1. Introduction
Apple (
Malus domestica), a perennial woody plant, is one of the most widely cultivated fruit trees worldwide, with China ranking first in both planting area and production [
1]. This economically significant crop is highly valued by consumers for its pleasant sweet-sour taste and rich nutritional content [
2].
Magnesium ions (Mg
2+) serve as an essential element for plant growth and development, being a vital component of both chlorophyll and various enzymes [
3,
4]. In apple trees, magnesium deficiency not only impairs tree growth and development but also adversely affects fruit quality and yield. Numerous studies have demonstrated that magnesium deficiency in plants leads to leaf chlorosis, resulting in reduced chlorophyll content and decreased photosynthetic rate [
5]. Furthermore, as the most abundant free divalent cation in plant cells, Mg
2+ plays crucial roles in membrane stabilization, ion balance maintenance, and root growth [
6]. Magnesium exists in plant systems predominantly as Mg
2+ ions [
7]. Beyond its crucial function in facilitating carbon assimilation during photosynthesis, Mg
2+ serves three key physiological roles in chloroplasts—modulation of excitation energy distribution between photosystems II and I, participation in grana stacking processes, and contribution to thylakoid membrane assembly—collectively maintaining chloroplast structural integrity. Furthermore, while essential for photosynthetic efficiency and root absorption, Mg
2+ dynamics exhibit additional complexity through its bidirectional transport within vascular systems, involving both xylem and phloem networks [
8].
Magnesium ions (Mg
2+) play vital roles in biochemical metabolism, and their selective permeability across biological membranes necessitates specialized transport systems. The uptake and translocation of Mg
2+ are mediated by complex membrane protein systems, primarily through active transport mechanisms [
9]. Current research has identified multiple Mg
2+ transport systems in plants, including the cobalt resistance A protein family (CorA), Mg
2+/H
+ exchangers (
Arabidopsis thaliana magnesium-proton exchanger,
AtMHX), P-type phosphatases, the MgtE transporter family, and various ion channels [
10]. Of these, CorA/MRS2-type transporters are particularly crucial for maintaining Mg
2+ homeostasis in plants [
11].
Following the completion of
Arabidopsis thaliana genome sequencing, researchers identified the AtMRS2/AtMGT family in 2000, showing both structural and sequence homology to bacterial CorA proteins [
12]. CorA-type homologs are evolutionarily conserved across all domains of life, and are present in archaea, eubacteria, and eukaryotes [
13]. Bacterial systems employ three coordinated Mg
2+ transporters, CorA, MgtA, and MgtB, with CorA demonstrating the highest Mg
2+ affinity and serving as the primary membrane-localized transporter in prokaryotes [
14]. The conserved GMN motif in CorA proteins has been experimentally confirmed as essential for Mg
2+ transport functionality [
14].
The
MRS2 gene, a eukaryotic homolog of bacterial CorA, was first characterized in yeast, where it encodes the Mrs2p protein—an integral component of the inner mitochondrial membrane [
15] Subsequent research has revealed that CorA/MRS2-type magnesium transporters are widely distributed across plant species. In Arabidopsis thaliana, genomic studies have identified 11 family members, comprising 10 functional CorA/MRS2-type magnesium transporters and 1 pseudogene (
AtMRS2-9) [
16] Functional analyses in Arabidopsis have demonstrated that these transporters exhibit broad substrate specificity capable of mediating not only Mg
2+ but also other divalent cations, including Fe
2+ [
14]
Among these, the
AtMRS2-1 gene represents one of the best-characterized members. This plasma membrane-localized transporter plays a crucial role in root magnesium acquisition, displaying both high affinity and specificity for Mg
2+ under physiological soil concentrations. While capable of transporting other divalent cations, this activity requires concentrations significantly exceeding typical soil levels [
14].
In rice (
Oryza sativa), nine CorA/MRS2-type magnesium transporter proteins have been identified. Among these,
OsMGT1 demonstrates plasma membrane localization and exhibits significant alleviation effects on aluminum toxicity in rice roots [
17]. This gene shows predominant expression in shoots and callus tissues [
18]. Similarly, six CorA/MRS2-type magnesium transporter proteins were identified in grapevine (
Vitis vinifera), with predicted involvement in chloroplast metabolism [
8].
Multiple CorA/MRS2-type Mg
2+ transporters have also been characterized in various horticultural crops, including tobacco (
Nicotiana tabacum) and peach (
Prunus persica). These transporters collectively regulate root Mg
2+ uptake and enhance plant adaptation to Mg
2+-deficient environments [
19]. Specifically, seven MRS2/MGT family members were identified in the tobacco genome.
NtMGT1 shows root-specific expression, while
NtMGT2,
NtMGT4, and
NtMGT5 display leaf-specific expression with light-inducible characteristics [
20].
The presence of multiple magnesium stressors in the natural environment poses a serious threat to plant growth and development, thereby limiting the sustainable development of agriculture [
21]. Magnesium ion transporters can reduce the toxicity of aluminum (Al) in plants.
AtMRS2-1, located on the vacuole, is insensitive to Al stress, while
AtMRS2-10 and
AtMRS2-11 show high sensitivity to Al toxicity [
22]. It also has an impact on the development of pollen.
PbrMGT7 was also expressed in the pollen, and acted on the mitochondria to keep the homeostasis of Mg
2+ in pollen development [
23]. Although it is known that a large amount of research has been conducted on magnesium ion transporters in other species, as far as we know, they have not yet been studied in apples. Therefore, an integrated approach was adopted in this study: the
MdMRS2 gene was identified through apple genome screening, and systematic bioinformatics analysis was performed to reveal the function of the
MdMRS2 gene in magnesium ion transport. The experimental treatments under magnesium-deficient and over-magnesium conditions were designed using apple isolate seedlings to determine the expression pattern of the
MdMRS2 gene in response to different magnesium concentrations to provide targets for the subsequent genetics, to verify its role in apple magnesium utilization efficiency and stress tolerance, and to improve the quality of apple seedlings by optimizing magnesium fertilizer application strategies and combining key gene expression patterns.
