Genetic Diversity and Phenotypic Variation of Indigenous Wild Cherry Species in Kazakhstan and Uzbekistan
Abstract
1. Introduction
2. Results
2.1. Phenotypic Evaluation of Prunus Species from Kazakhstan and Uzbekistan
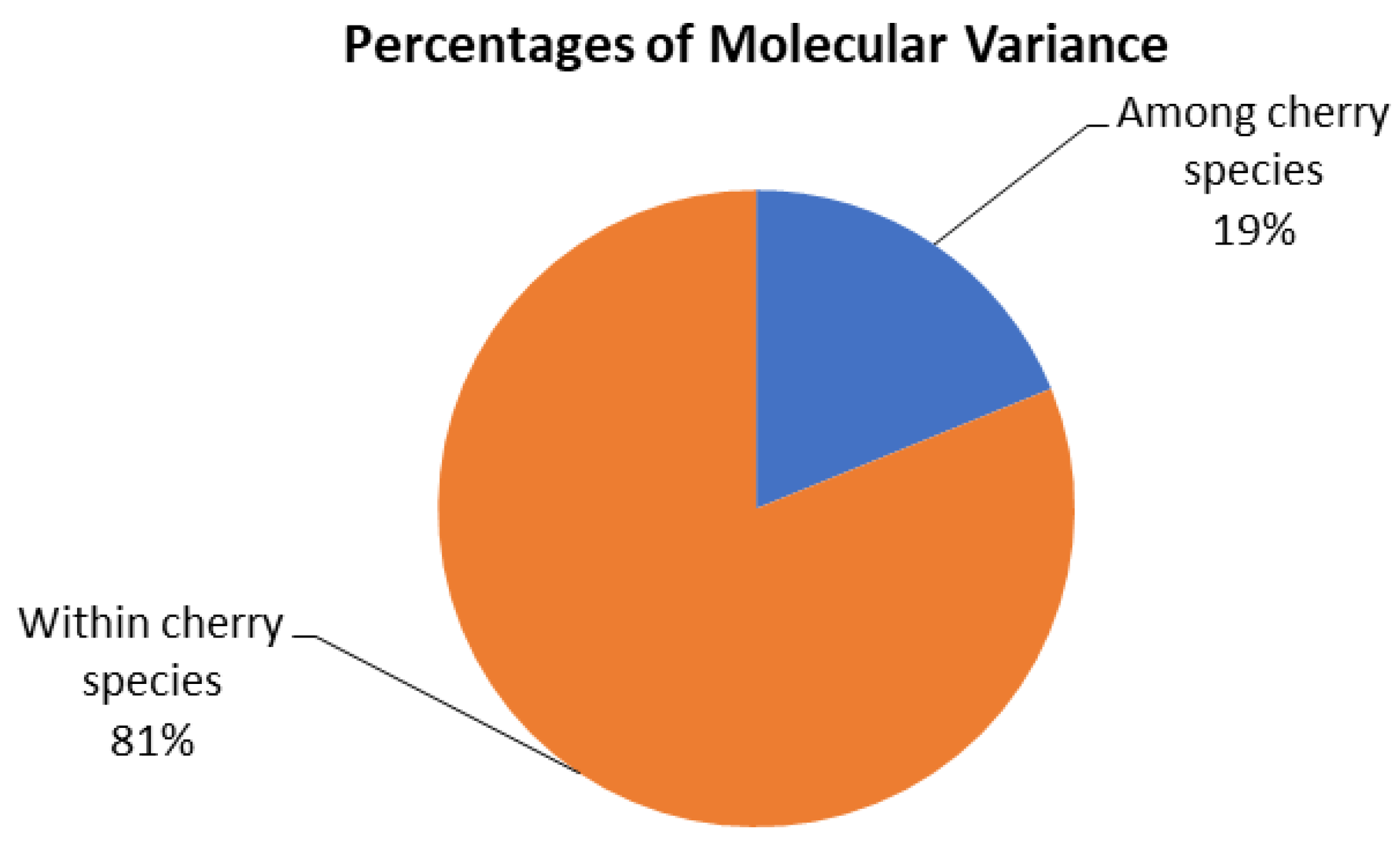
2.2. Genetic Diversity Parameters of the Prunus Species from Kazakhstan and Uzbekistan
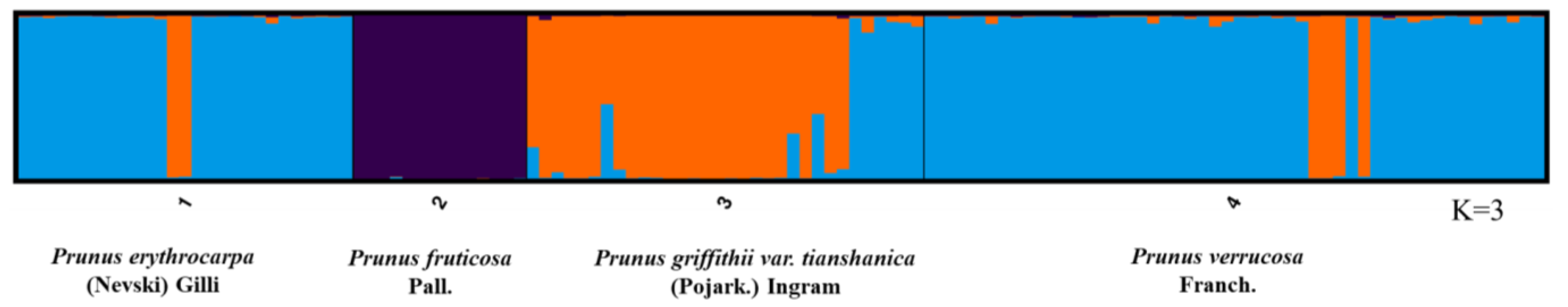
