (S2), and the blooming stage (S3)) was quantified (Figure 8). In white flowers, RpbHLH58, RpbHLH71, RpbHLH36, RpbHLH15, RpbHLH14, RpbHLH4, RpbHLH90, RpbHLH10, and RpbHLH26 were highly expressed in the bud stage, while RpbHLH17 and RpbHLH46 were highly expressed in the early flowering stage, and RpbHLH6, RpbHLH70, and RpbHLH47 were highly expressed in the blooming stage. Eleven genes (RpbHLH57, RpbHLH71, RpbHLH6, RpbHLH36, RpbHLH14, RpbHLH4, RpbHLH90, RpbHLH70, RpbHLH47, RpbHLH10, and RpbHLH26) exhibited an expression pattern of initially decreasing and subsequently increasing during different developmental stages, while the expression of RpbHLH17 and RpbHLH46 displayed the opposite pattern; the expression of RpbHLH15 consistently decreased during the developmental process. In pink flowers, RpbHLH14 and RpbHLH4 were highly expressed in the bud stage; RpbHLH70, RpbHLH47, and RpbHLH10 were highly expressed in the early flowering stage; and RpbHLH58, RpbHLH17, RpbHLH71, RpbHLH6, RpbHLH36, RpbHLH15, RpbHLH90, and RpbHLH26 were highly expressed in the blooming stage. Ten genes (RpbHLH58, RpbHLH17, RpbHLH71, RpbHLH6, RpbHLH36, RpbHLH15, RpbHLH14, RpbHLH4, RpbHLH90, and RpbHLH26) exhibited an expression pattern of initially decreasing and subsequently increasing during the developmental process, while RpbHLH10 and RpbHLH47 displayed the opposite pattern.
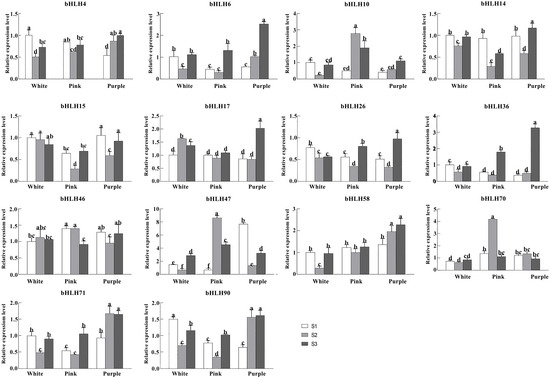
Figure 8.
The expression of RpbHLHs in flowers of different colors and different stages in Rhdodendron pulchrum. RT-qPCR analysis of 14 genes (RpbHLH4, RpbHLH6, RpbHLH10, RpbHLH14, RpbHLH15, RpbHLH17, RpbHLH26, RpbHLH36, RpbHLH46, RpbHLH47, RpbHLH58, RpbHLH70, RpbHLH71, and RpbHLH90) across various flower colors and developmental stages (the bud stage [S1], the early flowering stage [S2], and the peak flowering stage [S3]). The experiments were independently replicated a minimum of three times. The data are presented as the mean ± standard error (n = 3), with distinct letters signifying statistically significant differences (p < 0.05) as determined by Duncan’s multiple range test.
In purple flowers, RpbHLH46, RpbHLH15, and RpbHLH70 exhibited high expression levels during the bud stage. RpbHLH70 maintained high expression in the early flowering stage, while RpbHLH58, RpbHLH17, RpbHLH6, RpbHLH36, RpbHLH14, RpbHLH4, RpbHLH90, RpbHLH10, and RpbHLH26 demonstrated elevated expression during the blooming stage. The expression levels of RpbHLH58, RpbHLH71, RpbHLH4, RpbHLH90, and RpbHLH10 increased throughout the flowering process. RpbHLH70 displayed an initial increase followed by a decrease in expression. In contrast, RpbHLH46, RpbHLH6, RpbHLH36, RpbHLH15, RpbHLH14, RpbHLH70, and RpbHLH26 showed an initial decrease succeeded by an increase in expression. Overall, the data suggest that bHLH genes have dynamic and color-specific expression profiles that may regulate both the developmental processes and pigmentation pathways in different flower colors.