3. Discussion
The MRS2 gene family is a typical magnesium transporter [
24]. Plant magnesium transporters play an important role in the absorption and transport of magnesium in plants [
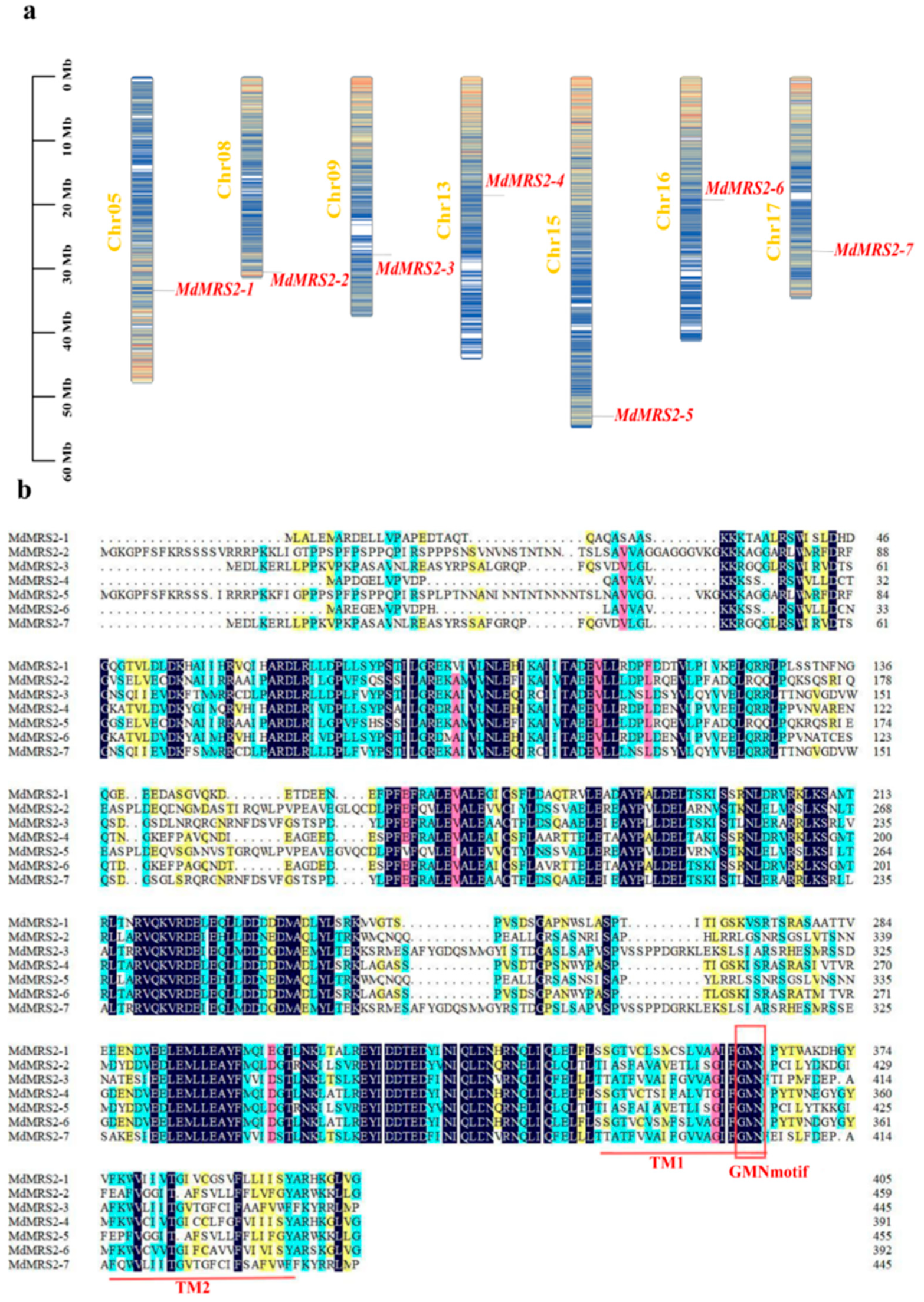
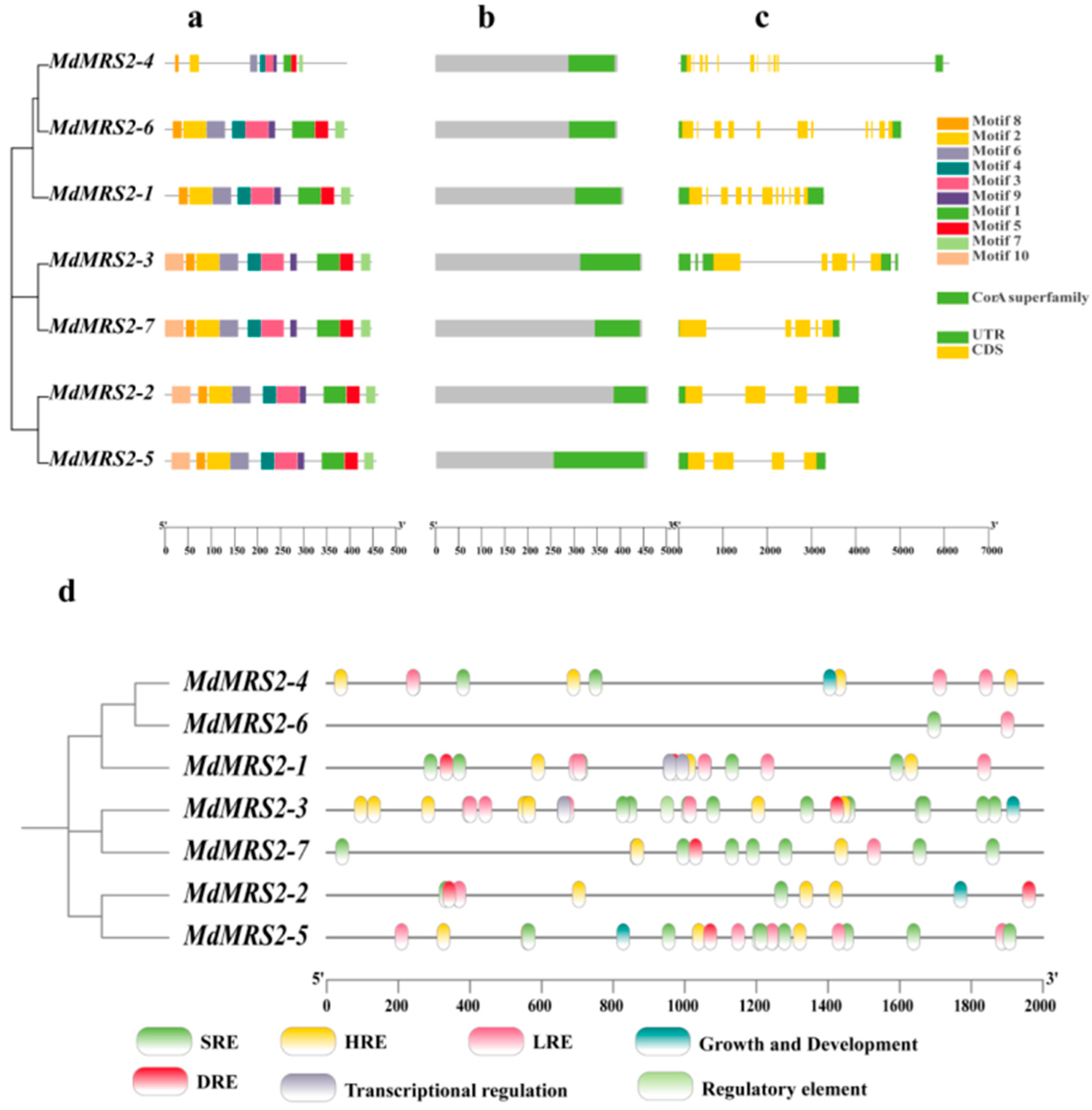
25]. The identification and cloning of the MRS2 gene family have been reported in many plants. In this study, seven apple
MRS2 genes on different chromosomes were screened by bioinformatics methods and named as
MdMRS2-1 ~
MdMRS2-7, according to the chromosome position. These members all contain a transmembrane structure and a conserved GMN characteristic motif. In this motif, G is necessary for magnesium uptake, M maintains the integrity of the ion channel conformation, and N stabilizes these two functions [
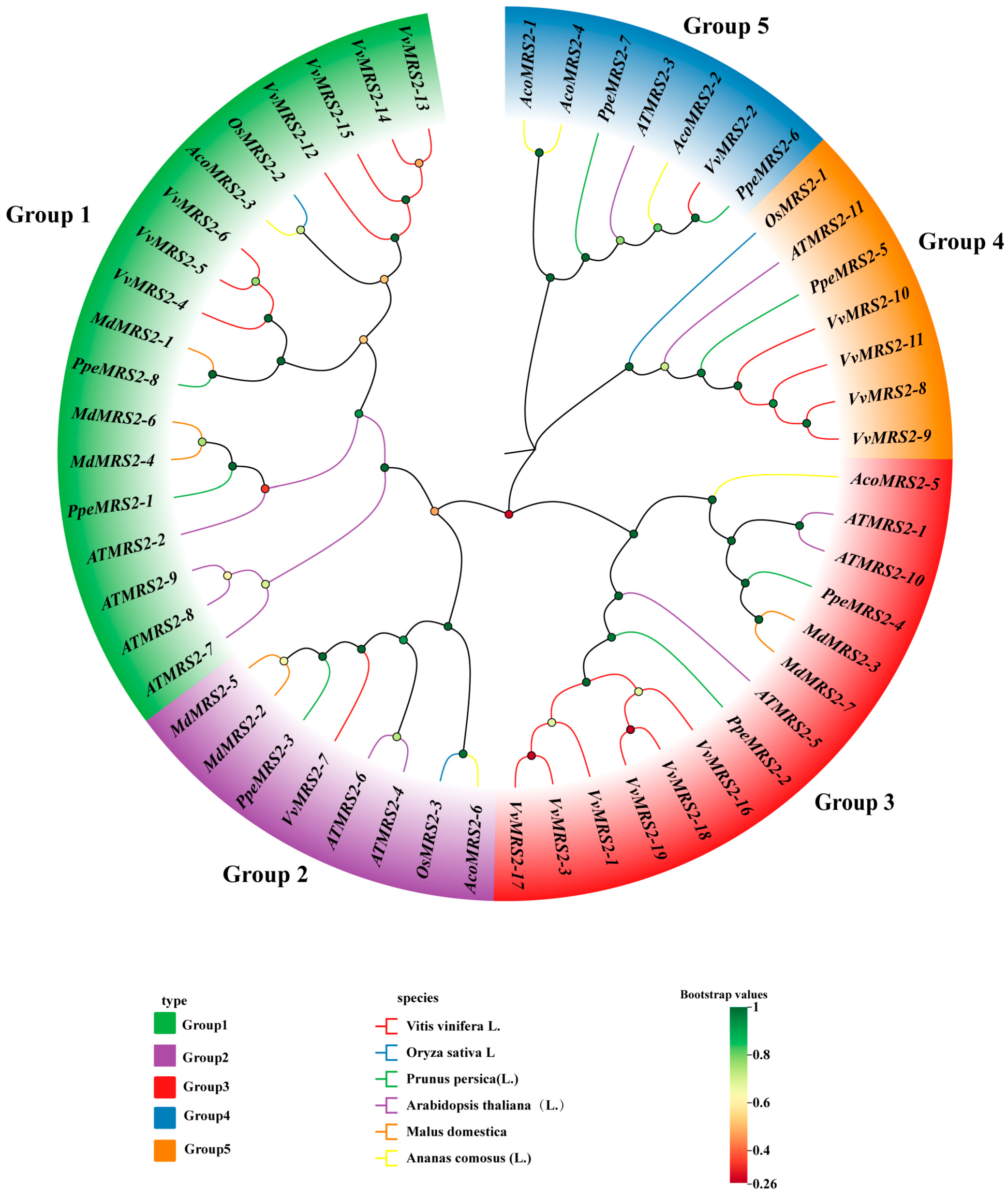
26]. In this paper, the evolutionary relationship of the
MRS2 system of six species was analyzed. The results show that the MRS2 gene family has five subfamilies, among which
MRS2 has the closest genetic relationship with dicotyledonous plants of the Rosaceae family, which is similar to the research results of Zhao et al. [
22]. And the same branch may have similar functions. For example, the
AtMRS2-7 gene and the
MdMRS2-1,
MdMRS2-4, and
MdMRS2-6 sequences belong to the same branch. In
Arabidopsis thaliana, the
AtMRS2-7 gene is a key gene to ensuring the survival of Arabidopsis thaliana under low magnesium conditions [
27]. Therefore, it is speculated that