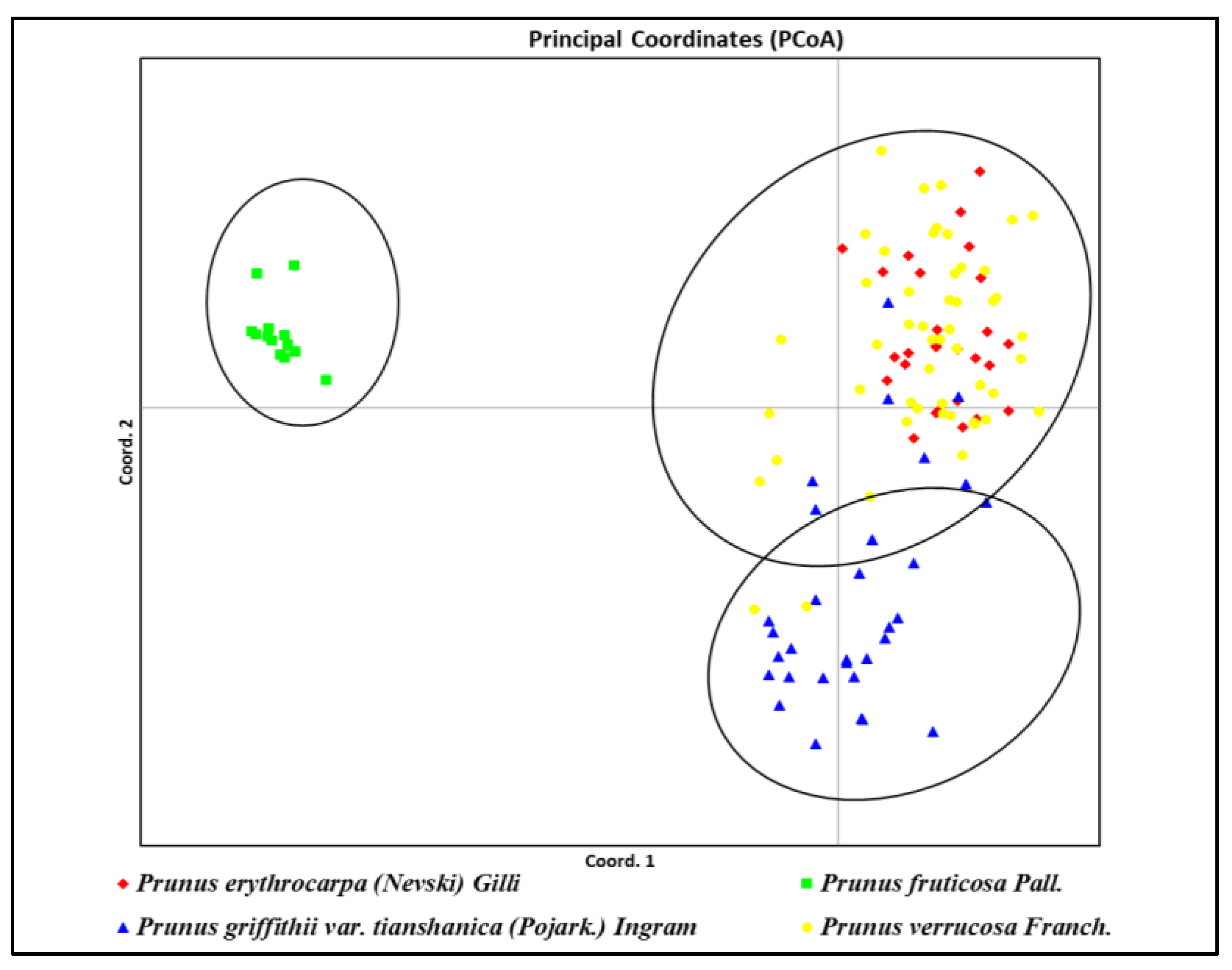
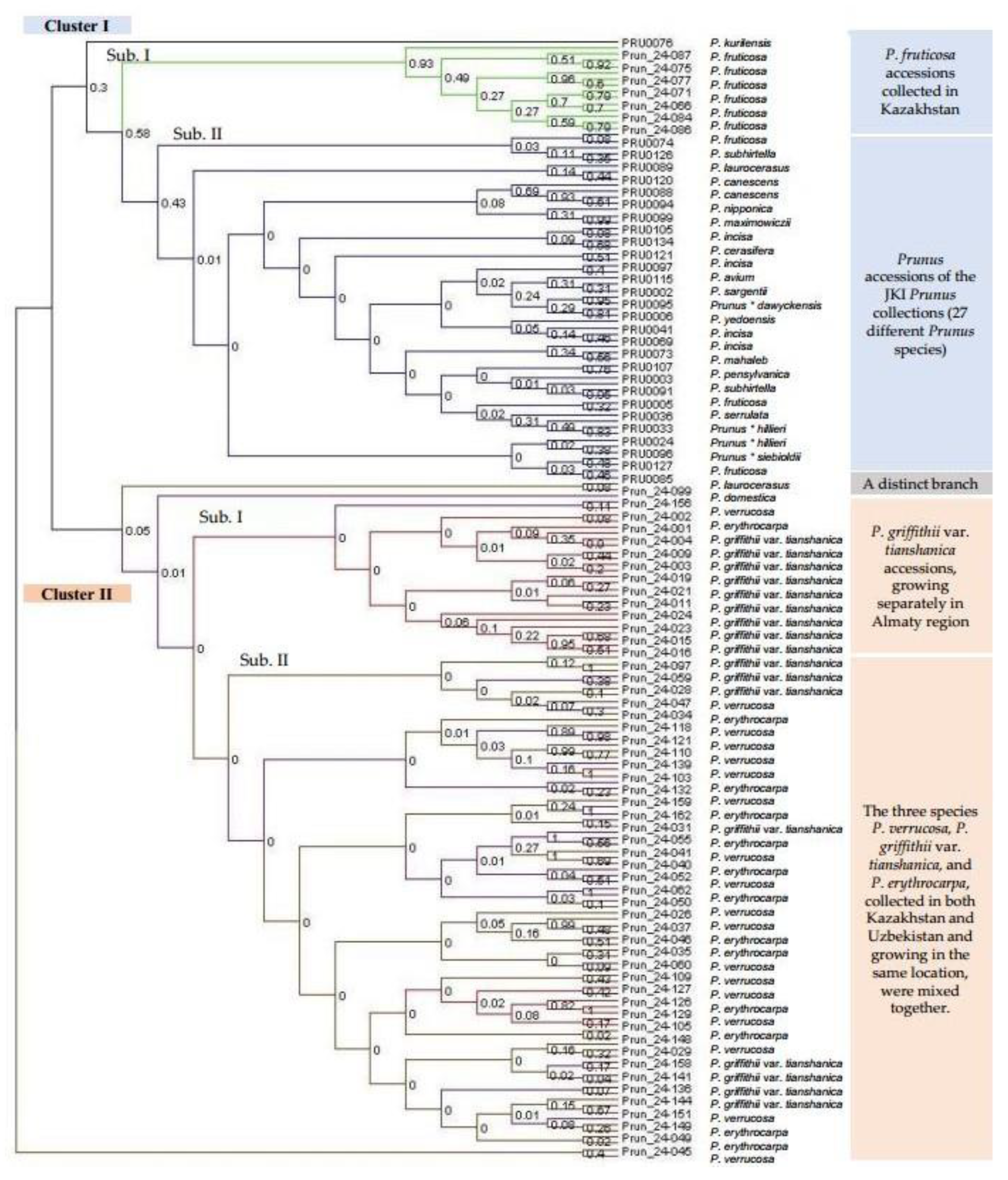
2.3. Genetic Clustering and Phylogenetic Tree
3. Discussion
3.1. Phenotypic Evaluation of Prunus Species
3.2. Genetic Diversity Parameters of the Prunus Species from Kazakhstan and Uzbekistan
3.3. Genetic Clustering and Phylogenetic Tree
4. Materials and Methods
4.1. Plant Material and Collection Sites
4.2. Phenotypic Assessment
4.3. DNA Extraction and SSR Analysis
4.4. Genetic Diversity Parameters
4.5. Genetic Structure and Phylogenetic Relationships Among the Prunus Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Statista. Cherry Production Worldwide 2023. Available online: https://www.statista.com/statistics/577489/world-cherry-production/ (accessed on 3 February 2025).