3. Discussion
Studies have demonstrated that the bHLH gene family is widely present in higher plants and plays a crucial role in plant growth, development, stress responses, and defense reactions [15]. A total of 109 bHLH genes (RpbHLHs) were identified from the R. pulchrum genome, and their physicochemical properties and subcellular localization were analyzed further. Through a phylogenetic analysis, we compared 109 RpbHLHs with 166 bHLH genes from Arabidopsis thaliana and constructed an evolutionary tree. The results revealed significant evolutionary divergence between R. pulchrum and Arabidopsis, which may be linked to the unique flower color traits of R. pulchrum [16]. These genes were grouped into 31 subfamilies, with subfamily 25 containing the highest number of genes (14), while subfamily 19 contained only a single gene (RpbHLH). Notably, no RpbHLHs were found in subfamilies 6, 8, 18, 20, 21, 22, or 29, suggesting the potential loss of these subfamilies during the evolution of R. pulchrum [8]. Some subfamilies contained only AtbHLH genes, while others included both RpbHLHs and AtbHLH genes, indicating species-specific differences in gene family evolution [10].
Basic helix–loop–helix (bHLH) transcription factors in ornamental plants exhibit highly conserved structural features that underpin their roles in floral pigmentation and morphogenesis. The results show that nearly all bHLH proteins harbor core helix–loop–helix motifs (motif 1 and motif 2) across species, reflecting a preserved domain architecture crucial for DNA binding and dimerization [17]. Gene structure is likewise conserved within bHLH subfamilies; members of the same clade tend to share similar exon–intron organization [18]. Notably, many anthocyanin-related bHLHs fall into intron-poor subgroups (with some being completely intronless), indicating an evolutionary streamlining that is consistent within these clades [18]. Such consistency in the motif composition and gene structure across diverse ornamental species suggests strong purifying selection maintaining these features over time [19,20]. The intact bHLH domain is almost universally maintained, ensuring that bHLH proteins can form the MYB–bHLH–WD40 (MBW) complexes that activate anthocyanin biosynthetic genes [21,22,23]. For example, transcriptomic studies in R. pulchrum have identified bHLH regulators whose high expression correlates with intense petal pigmentation, acting as positive regulators of anthocyanin accumulation [23]. Beyond pigment production, the cis-elements associated with bHLH genes are enriched in developmental and light-responsive motifs, hinting at broader roles in floral organ development and environmental response [17].
Flower colors such as red, pink, purple, and blue are largely determined by flavonoid pigments (especially anthocyanins) whose biosynthesis is tightly controlled by transcription factors [24]. A well-conserved regulatory module is the MYB-bHLH-WD40 (MBW) complex, consisting of an R2R3-MYB activator, a bHLH co-factor, and a WD40 repeat protein [25]. Extensive studies in diverse plants show that the role of bHLH factors in anthocyanin regulation is functionally conserved across species. In maize, the founding MBW components (C1/Lc MYB and R/B bHLHs) were first linked to pigment regulation, and similar players have since been identified in many angiosperms [24]. R. pulchrum exhibits cultivars with white, pink, light pink, and purple flowers, making it an excellent system for studying color regulation [23]. Transcriptome analyses of these differently colored petals have identified several bHLH genes whose expression correlates with anthocyanin accumulation. Notably, five R. pulchrum bHLH homologs (designated RpbHLH4, RpbHLH14, RpbHLH17, RpbHLH58, and RpbHLH70) show elevated transcript levels in pigmented pink/purple flowers compared to those in white flowers (which lack anthocyanin). In particular, one bHLH gene (Rhsim08G0230000, corresponding to RpbHLH17) was significantly upregulated in deep purple petals (light pink or white), indicating that it acts as a positive regulator of anthocyanin biosynthesis in this species [23,26].
bHLH transcription factors generally function as part of the conserved MYB-bHLH-WD40 complex, where they stabilize and enhance MYB activator binding to promoters of anthocyanin biosynthetic genes [22]. When an appropriate bHLH partner is present and highly expressed, the MBW complex efficiently activates enzymes in the anthocyanin pathway, leading to the accumulation of red, pink, or purple pigments in the petals [24]. Conversely, the absence or low expression of a functional bHLH gene can result in diminished anthocyanin production and thus lighter or white flowers [24,27]. The functional conservation of this mechanism is evident in R. pulchrum, where bHLH genes like RpbHLH17, RpbHLH58, and others are highly expressed in richly colored flowers, correlating with increased anthocyanin contents (Figure 1 and Figure 8).