MdMRS2-1,
MdMRS2-4, and
MdMRS2-6 genes in apple have similar functions to ensure the survival of plants under low magnesium conditions. In phylogenetic tree and collinearity analysis, it was found that the closeness between apples and dicotyledonous plants is greater than that between monocotyledonous plants. The relationship between tomatoes (dicotyledonous plants) and
Arabidopsis thaliana is closer than that between tomatoes and rice. However, Tong Mengying [
28] found in her research on MaMRS2 that bananas (monocotyledonous plants) have a relatively distant genetic relationship with Arabidopsis thaliana (dicotyledonous plants) but a relatively close relationship with the monocotyledonous plant rice; the above results can indicate that the similarity between the CorA/MRS2 genes in plants is species-related. Zhao et al. found in the study of the grape MRS2 family that the Ka/Ks of all members of the VvMRS2 family were significantly lower than 1 [
8]. In this study, it was also found that the Ka/Ks of all members of the MdMRS2 family were also significantly lower than 1, indicating that the nonsynonymous substitution rate is much lower than the synonymous substitution rate. These results indicate that the members of the MRS2 gene family have mainly undergone purification selection during evolution.
In papaya, CpMGT is mainly distributed within membrane cells [
29]; in corn ZmMGT and Brazilian rubber tree HbMGT are mainly distributed in chloroplasts [