- Blando, F.; Oomah, B.D. Sweet and sour cherries: Origin, distribution, nutritional composition and health benefits. Trends Food Sci. Technol. 2019, 86, 517–529. [Google Scholar] [CrossRef]
- Fonseca, L.R.S.; Silva, G.R.; Luís, Â.; Cardoso, H.J.; Correia, S.; Vaz, C.V.; Duarte, A.P.; Socorro, S. Sweet Cherries as Anti-Cancer Agents: From Bioactive Compounds to Function. Molecules 2021, 26, 2941. [Google Scholar] [CrossRef]
- Pavlov, N.V. (Ed.) Flora of Kazakhstan, 3rd ed.; Alma-Ata, AN KazSSR: Almaty, Kazakhstan, 1961; pp. 510–516. [Google Scholar]
- Dzhangaliev, A.D.; Salova, T.N.; Turekhanova, R.M. Wild Fruit Plants of Kazakhstan; KazGosINTI: Almaty, Kazakhstan, 2001. [Google Scholar]
- Sitpayeva, G.T.; Kudabayeva, G.M.; Dimeyeva, L.A.; Gemejiyeva, N.G.; Vesselova, P.V. Crop wild relatives of Kazakhstani Tien Shan: Flora, vegetation, resources. Plant Divers. 2020, 42, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Abduraimov, O.S.; Maxmudov, A.V.; Kovalenko, I.; Allamurotov, A.L.; Mavlanov, B.J.; Shakhnoza, S.U.; Mamatkasimov, O.T. Floristic diversity and economic importance of wild relatives of cultivated plants in Uzbekistan (Central Asia). Biodivers. J Biol. Divers. 2023, 24, 1668–1675. [Google Scholar] [CrossRef]
- Vvedensky, A.I. (Ed.) Flora of Uzbekistan, 3rd ed.; Tashkent Publishing House of the Academy of Sciences of the Uzbek SSR: Tashkent, Uzbekistan, 1955; pp. 367–372. [Google Scholar]
- Rehder, A. Bibliography of Cultivated Trees and Shrubs. 1949. Bibliography of Cultivated Trees and Shrubs Hardy in the Cooler Temperate Regions of the Northern Hemisphere. Ar-Nold Arboretum of Harvard Univ. Available online: https://agris.fao.org/search/en/providers/122376/records/6511a07b60f8dcc51c5fecc3 (accessed on 3 February 2025).
- Dzhangaliev, A.D.; Salova, T.N.; Turekhanova, P.M. The Wild Fruit and Nut Plants of Kazakhstan. In Horticultural Reviews; Wild Apple and Fruit Trees of Central Asia; Janick, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2002; Volume 29, pp. 328–329. ISBN 9780471463375. [Google Scholar]
- Lacis, G.; Rashal, I.; Ruisa, S.; Trajkovski, V.; Iezzoni, A.F. Assessment of genetic diversity of Latvian and Swedish sweet cherry (Prunus avium L.) genetic resources collections by using SSR (microsatellite) markers. Sci. Hortic. 2009, 121, 451–457. [Google Scholar] [CrossRef]
- Antonius, K.; Aaltonen, M.; Uosukainen, M.; Hurme, T. Genotypic and phenotypic diversity in Finnish cultivated sour cherry (Prunus cerasus L.). Genet. Resour. Crop Evol. 2012, 59, 375–388. [Google Scholar] [CrossRef]
- Stanys, V.; Baniulis, D.; Morkunaite-Haimi, S.; Siksnianiene, J.B.; Frercks, B.; Gelvonauskiene, D.; Stepulaitiene, I.; Staniene, G.; Siksnianas, T. Characterising the genetic diversity of Lithuanian sweet cherry (Prunus avium L.) cultivars using SSR markers. Sci. Hortic. 2012, 142, 136–142. [Google Scholar] [CrossRef]
- Kato, S.; Matsumoto, A.; Yoshimura, K.; Katsuki, T.; Iwamoto, K.; Tsuda, Y.; Ishio, S.; Nakamura, K.; Moriwaki, K.; Shiroishi, T.; et al. Clone identification in Japanese flowering cherry (Prunus subgenus Cerasus) cultivars using nuclear SSR markers. Breed Sci. 2012, 62, 248–255. [Google Scholar] [CrossRef]
- Barreneche, T.; Cárcamo de la Concepción, M.; Blouin-Delmas, M.; Ordidge, M.; Nybom, H.; Lacis, G.; Feldmane, D.; Sedlak, J.; Meland, M.; Kaldmäe, H.; et al. SSR-Based Analysis of Genetic Diversity and Structure of Sweet Cherry (Prunus avium L.) from 19 Countries in Europe. Plants 2021, 10, 1983. [Google Scholar] [CrossRef]
- Clarke, J.B.; Sargent, D.J.; Bošković, R.I.; Belaj, A.; Tobutt, K.R. A cherry map from the inter-specific cross Prunus avium ‘Napoleon’ × P. nipponica based on microsatellite, gene-specific and isoenzyme markers. Tree Genet. Genomes 2009, 5, 41–51. [Google Scholar] [CrossRef]
- World Weather. Available online: https://world-weather.ru/ (accessed on 23 February 2025).