The enrichment analysis suggests that the target RpbHLHs are involved in regulating a range of critical processes, such as hormonal signaling, energy metabolism, and stress responses. Plant hormones such as auxins, gibberellins, and ethylene are known to influence the synthesis of flavonoids and anthocyanins, which are key contributors to flower pigmentation [28]. The involvement of RpbHLHs in regulating plant hormone networks thus suggests an indirect but significant role in the control of pigment biosynthesis, further supporting their importance in flower color regulation. Furthermore, the flavonoid biosynthesis pathway, which directly influences flower color, highlights the role of RpbHLHs in regulating the synthesis of the compounds responsible for pigmentation [29]. The KEGG pathway analysis revealed that genes involved in the flavonoid biosynthesis pathway (ko00941) are crucial in regulating flower color. The flavonoid biosynthesis pathway includes essential enzymes like chalcone synthase (CHS), which are involved in the production of anthocyanins and other pigments, directly influencing flower color. Genes such as RpbHLH17, RpbHLH61, and other bHLH family members have been identified as similar to chalcone synthases (CHSs), confirming their role in the flavonoid biosynthesis pathway. According to the SWISS database, RpbHLH17/19/46/58/61 are classified as chalcone synthases, which play a pivotal role in the synthesis of anthocyanins and other pigments, thus contributing to flower color regulation [30]. The bHLH genes RpbHLH4/76/86 share homology with AtbHLH11/34/47/104/105/115/121, all belonging to the Myc-like bHLH family. This family enhances the efficiency of the amino acid residues involved in anthocyanin biosynthesis, thus playing a crucial role in flower pigmentation. Studies have shown that AtbHLH011 (bHLH168) regulates anthocyanin biosynthesis in purple cabbage, and AtbHLH034 (EGL1) controls pigment synthesis and accumulation in plants, thereby influencing flower color [31,32].
The R. pulchrum transcriptome data reveal that several RpbHLHs (RpbHLH14, 58, 65, 10, 70, and 26) show peak expression at the floral bud, early flowering, or full-bloom stages, suggesting that they perform stage-specific regulatory functions in flower development. The enrichment of genes in the plant hormone signal transduction pathway suggested that the RpbHLHs were closely involved in regulating plant hormone networks. This is particularly relevant to flower development, as plant hormones such as auxins, gibberellins, ethylene, and jasmonic acid regulate various aspects of flower growth, maturation, and pigmentation [33]. For instance, RpbHLH10 and RpbHLH26 are highly expressed in floral buds and early flowers, which might relate to initiating pollen and anther development during early floral bud formation. The high bud-stage expression of RpbHLH14 and RpbHLH58 points to their roles in floral initiation and growth, possibly analogous to bHLH factors in hormone signaling (such as MYC2/JAM family members) that activate JA- or auxin-responsive genes to promote bud outgrowth and stamen maturation [34]. By full bloom, RpbHLH65 and RpbHLH70 are strongly upregulated, which may reflect functions in flower opening and organ expansion—comparable to bHLH genes like BIGPETAL that constrain petal size or those integrating GA and light signals into timing anthesis [35,36]. Integrating these findings on the R. pulchrum transcriptome with the cross-species literature suggests that these bHLH transcription factors likely coordinate flower development by controlling meristem identity, organ identity and growth, and the timing of flowering events via hormonal and developmental pathways [35,37]. Other enriched pathways, such as glycolysis/gluconeogenesis and the MAPK signaling pathway, indicate that RpbHLHs also play a role in managing energy metabolism and stress responses, which are essential for optimal flowering.