30,
31]. Liu et al. found that the subcellular localization prediction of tomatoes showed that
SlMRS2-1,
SlMRS2-5, and
SlMRS2-11 were located on the plasma membrane;
SlMRS2-2 and
SlMRS2-3 were located on the chloroplast membrane;
SlMRS2-1 was located on the cytoplasm; and
SlMRS2-4 was located on the nucleus and plasma membrane [
19]. Wang Yongjun’s research found that eight MGT members in sugarcane are located on chloroplasts, while the remaining two are predicted to be located on mitochondria (
SsMGT3) and the plasma membrane (
SsMGT5) [
32]. In his research on the magnesium ion transporter protein of pineapple, Hu Bingyan made transient expression of the
AcoMRS2-3 gene and found that its localization in tobacco cells was in the Golgi apparatus, endoplasmic reticulum, and plastids [
33]. In
MdMRS2,
MdMRS2-1 is located in the nucleus and chloroplasts;
MdMRS2-2 is located on the cell membrane and in the nucleus;
MdMRS2-3 is located in the cell membrane, nucleus, and chloroplasts;
MdMRS2-4 is located in the cytoplasm and chloroplasts;
MdMRS2-5 is located in the chloroplasts;
MdMRS2-6 and
MdMRS2-7 are located in the cell nucleus. The transient transformation of the
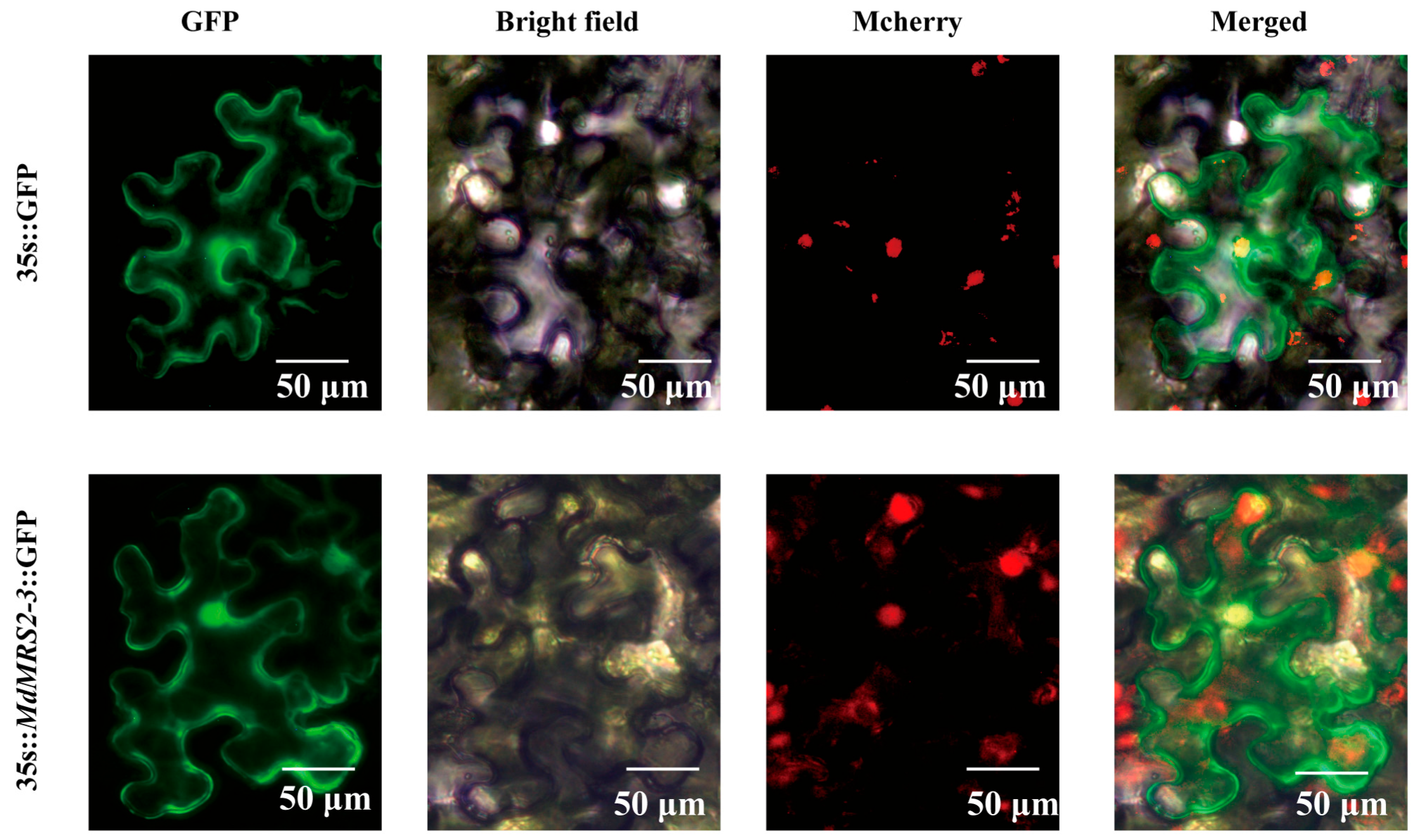
MdMRS2-3 gene revealed that it is located in the nucleus, cytoplasm, and chloroplast of tobacco, indicating that
MdMRS2 is likely to maintain the dynamic balance of magnesium ions in tissue cells in a multi-member cooperative manner.