- Wang, X.; Wang, L.; Sun, Y.; Chen, J.; Liu, Q.; Dong, S. Genetic diversity and conservation of Siberian apricot (Prunus sibirica L.) based on microsatellite markers. Sci. Rep. 2023, 13, 11245. [Google Scholar] [CrossRef] [PubMed]
- Struss, D.; Ahmad, R.; Southwick, S.M.; Boritzki, M. Analysis of sweet cherry (Prunus avium L.) cultivars using SSR and AFLP markers. Am. Soc. Hortic. Sci. 2003, 128, 904–909. [Google Scholar] [CrossRef]
- Patzak, J.; Henychová, A.; Paprštein, F.; Sedlák, J. Evaluation of genetic variability within sweet cherry (Prunus avium L.) genetic resources by molecular SSR markers. Acta Sci. Pol. Hortorum Cultus 2019, 18, 157–165. [Google Scholar] [CrossRef]
- Chen, T.; Huang, X.J.; Zhang, J.; Chen, Q.; Liu, Y.; Tang, H.R.; Pan, X.; Wang, X.R. Genetic diversity and population structure patterns in Chinese cherry (Prunus pseudocerasus Lindl) landraces. Plant Mol. Biol. Report. 2016, 34, 440–453. [Google Scholar] [CrossRef]
- Reim, S.; Lochschmidt, F.; Proft, A.; Höfer, M. Genetic integrity is still maintained in natural populations of the indigenous wild apple species Malus sylvestris (Mill.) in Saxony as demonstrated with nuclear SSR and chloroplast DNA markers. Ecol. Evol. 2020, 10, 11798–11809. [Google Scholar] [CrossRef]
- Macková, L.; Vít, P.; Urfus, T. Crop-to-wild hybridization in cherries—Empirical evidence from Prunus fruticosa. Evol. Appl. 2018, 11, 1748–1759. [Google Scholar] [CrossRef]
- Barać, G.; Ognjanov, V.; Vidaković, D.O.; Dorić, D.; Ljubojević, M.; Dulić, J.; Miodragović, M.; Gašić, K. Genetic diversity and population structure of European ground cherry (Prunus fruticosa Pall.) using SSR markers. Sci. Hortic. 2017, 224, 374–383. [Google Scholar] [CrossRef]
- Reim, S.; Schiffler, J.; Braun-Lüllemann, A.; Schuster, M.; Flachowsky, H.; Höfer, M. Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank. Plants 2023, 12, 205. [Google Scholar] [CrossRef]
- Turkoglu, Z.; Bilgener, S.; Ercisli, S.; Yildirim, N. Simple sequence repeat (SSR) analysis for assessment of genetic variability in wild cherry germplasm. J. Appl. Bot. Food Qual. 2012, 85, 229–233. [Google Scholar]
- Li, M.M.; Cai, Y.L.; Qian, Z.Q.; Zhao, G.F. Genetic diversity and differentiation in Chinese sour cherry Prunus pseudocerasus Lindl., and its implications for conservation. Genet. Resour. Crop Evol. 2009, 56, 455–464. [Google Scholar] [CrossRef]
- Determinant of the Plants of Central Asia (Critical Flora Synopsis); FAN: Tashkent, Uzbekistan, 1976; Volume 5, pp. 238–245.