This study focused on identifying the RpbHLH family in R. pulchrum and analyzing its potential roles in flower color regulation. Flower color metabolite identification was not included in this study due to time and resource constraints. However, future research will aim to use metabolomics or chromatography to identify and quantify the specific compounds involved in flower color formation, providing a more comprehensive understanding of how these genes contribute to pigmentation.
4. Materials and Methods
4.1. Plant Materials
In this study, 45 healthy and stable potted R. pulchrum (Rhododendron × pulchrum Sweet) plants were selected from two-year-old specimens and raised in an artificial climate incubator. The plants were transferred into incubator conditions two weeks prior to sampling to acclimate. The artificial climate incubator maintained a photoperiod of 16 h of light/8 h of dark, with a light intensity of 600 μmol m−2 s−1, temperatures of 22 °C during the day and 18 °C at night, and a humidity level of 70%. The plants were categorized into three groups based on flower color: white, pink, and purple. All three groups belonged to the same variety and were grown under identical cultivation conditions.
Although the plants belong to the same variety, variations in flower color may arise from genetic differences, environmental influences, or developmental stages. To minimize potential environmental effects, all plants were maintained under identical conditions in the climate incubator. Each flower color group was further subdivided into three biological replicates, each containing five plants. Samples were collected at three distinct developmental stages: the bud stage (S1) on 23 March 2023; the early flowering stage (S2) on 15 April, just prior to full bloom; and the blooming stage (S3) on 25 April, when the flowers were fully open. At each time point, three biological replicates, each weighing approximately 0.1 g, were randomly selected from the five plants and immediately frozen in liquid nitrogen for storage at −80 °C. In total, 27 samples were collected and categorized into different groups: the white flower bud stage (whiteS1-1, whiteS1-2, whiteS1-3), the early white flowering stage (whiteS2-1, whiteS2-2, whiteS2-3), and the white flower blooming stage (whiteS3-1, whiteS3-2, whiteS3-3); the pink flower bud stage (pinkS1-1, pinkS1-2, pinkS1-3), the early pink flowering stage (pinkS2-1, pinkS2-2, pinkS2-3), and the pink flower blooming stage (pinkS3-1, pinkS3-2, pinkS3-3); and the purple flower bud stage (purpleS1-1, purpleS1-2, purpleS1-3), the early purple flowering stage (purpleS2-1, purpleS2-2, purpleS2-3), and the purple flower blooming stage (purpleS3-1, purpleS3-2, purpleS3-3) (Figure 1).
4.2. Determination of Flavonoids and Anthocyanins
The contents of flavonoids and anthocyanins were determined using ultraviolet–visible spectrophotometry [38,39], using a Spark instrument (Tecan Austria GmbH, Grödig, Austria).
4.3. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR Analysis
Total RNA was extracted from the plant samples using the RNAprep Pure Kit (Tiangen, Hangzhou, China). To remove trace amounts of DNA, the RNA samples were treated with RNase-free DNase I. The concentration and purity of the RNA were assessed using a micro-spectrophotometer. For the optimal results, the RNA concentration should exceed 200 ng/μL. To measure the concentration, 1 μL of RNA was analyzed using a micro-spectrophotometer. Typically, a concentration above 100 ng/μL is desired, and gel electrophoresis of the RNA should reveal two distinct bands, representing the 28S and 18S ribosomal RNA subunits (Figure S1).