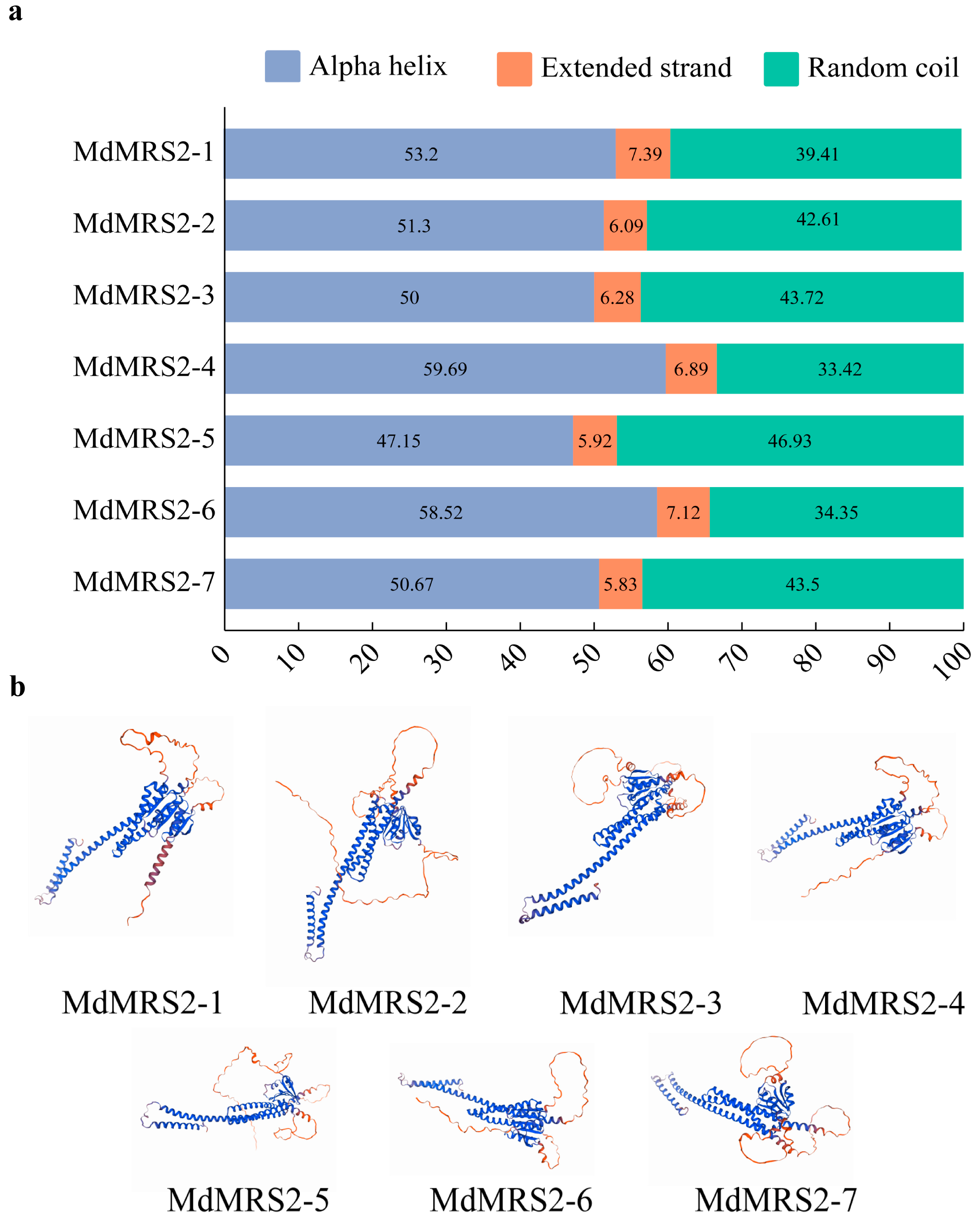
By analyzing the primary structure of MdMRS2 family proteins, a total of 10 conserved motifs were identified, among which motif 1, motif 2, motif 3, motif 4, motif 5, motif 6, motif 7, and motif 8 existed in all the protein members, and it was speculated that these conserved motifs were important for the function of MdMRS2 family proteins. Tan et al. found in MeMGT that the proteins of this family are mainly composed of α-helix, extended strand, β-turn, and random curl, among which the proportion of β-turn is the lowest, ranging from 1.21% to 4.26% [
34]. In the analysis of the secondary and tertiary structures of the MdMRS2 family proteins, it was revealed that the secondary structures of the seven proteins predominantly consisted of α-helices, extended strands, and random coils. Notably, β-sheets were absent, which may potentially be attributed to natural selection or evolutionary pressures. Upon examining their tertiary structures, it was observed that all seven proteins exhibited highly similar spatial conformations.