- Höfer, M.; Flachowsky, H. Erhaltungskonzept der Wildartensammlungen der Obstgenbank Dresden-Pillnitz–Aktivsammlung, Kryokonservierung, Global Seed Vault. J. Kult. 2020, 72, 466–472. [Google Scholar]
- Schmidt, H.; Vittrup-Christensen, J.; Watkins, R.; Smith, R.A. International Board for Plant Genetic Resources (IBPGRI); Commission of the European Communities (CEC): Rome, Italy, 1985; p. 33. [Google Scholar]
- Monika, H.; Giovannini, D. Phenotypic Characterization and Evaluation of European Cherry Collections: A Survey to Determine the Most Commonly used Descriptors. Sci. Pages Hortic. 2017, 1, 7–12. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software; Online Database. 2006. Available online: https://darwin.cirad.fr/ (accessed on 4 April 2025).
- Gascuel, O.; Mirkin, B.; McMorris, F.; Roberts, F.; Rzhetsky, A. Concerning the NJ algorithm and its unweighted version, UNJ. In Mathematical Hierarchies and Biology; DIMACS Series in Discrete Mathematics and Theoretical Computer Science; RI American Mathematical Society: Providence, RI, USA, 1997; pp. 149–170. [Google Scholar]
- Huson, D.H.; Richter, D.C.; Rausch, C.; Dezulian, T.; Franz, M.; Rupp, R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinform. 2007, 8, 460. [Google Scholar] [CrossRef]

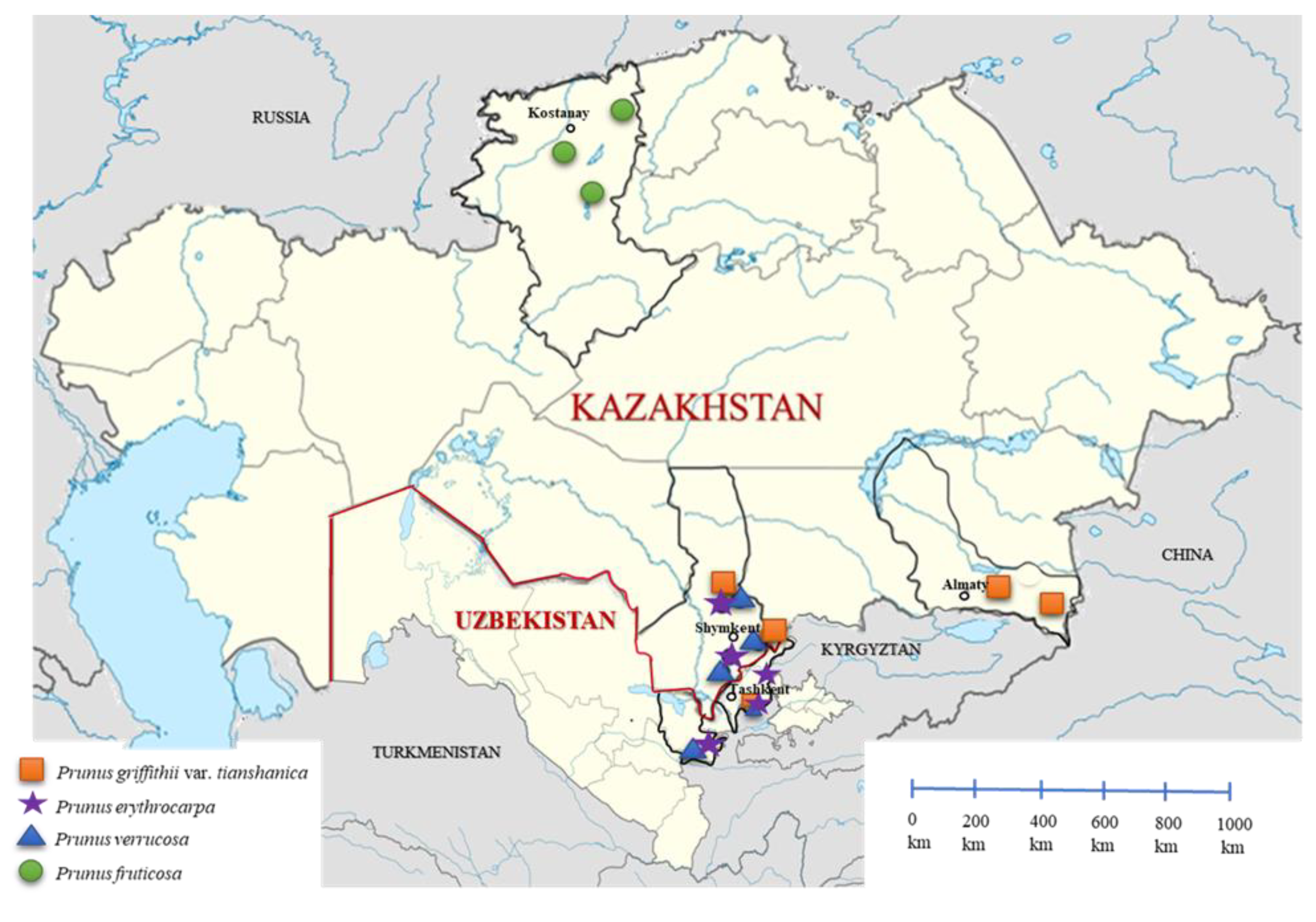
| Species | Location | Population | GPS Coordinates (WGS84) | Number of Accessions | ||
|---|---|---|---|---|---|---|
| Latitude | Longitude | Altitude Above Sea Level, m | ||||
| Prunus griffithii var. tianshanica | Kazakhstan, Almaty region | Tian 1 | 43°27′09.41″ | 78°39′16.94″ | 1117–1171 | 10 |
| Tian 2 | 43°21′01.28″ | 79°37′10.40″ | 1438–1488 | 15 | ||
| Kazakhstan, Turkestan region, Sairam-Ugam National Park | Tian 3 | 43°04′14.19″ | 69°54′01.76″ | 695–730 | 9 | |
| Tian 4 | 42°57′19.06″ | 70°02′58.11″ | 836–864 | 7 | ||
| Uzbekistan, Tashkent region, Ugam-Chatkal National Park | Tian 5 | 41°31′05.16″ | 70°01′05.03″ | 1689–1798 | 4 | |
| Total | 45 | |||||
| Prunus erythrocarpa | Kazakhstan, Turkestan region, Aksu-Zhabagly Nature Reserve | Eryth 1 | 42°24′48.36″ | 70°28′14.82″ | 1252–1534 | 16 |
| Kazakhstan, Turkestan region, Sairam-Ugam National Park | Eryth 2 | 44°11′21.00″ | 70°31′34.80″ | 716 | 3 | |