The extracted RNA was utilized as the template to generate cDNA using the Evo M-MLV One Step RT-PCR Kit (AG11606, ACCURATE BIOTECHNOLOGY (HUNAN), Changsha, China) following the manufacturer’s instructions. The reaction system consisted of 2 µL of gDNA Clean Reaction Mix Ver.2, 4 µL of 5× Evo M-MLV RT Reaction Mix Ver.2, 2 µL of RNA, and RNase Free ddH2O to a total volume of 20 µL. The reaction conditions were set at 37 °C for 15 min and 85 °C for 5 s and then maintained at 4 °C. The resulting cDNA was stored at −20 °C for subsequent use. The primers for quantitative reverse transcription polymerase chain reaction (qRT-PCR) were designed using Primer3 (https://primer3.ut.ee, accessed on 28 June 2024) and synthesized by Qingke Biotechnology Co., Ltd. (Hangzhou, China). RpUBQ was selected as the internal reference gene [40]. For qRT-PCR, 10-fold-diluted cDNA was employed as the template, and the reaction was performed using the SYBR Green Premix Pro Taq HS qPCR Kit (AG11701, ACCURATE BIOTECHNOLOGY (HUNAN), Changsha, China) and analyzed using the LightCycler®480 II (Roche Diagnostics, Mannheim, Germany). The reaction system of 10 µL included 5 µL of 2× SYBR Green Pro Taq HS Premix, 1 µL of the cDNA template, 0.2 µL of the primer (at a final concentration of 0.2 mM), and 3.6 µL of RNase Free ddH2O (Table S8). The reaction program consisted of pre-denaturation at 95 °C for 30 s, denaturation at 95 °C for 5 s, and annealing and extension at 60 °C for 30 s, with a total of 40 cycles. Three biological replicates were set for each sample, and the relative expressions of the candidate genes were calculated using the 2−ΔΔCt method [41].
All experimental data were analyzed using a one-way ANOVA in SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). Before conducting the ANOVA, the assumption of normality was assessed using the Shapiro–Wilk test. When p > 0.05, the data were considered to follow a normal distribution. The assumption of homogeneity of variances was tested using Levene’s test, and when p > 0.05, the variances among the treatment groups were considered equal. Duncan’s test was employed to determine significant differences among the samples at a threshold of p < 0.05. The data were then visualized using GraphPad Prism 9.0.0 (GraphPad Software, San Diego, CA, USA). To ensure reliability, each sample was tested with three biological replicates.
4.4. Identification and Characterization of the Physicochemical Properties of the RpbHLHs
The RpbHLH gene sequences were downloaded from the R. pulchrum genomic library (https://figshare.com, accessed on 15 March 2024) [42]. The bHLH domain PF00010.26 was obtained from the PFAM database (https://pfam.xfam.org, accessed on 18 March 2024). The presence of the bHLH conserved domain in the R. pulchrum proteome was identified using hmmsearch (http://hmmer.janelia.org, accessed on 18 March 2024). Subsequently, the R. pulchrum protein sequence was compared to the PFAM database [43] using hmmscan [44] to determine whether the protein sequence contained the bHLH domain. The number of amino acids was counted using the SeqKit 2.10.0 (https://github.com, accessed on 5 April 2024). The ExPASy-ProtParam tool (https://web.expasy.org, accessed on 6 April 2024) was employed to analyze the molecular weight (MW) and isoelectric point (PI) of the protein. Finally, subcellular localization predictions were performed using WoLF PSORT 1.0 [45] (https://wolfpsort.hgc.jp, accessed on 7 April 2024).
4.5. Chromosomal Localization, Collinearity Analysis, and Ka/Ks Calculation of RpbHLHs
Homologous genes were identified using OrthoFinder 2.5.2 (https://github.com, accessed on 9 April 2024) [46], and TBtools 2.310 [47] (https://github.com, accessed on 12 April 2024) was used to draw a collinearity map of these genes on the chromosome. The synonymous substitution rate (Ks) and the non-synonymous substitution rate (Ka) of homologous genes were calculated using KaKs_Calculator 2.0 (https://ngdc.cncb.ac.cn, accessed on 15 April 2024) [48].
4.6. Prediction of the Promoter’s Regulatory Elements
To predict the regulatory elements of the RpbHLH promoter, a 2000-base-pair (bp) upstream region of the RpbHLH starting codon was extracted and defined as the promoter region. The PlantCARE database [49] (http://www.biodata-tech.com, accessed on 20 April 2024) and the plant cis-acting regulatory DNA elements (PLACE) database [50] (https://www.dna.affrc.go.jp, accessed on 21 April 2024) were utilized to predict regulatory elements within the promoter sequence. A physical map illustrating the positions of these elements was generated, and the frequency of each element was quantified.