The
cis-acting elements of the promoter play an important role in the transcription level of the gene, and the analysis of the
cis-acting elements provides a certain reference value for predicting gene function [
35]. By analyzing the promoter sequence of the gene, its role in intracellular and intercellular signaling pathways can be predicted to determine its function in cells. The homeostatic elements in chili peppers include light reaction elements and anaerobic induction response elements, while other components are restricted to one or more members of the chili MGT. For example, circadian rhythm response elements, endosperm expression response elements, seed-specific regulatory elements, wound response elements, and hypoxia-specific induction elements are, respectively, present only in the promoter regions of
MGT3/10,
MGT10,
MGT9,
MGT3, and
MGT9 in chili peppers [
8]. Wang et al. conducted an expression analysis of the MGT gene in the diurnal cycle of two species of sugar plants.
MGT9 and
MG10 were observed to have a peak expression in the middle of the night period in
S. spontaneum, but they showed no diurnal expression in
S. officinarum, indicating diurnal rhythms regulate these two
MGTs in
S. spontaneum rather than
S. officinarum [
32]. In this study, the promoter region of the MdMRS2 gene family was analyzed. The results showed that the promoter of the apple MdMRS2 gene family had
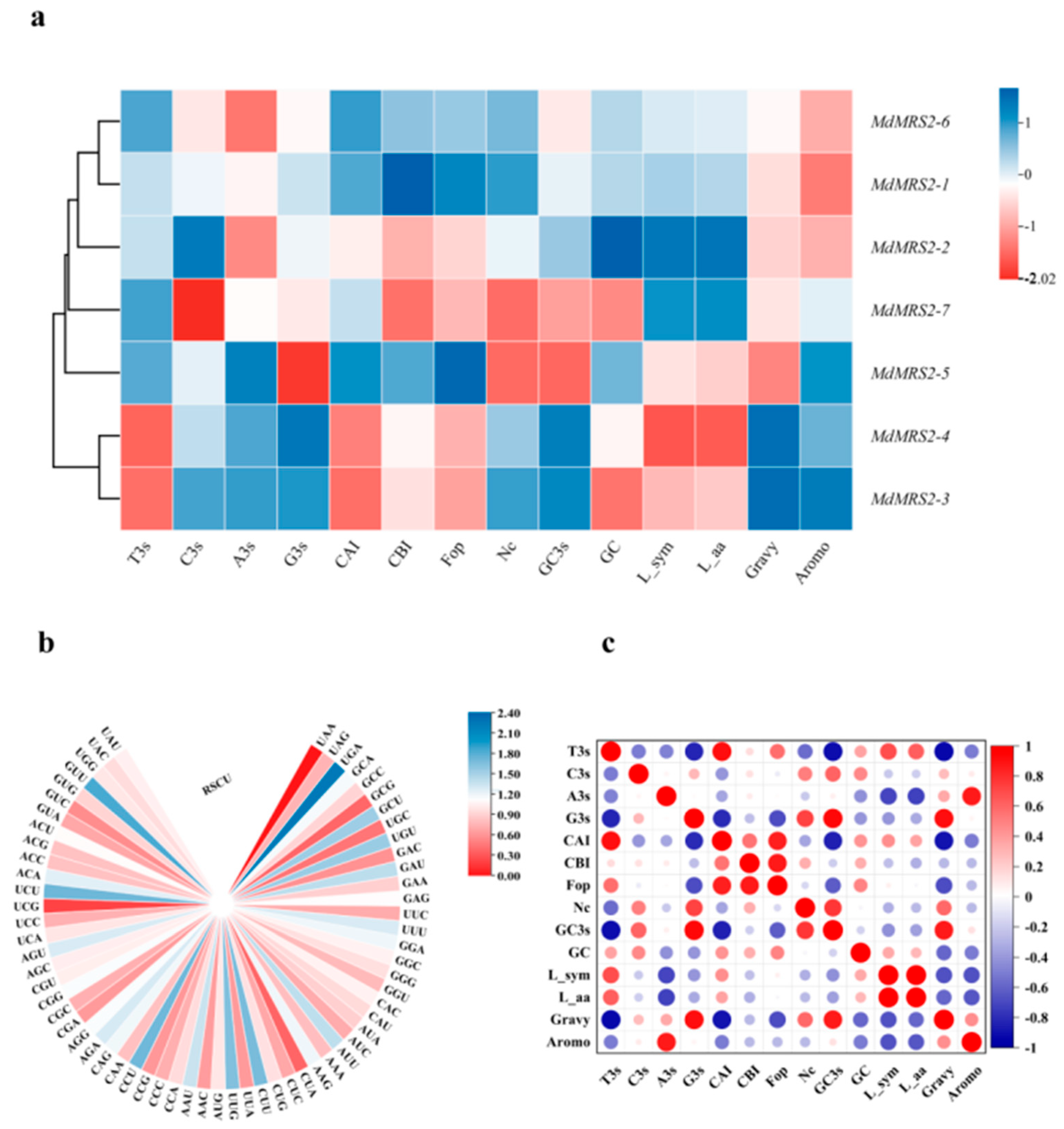
cis-acting elements related to light response, hormone response, drought, and low-temperature response. Especially, the light response elements and gibberellin response elements appeared frequently; this indicates that the MdMRS2 gene family may play an important role in the growth and development of the apple. It plays an important role in the photosynthesis of plants by regulating the transport of magnesium ions by combining with light response elements. Three of the thirty translated amino acids of the MdMRS2 family have Ter, one of the triplet amino acids generated by a method with shortening, and all of their translated codons are termination codons. Codon bias analysis of the gene showed that T3s was negatively correlated with G3s, Gc3s, Gravy, G3s with CAI, and Fop. Fop was positively correlated with CAI and CBI, and G3s with GC3s and Gravy. (
Figure 6) This suggests that the base type at position 3 of the synonymous codon influences the degree of codon usage preference.