| Uzbekistan, Tashkent region, Ugam-Chatkal National Park | Eryth 3 | 41°36′00.00″ | 70°06′16.00″ | 1690–1756 | 9 | |
| Eryth 4 | 41°13′46.00″ | 70°14′40.00″ | 1543–2069 | 5 | ||
| Uzbekistan, Jizzakh region, Zaamin National Park | Eryth 5 | 39°52′38.00″ | 68°28′27.00″ | 2060 | 1 | |
| Total | 34 | |||||
| Prunus verrucosa | Kazakhstan, Turkestan region, Aksu-Zhabagly Nature Reserve | Ver 1 | 42°19′49.86″ | 70°22′22.26″ | 1123–1598 | 24 |
| Kazakhstan, Turkestan region, Sairam-Ugam National Park | Ver 2 | 42°40′07.21″ | 70°13′45.82″ | 848–854 | 5 | |
| Ver 3 | 43°05′45.43″ | 69°55′44.34″ | 715–733 | 9 | ||
| Uzbekistan, Tashkent region, Ugam-Chatkal National Park | Ver 4 | 41°10′03.00″ | 70°14′13.00″ | 1572 | 2 | |
| Uzbekistan, Jizzakh region, Zaamin National Park | Ver 5 | 39°44′24.00″ | 68°38′03.00″ | 2029–2233 | 13 | |
| Total | 34 | |||||
| Prunus fruticosa | Kazakhstan, Kostanay region | Frut 1 | 53°02′14.40″ | 63°40’35.22″ | 187–199 | 11 |
| Frut 2 | 53°17’18.00″ | 64°12’31.80″ | 190–202 | 7 | ||
| Frut 3 | 52°26’35.40″ | 64°18’30.00″ | 153–169 | 13 | ||
| Total | 31 | |||||
| Total | 163 | |||||
| Species | N | Na | Ne | I | Ho | He | uHe | F |
|---|---|---|---|---|---|---|---|---|
| Prunus erythrocarpa | 15.19 | 8.96 | 5.82 | 1.63 | 0.52 | 0.66 | 0.70 | 0.21 |
| Prunus fruticosa | 8.12 | 2.19 | 1.71 | 0.54 | 0.42 | 0.30 | 0.32 | −0.41 |
| Prunus griffithii var. tianshanica | 17.73 | 7.81 | 4.57 | 1.55 | 0.51 | 0.68 | 0.71 | 0.27 |
| Prunus verrucosa | 28.62 | 14.04 | 8.53 | 2.10 | 0.57 | 0.80 | 0.84 | 0.27 |
| Mean | 17.41 | 8.25 | 5.16 | 1.46 | 0.51 | 0.61 | 0.64 | 0.15 |
| SE | 1.45 | 0.71 | 0.41 | 0.09 | 0.04 | 0.03 | 0.03 | 0.05 |
| Prunus erythrocarpa | Prunus fruticosa | Prunus griffithii var. tianshanica | Prunus verrucosa | |
|---|---|---|---|---|
| 1.00 | Prunus erythrocarpa | |||
| 0.09 | 1.00 | Prunus fruticosa | ||
| 0.44 | 0.03 | 1.00 | Prunus griffithii var. tianshanica | |
| 0.50 | 0.09 | 0.46 | 1.00 | Prunus verrucosa |
| Species | Number of Accessions in the JKI Gene Bank Collection |
|---|---|
| Prunus × cistena | 1 |
| Prunus × dawyckensis | 3 |
| Prunus × hillieri | 3 |
| Prunus × siebioldii | 1 |
| Prunus avium | 2 |
| Prunus brigantina | 1 |
| Prunus canescens | 4 |
| Prunus cerasifera | 2 |
| Prunus domestica | 3 |
| Prunus fruticosa | 2 |
| Prunus incisa | 6 |
| Prunus kurilensis | 2 |