4.7. Multiple Gene Alignment and Phylogenetic Analysis of the RpbHLHs
The RpbHLH proteins were aligned with Arabidopsis AtbHLH proteins. The Arabidopsis protein sequences were obtained from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org, accessed on 7 May 2024). The alignment was performed using the default parameters in MAFFT 7.313 [51]. The maximum likelihood (ML) phylogenetic tree of the RpbHLH family was constructed using FastTree [52] using the general time-reversible (GTR) model and the Gamma20 model of rate heterogeneity. The phylogenetic tree was assessed using the Shimodaira–Hasegawa test.
4.8. Structure and Motif Analyses of RpbHLHs
The gene structures of the RpbHLHs were visualized using the Gene Annotation GFF library [42] (https://figshare.com, accessed on 11 May 2024). The motifs in the RpbHLH family proteins were analyzed using the MEME software [53] (http://meme-suite.org, accessed on 12 May 2024), with the parameter for identifying the motifs set to 6, while the other parameters were maintained at their default values. The motif structure and gene structure diagrams were generated based on the motif prediction results and reference genome annotation data.
4.9. Prediction and Enrichment Analysis of Target Genes of RpbHLHs
PlantRegMap [54] was employed to predict the target genes of the RpbHLHs by identifying bHLH binding sites within the 2 kb promoter region of the entire genome, utilizing a stringent significance threshold of p ≤ 1 × 10−8. Following the identification of the target genes, a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was conducted to ascertain significant biological pathways.
4.10. The Spatiotemporal-Specific Expression Analysis of the RpbHLHs
Total RNA was extracted from flowers of different colors using an RNAprep Pure Plant Plus kit (DP441, Tiangen Biotech Co., Ltd., Beijing, China). The RNA library was prepared using an Illumina (San Diego, CA, USA) TruSeq RNA kit 2.0 and subsequently sequenced on the Illumina NovaSeq 6000 platform in PE150 mode according to Li et al. [55]. The RNA-seq data were deposited into the NCBI Sequence Read Archive (accession number: PRJNA1265659). The RNA-seq data for various developmental stages were obtained from the NCBI bioprojects PRJNA485857. The RPKM values of these RpbHLHs were calculated in transcripts per million (TPM), and heatmaps of the results were generated using R 3.5.2, with the colors representing log2(TMP+1).
5. Conclusions
This study highlights the significant role of the bHLH gene family in R. pulchrum, particularly in regulating flower color and development. We identified 109 RpbHLHs, which exhibited diverse characteristics in terms of the amino acid count, molecular weight, and isoelectric point. The evolutionary analysis revealed notable differences between the RpbHLHs in R. pulchrum and Arabidopsis thaliana, with some genes specific to R. pulchrum, suggesting their role in flower color formation and development. A further analysis of the gene structure showed variability in the number of exons, with conserved motifs across different subfamilies, indicating functional conservation. The presence of conserved motifs provides insight into the regulatory roles of these genes in flower color and development. RpbHLHs are distributed across several chromosomes and respond to environmental factors like light, mechanical stress, and drought. The promoter analysis further indicates these genes are involved in both flower color regulation and hormone signaling pathways. The expression analysis showed significant differences in the RpbHLH expression across different flower colors (white, pink, and purple) and developmental stages (bud, early flowering, and blooming). For example, genes like RpbHLH14, RpbHLH58, and RpbHLH65 were highly expressed in white flowers, particularly during the bud and blooming stages. In purple flowers, genes such as RpbHLH17 and RpbHLH58 showed significantly higher expression. The KEGG pathway analysis revealed that RpbHLHs are involved in multiple important pathways, such as plant hormone signaling, glycolysis/gluconeogenesis, and MAPK signaling, highlighting their role in flower color regulation and development, especially in flavonoid biosynthesis, which is closely linked to flower pigmentation. Collectively, these findings enhance our understanding of the structure of the RpbHLH family and provide a foundation for further investigation into the regulatory function of bHLH genes in flower color and development in R. pulchrum splendida.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants14111713/s1. Figure S1: The electrophoresis gel imaging of the 28s and 18s ribosomal RNA subunits. Table S1: The analysis of the physicochemical properties of proteins in the RpbHLH family. Table S2: RpbHLH gene and protein sequence data. Table S3: RpbHLH pair KaKs ratio analysis. Table S4: Prediction of the cis-acting elements in the RpbHLH promoter region. Table S5: PlantRegMap-predicted target gene IDs. Table S6: Flower color and RpbHLH expression (FPKM). Table S7: ANOVA results for the RpbHLHs. Table S8: qRT-PCR primer sequences for bHLH genes.