The fluorescence quantitative results showed that under the conditions of Mg
2+ deficiency and Mg
2+ excess, the relative expression level would be significantly upregulated only under the condition of Mg
2+ excess. Under other conditions, it was upregulated or downregulated to varying degrees compared with CK, but none were significant. This was consistent with the results for tomato [
14]. Yan et al. [
36] observed that the expression of the
MGT6 gene in Arabidopsis thaliana responded to low Mg
2+ in root tissues and showed a corresponding response in high Mg
2+, but the difference was not significant. When observing the phenotype of Arabidopsis thaliana under ultra-high Mg
2+ conditions, it was found that there were serious phenotypic defects. These data suggested that the mgt6 mutant is not only compromised under low Mg
2+ levels but also hypersensitive to high-Mg
2+ stress. This gene family solves the problem of ionic toxicity by regulating magnesium ions. Therefore, we speculate that the regulation of magnesium ions in apples plays an important role in addressing ionic toxicity.
4. Materials and Methods
4.1. Materials and Treatments
The experiment was conducted in 2024 at the Laboratory of Fruit Tree Physiology and Biotechnology, College of Horticulture, Gansu Agricultural University. The plant materials consisted of in vitro-grown ‘Orin’ apple (Malus domestica) plantlets cultured on MS medium (4.42 g·L−1 MS basal salts, 30 g·L−1 sucrose, 6 g·L−1 agar, 0.1 mg·L−1 6-Benzylaminopurine (6-BA), and 0.2 mg·L−1 Indole acetic acid (IAA), pH 5.8–6.0). The plants were maintained in a growth chamber under controlled conditions: 25 °C with 16 h light/20 °C with 8 h darkness.
After 40 days of culture, uniformly growing, healthy, and contamination-free plantlets were selected and transferred to a hydroponic system containing modified MS nutrient solution with varying magnesium concentrations. Four MgSO4 treatment groups were established: CK (control), 1 mmol·L−1 MgSO4; M1 (Mg2+-deficient), 0 mmol·L−1 MgSO4; M2 (moderate Mg2+), 2 mmol·L−1 MgSO4; M3 (high Mg2+), 4 mmol·L−1 MgSO4. Each treatment was replicated three times. Following 48 h of treatment, leaf samples were collected, immediately wrapped in aluminum foil, flash-frozen in liquid nitrogen, and stored at −80 °C for subsequent analysis.
4.2. Identification of the Apple MRS2 Gene Family
The conserved amino acid sequences of
MRS2 genes from Arabidopsis thaliana were employed as queries to identify putative apple MRS2 family members through BLAST homology searches on Phytozome13 (
https://phytozome-next.jgi.doe.gov/ accessed on 15 August 2024). Candidate genes were initially screened for redundancy using DNAMAN software v6.0, followed by domain validation through HMMER online analysis (
https://www.ebi.ac.uk/Tools/hmmer/ accessed on 15 August 2024) to confirm the presence of characteristic
MRS2 domains (PF01544.21). Physicochemical properties, including amino acid length, molecular weight, and isoelectric point, were determined using the Expasy ProtParam tool (
https://web.expasy.org/protparam/ accessed on 19 August 2024). Subcellular localization predictions were performed using both WoLF PSORT (
https://wolfpsort.hgc.jp/ accessed on 19 August 2024) and CELLO v.2.5 (
http://cello.life.nctu.edu.tw/ accessed on 19 August 2024) bioinformatics platforms.
4.3. Phylogenetic Tree, Synteny Analysis
The MRS2 family members from Arabidopsis thaliana, apple (
Malus domestica), grape (
Vitis vinifera), rice (
Oryza sativa), peach (
Prunus persica), and pineapple (
Ananas comosus) were identified through Phytozome v13 (
https://phytozome-next.jgi.doe.gov/ accessed on 24 August 2024). The amino acid sequences of these
MRS2 genes were converted to FASTA format using ClustalX v1.83, followed by phylogenetic tree construction with MEGA 7.0 software. The resulting phylogenetic tree was visualized and annotated using TVBOT (
https://www.chiplot.online/tvbot.html accessed on 24 August 2024).
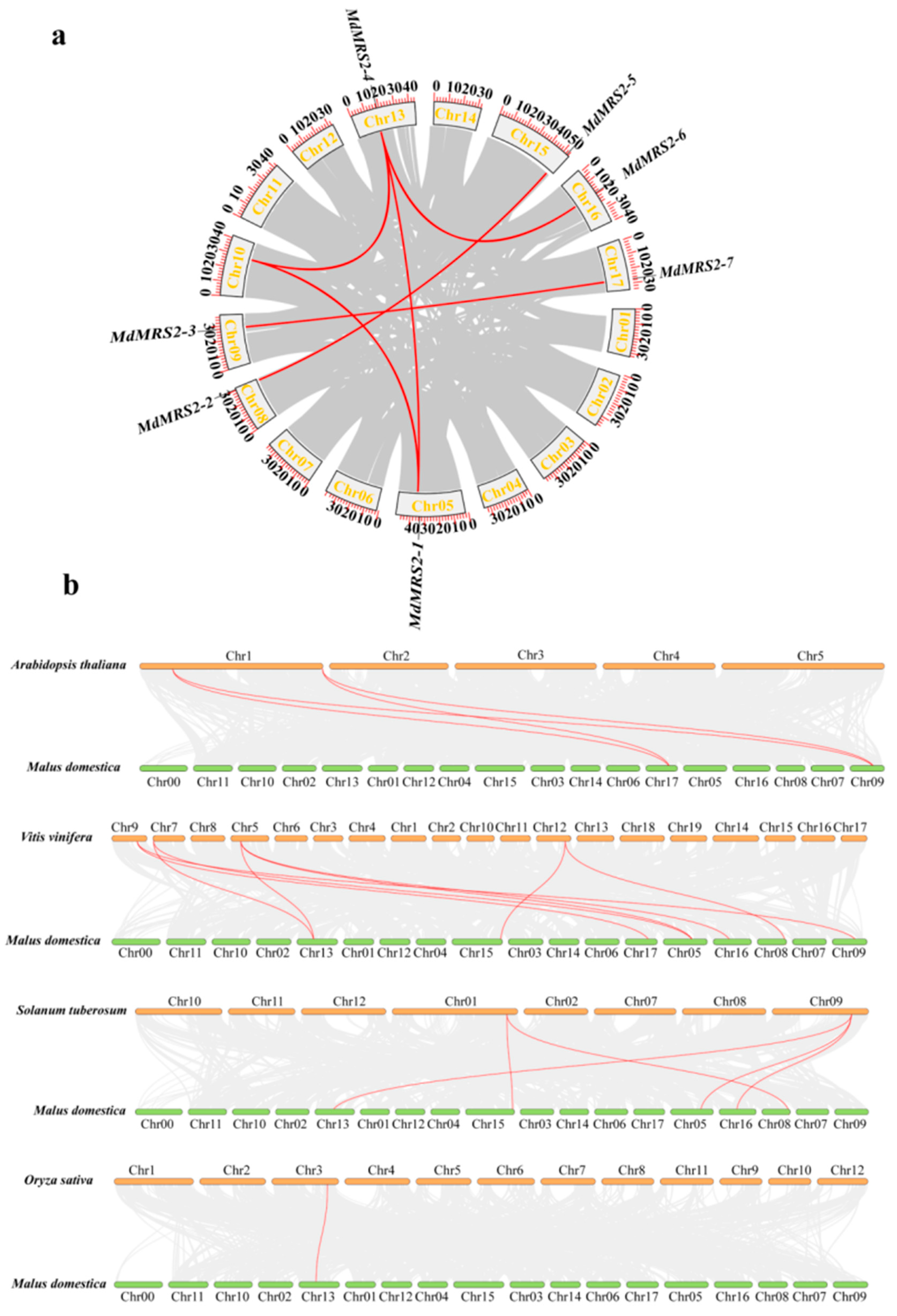
The whole-genome sequences of Arabidopsis thaliana, apple (
Malus domestica), grape (
Vitis vinifera), rice (
Oryza sativa), and potato (
Solanum tuberosum) were obtained from Phytozome v13 (