| Prunus laurocerasus | 2 |
| Prunus maackii | 1 |
| Prunus mahaleb | 3 |
| Prunus maximowiczii | 2 |
| Prunus mollis | 1 |
| Prunus nipponica | 2 |
| Prunus padus | 1 |
| Prunus pensylvanica | 1 |
| Prunus sargentii | 2 |
| Prunus serrula | 1 |
| Prunus serrulata | 4 |
| Prunus subhirtella | 2 |
| Prunus tomentosa | 2 |
| Prunus virginiana | 1 |
| Prunus yedoensis | 2 |
| Accessions total | 57 |
| Primer Mix | Primer | Dye | Size Range | PCR Annealing Temp. |
|---|---|---|---|---|
| Primer Mix 1 | EMPA002 | At532 | 98–133 | 55 °C |
| EMPA003 | At550 | 161–181 | 55 °C | |
| CPPCT006 | FAM | 172–205 | 55 °C | |
| EMPaSO2 | At565 | 130–187 | 55 °C | |
| Primer Mix 3 | CPPCTO22 | FAM | 219–235 | 55 °C |
| EMPaS14 | At565 | 164–212 | 55 °C | |
| UDP98-412 | At532 | 83–155 | 55 °C | |
| EMPaSO1 | At532 | 199–250 | 48 °C | |
| EMPaS12 | FAM | 100–150 | 55 °C | |
| EMPaS10 | At565 | 135–207 | 55 °C | |
| Primer Mix 4 | EMPA017 | FAM | 218–244 | 55 °C |
| EMPA026 | At532 | 179–258 | 55 °C | |
| PSO5CO3 | At550 | 105–168 | 48 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manapkanova, U.; Rymkhanova, N.; Reim, S.; Fritzsche, E.; Höfer, M.; Beshko, N.; Satekov, Y.; Kushnarenko, S.V. Genetic Diversity and Phenotypic Variation of Indigenous Wild Cherry Species in Kazakhstan and Uzbekistan. Plants 2025, 14, 1676. https://doi.org/10.3390/plants14111676
Manapkanova U, Rymkhanova N, Reim S, Fritzsche E, Höfer M, Beshko N, Satekov Y, Kushnarenko SV. Genetic Diversity and Phenotypic Variation of Indigenous Wild Cherry Species in Kazakhstan and Uzbekistan. Plants. 2025; 14(11):1676. https://doi.org/10.3390/plants14111676
Chicago/Turabian StyleManapkanova, Ulzhan, Nazgul Rymkhanova, Stefanie Reim, Eric Fritzsche, Monika Höfer, Natalya Beshko, Yeskendir Satekov, and Svetlana V. Kushnarenko. 2025. "Genetic Diversity and Phenotypic Variation of Indigenous Wild Cherry Species in Kazakhstan and Uzbekistan" Plants 14, no. 11: 1676. https://doi.org/10.3390/plants14111676
APA StyleManapkanova, U., Rymkhanova, N., Reim, S., Fritzsche, E., Höfer, M., Beshko, N., Satekov, Y., & Kushnarenko, S. V. (2025). Genetic Diversity and Phenotypic Variation of Indigenous Wild Cherry Species in Kazakhstan and Uzbekistan. Plants, 14(11), 1676. https://doi.org/10.3390/plants14111676