Author Contributions
Conceptualization: S.J. and H.W.; methodology: J.S. (Jiaran Sheng), J.S. (Jianshang Shen), and X.L.; software: J.S. (Jiaran Sheng); validation: X.C.; formal analysis: Y.S.; investigation: J.S. (Jiaran Sheng) and J.S. (Jianshang Shen); resources: S.J. and X.L.; data curation: J.S. (Jiaran Sheng) and Y.S.; writing—original draft preparation: J.S. (Jiaran Sheng) and J.S. (Jianshang Shen); writing—review and editing: X.L., S.J., and H.W.; supervision: X.L. and S.J.; project administration: S.J.; funding acquisition: S.J. and J.S. (Jianshang Shen). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32201608), Zhejiang Provincial Natural Science Foundation of China (LGN22C160006), and the Research Foundation for Advanced Talents of Hangzhou Vocational & Technical College (RCXY202303).
Data Availability Statement
The RNA-seq data in this study were deposited into the NCBI Sequence Read Archive under accession number PRJNA1265659. The other datasets generated during and/or analyzed during the current study can be made available by the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lan, X.; Zhang, L.; Zhang, J.; Cui, H.; Jiang, C.; Shi, L. Research progress of Rhododendron breeding. Acta Hortic. Sin. 2012, 39, 1829–1838. [Google Scholar]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, T.; Fass, D.; Arnaud, M.; Harrison, S.C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994, 8, 970–980. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y.; et al. Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 2023, 225, 389–403. [Google Scholar] [CrossRef]
- Toiber, D.; Sebastian, C.; Mostoslavsky, R. Characterization of nuclear sirtuins: Molecular mechanisms and physiological relevance. Handb. Exp. Pharmacol. 2011, 206, 189–224. [Google Scholar]
- Sun, H.; Fan, H.J.; Ling, H.Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, S.; Wang, X.; Mao, T.; Bao, M.; Zhang, J.; Zhang, J. Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mum. Hortic. Plant J. 2022, 8, 531–544. [Google Scholar] [CrossRef]
- Grotewold, E. The Science of Flavonoids; Springer: New York, NY, USA, 2006. [Google Scholar]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2010, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Moon, S.; Lee, Y.S.; Zhu, L.; Liang, W.; Zhang, D.; Jung, K.H.; An, G. Defective Tapetum Cell Death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 2016, 170, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Zhuang, Y.; Zhou, L.; Geng, L.; Jiang, L.; Sui, Y.; Luo, L.; Pan, H.; Zhang, Q.; Yu, C. Genome-wide identification of the bHLH transcription factor family in Rosa persica and response to low-temperature stress. PeerJ 2024, 12, e16568. [Google Scholar] [CrossRef]
- Dong, J.; Wu, Y.W.; Dong, Y.; Pu, R.; Li, X.J.; Lyu, Y.M.; Bai, T.; Zhang, J.L. Genome-Wide identification of the bHLH gene family in Rhododendron delavayi and its expression analysis in different floral tissues. Genes 2024, 15, 1256. [Google Scholar] [CrossRef]
- Leng, L.; Zhang, X.; Liu, W.; Wu, Z. Genome-wide identification of the MYB and bHLH families in Carnations and expression analysis at different floral development stages. Int. J. Mol. Sci. 2023, 24, 9499. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Wang, M.J.; Ou, Y.; Li, Z.; Zheng, Q.D.; Ke, Y.J.; Lai, H.P.; Lan, S.R.; Peng, D.H.; Liu, Z.J.; Ai, Y. Genome-Wide identification and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Cymbidium ensifolium. Int. J. Mol. Sci. 2023, 24, 3825. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhou, C. Transcriptomic analysis reveals the regulatory mechanism of color diversity in Rhododendron pulchrum Sweet (Ericaceae). Plants 2023, 12, 2656. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Tang, Y.; Wang, X.; Xu, C.; Tao, J.; Zhao, D. Tree peony R2R3-MYB transcription factor PsMYB30 promotes petal blotch formation by activating the transcription of the anthocyanin synthase gene. Plant Cell Physiol. 2022, 63, 1101–1116. [Google Scholar] [CrossRef]