https://phytozome-next.jgi.doe.gov/) (accessed on 19 August 2024). Subsequently, synteny analysis was performed using TBtools to identify conserved genomic blocks and homologous gene pairs among these species.
4.4. Ka/Ks, Motif, Gene Structure, and Cis-Acting Element Analysis
4.5. Codon Bias Analysis
Codon usage bias analysis was performed using CodonW v1.4.4 (
http://codonw.sourceforge.net, accessed on 26 September 2024). The resulting data were processed and organized using Microsoft Excel 2010 (Microsoft Corporation, Redmond, WA, USA). For statistical correlation analysis, the formatted data were imported into OriginPro 2021 (OriginLab Corporation). Relative synonymous codon usage (RSCU) analysis was conducted using RStudio 2022.07.2 (
https://www.rstudio.com/products/rstudio/download/, accessed on 4 December 2024) with the seqinr package.
4.6. RNA Extraction and qRT-PCR of MdMRS2-3 Gene
Total RNA was extracted from apple tissues using a plant RNA extraction kit (Acres Biotechnology Co., Ltd., Zhejiang, China) following the manufacturer’s protocol. RNA quality was assessed by measuring absorbance ratios (OD260/280) and concentration using spectrophotometry, with integrity verified by 1.2% agarose gel electrophoresis. Qualified RNA samples were stored at −80 °C until use.
For qRT-PCR analysis, gene-specific primers for
MRS2 genes were designed (
Table S1) and synthesized by Sangon Biotech (Shanghai, China) Co., Ltd. (
https://www.sangon.com accessed on 4 November 2024). First-strand cDNA was synthesized from 1 μg total RNA using the PrimeScript RT reagent Kit (Perfect Real Time; Takara Bio Inc., Kusatsu, Japan). Quantitative PCR was performed using SYBR Premix Ex Taq II (TakaraBio Inc.) on a LightCycler 96 Real-Time PCR System (Roche Diagnostics, Switzerland) with the following cycling conditions: initial denaturation at 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s. All reactions were performed in triplicate technical replicates.
4.7. Tobacco Transient Transformation
The full-length coding sequence of MdMRS2-3 was amplified by PCR using cDNA derived from ‘Gala’ apple seedlings as template. The purified PCR product was subsequently cloned into the pCAMBIA2300-GFP expression vector through restriction enzyme digestion and ligation. The recombinant plasmid was transformed into DH5α cells (TransGen Biotech, Beijing, China), with positive clones initially screened by colony PCR. Verified clones were sent to Sangon Biotech (Shanghai) for Sanger sequencing. Following sequence confirmation, the recombinant plasmid was extracted and transformed into Agrobacterium tumefaciens strain GV3101 via electroporation. Positive Agrobacterium transformants were selected for subsequent plant transformation experiments.
The recombinant Agrobacterium culture harboring the target gene was centrifuged at 5000× g for 10 min at 4 °C, and the bacterial pellet was collected. The cells were resuspended in infiltration buffer (50 mL ddH2O + 500 μL MES + 200 μL MgCl2 + 75 μL As) to an optical density (OD600) of 0.7–0.8. The bacterial suspension was pressure-infiltrated into the abaxial surface of 25-day-old Nicotiana benthamiana leaves using a 2 mL sterile syringe, with empty vector (pCAMBIA2300) serving as negative control. Infiltrated plants were maintained in darkness at 25 °C for 48 h followed by 24 h under normal illumination (16 h light/8 h dark).
The injected bacterial solution was cut into the leaves with a scalpel to obtain the bacterial solution, and the samples were collected by the laboratory. A 0.2 mm × 0.2 mm microscopic sample was cut with a scalpel from the leaf injected with the bacterial solution and placed under an AX10 fluorescence inverted microscope (ZEISS, Germany) for observation and photographing.
4.8. Data Statistics and Analysis
The experimental data were organized and categorized using Microsoft Excel 2010, and graphical representations were generated using Origin 2021. Statistical analyses, including analysis of variance (ANOVA) and multiple comparisons, were performed using SPSS Statistics 22.0. Duncan’s multiple range test was employed for post hoc analysis of significant differences at a significance level of p < 0.05.