- Bogs, J.; Jaffé, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine rranscription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Q.; Lang, L.; Jiang, C.; Wang, X.F.; Sun, H.M. Integrated metabolome and transcriptome analyses reveal the molecular mechanism of a color mutation in miniature roses. BMC Plant Biol. 2019, 20, 611. [Google Scholar]
- Wang, L.; Yang, S.; Ni, J.; Teng, Y.; Bai, S. Advances of anthocyanin synthesis regulated by plant growth regulators in fruit trees. Sci. Horti. 2023, 307, 111476. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef]
- Chen, C.T.; Yang, C.Y.; Tzen, J.T.C. Molecular characterization of polyphenol oxidase between small and large leaf tea cultivars. Sci. Rep. 2022, 12, 12870. [Google Scholar] [CrossRef]
- Jin, S.W.; Rahim, M.A.; Afrin, K.S.; Park, J.I.; Kang, J.G.; Nou, I.S. Transcriptome profiling of two contrasting ornamental cabbage (Brassica oleracea var. acephala) lines provides insights into purple and white inner leaf pigmentation. BMC Genom. 2018, 19, 797. [Google Scholar] [CrossRef]
- Zhou, Y.; Mumtaz, M.A.; Zhang, Y.; Shu, H.; Hao, Y.; Lu, X.; Cheng, S.; Zhu, G.; Wang, Z. Response of anthocyanin accumulation in pepper (Capsicum annuum) fruit to light days. Int. J. Mol. Sci. 2022, 23, 8357. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. The hormonal regulation of flower development. J. Plant Growth Regul. 2011, 30, 242–254. [Google Scholar] [CrossRef]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.K.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Xin, R.; Kim, D.H.; Sung, S.; Lange, T.; Huq, E. NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 2016, 143, 682–690. [Google Scholar]
- Szécsi, J.; Joly, C.; Bordji, K.; Varaud, E.; Cock, J.M.; Dumas, C.; Bendahmane, M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006, 25, 3912–3920. [Google Scholar] [CrossRef]
- Nakata, M.; Ohme-Takagi, M. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal. Behav. 2013, 8, e26473. [Google Scholar] [CrossRef]
- Jia, Z.; Tang, M.; Wu, J. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar]
- Lee, J.; Robert, W.D.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef]
- Da Silva, M.F.; Gonçalves, M.C.; dos Santos Brito, M.; Nóbile, P.M.; de Andrade, L.M.; Medeiros, C.N.; Creste, S.; Pinto, L.R. Reference genes for gene expression studies targeting sugarcane infected with Sugarcane mosaic virus (SCMV). BMC Res. Notes 2019, 12, 149. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Shen, J.S.; Lan, L.; Kan, S.L.; Cheng, H.F.; Peng, P.; Wan, Z.Y.; Hu, Y.; Huang, X.L.; Li, X.Q.; Ye, Y.J.; et al. A haplotype-resolved genome for Rhododendron × pulchrum and the expression analysis of heat shock genes. J. Syst. Evol. 2023, 62, 489–504. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, X.; Wang, Y.; Jin, S.; Wan, Z. Physiological and transcriptomic responses of two Rhododendron L. cultivars to drought stress: Insights into drought tolerance mechanisms. Agronomy 2025, 15, 1278. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).