Salt Stress Leads to Morphological and Transcriptional Changes in Roots of Pumpkins (Cucurbita spp.)
Abstract
1. Introduction
2. Results
2.1. Effect of NaCl on the Root Morphology of Pumpkins at the Germination Stage (Experiment 1)
2.2. Effect of NaCl on the Plant and Root Weight of PUMPKIns at the Seedling Stage (Experiment 2)
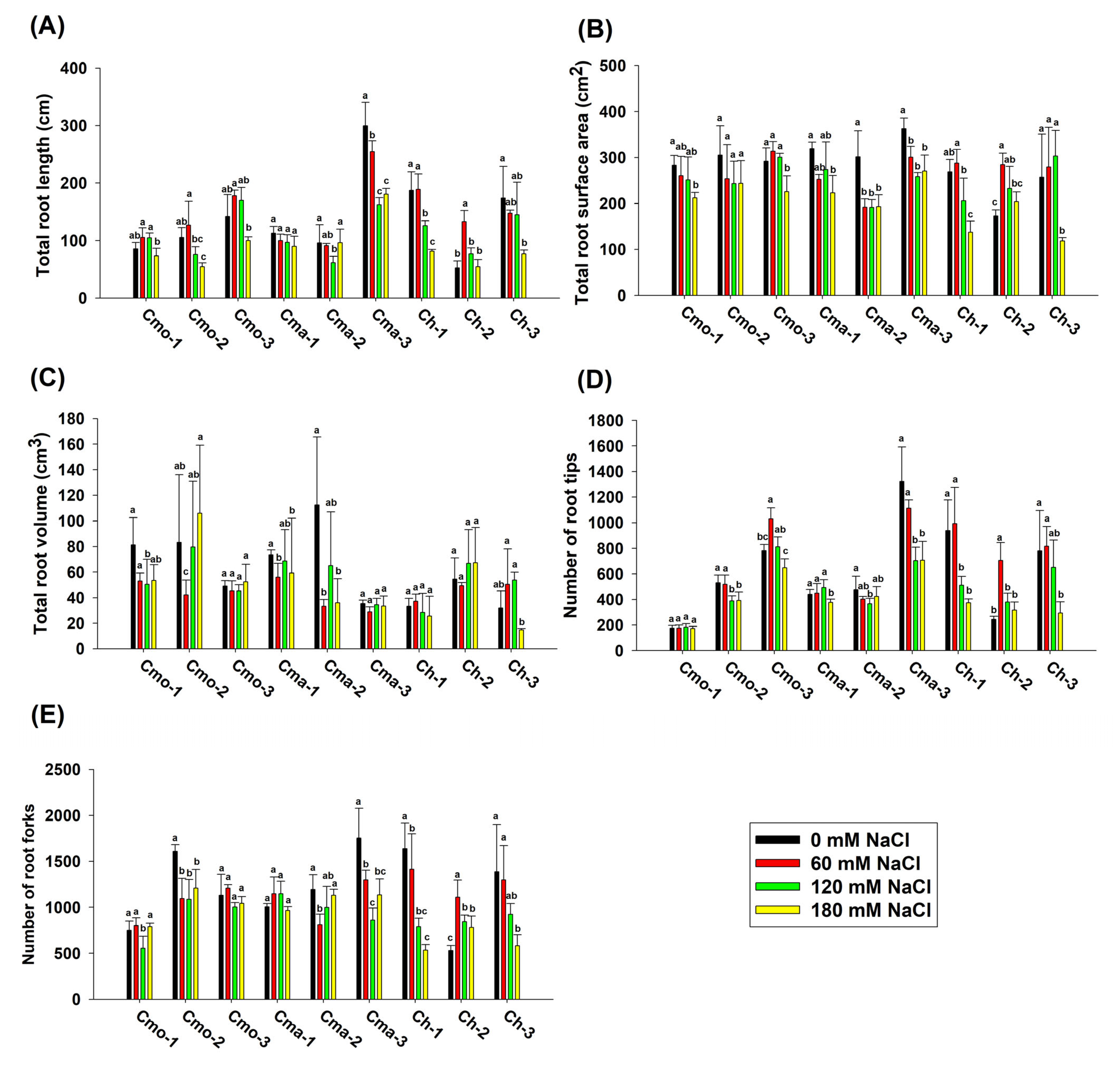
2.3. Effect of NaCl on the Root Morphology of Pumpkins at the Seedling Stage (Experiment 2)
2.4. Transcriptomic Differential Profiles of Roots Induced by Salt Stress (Experiment 3)
2.4.1. Identifying DEGs Involved in Salt Stress
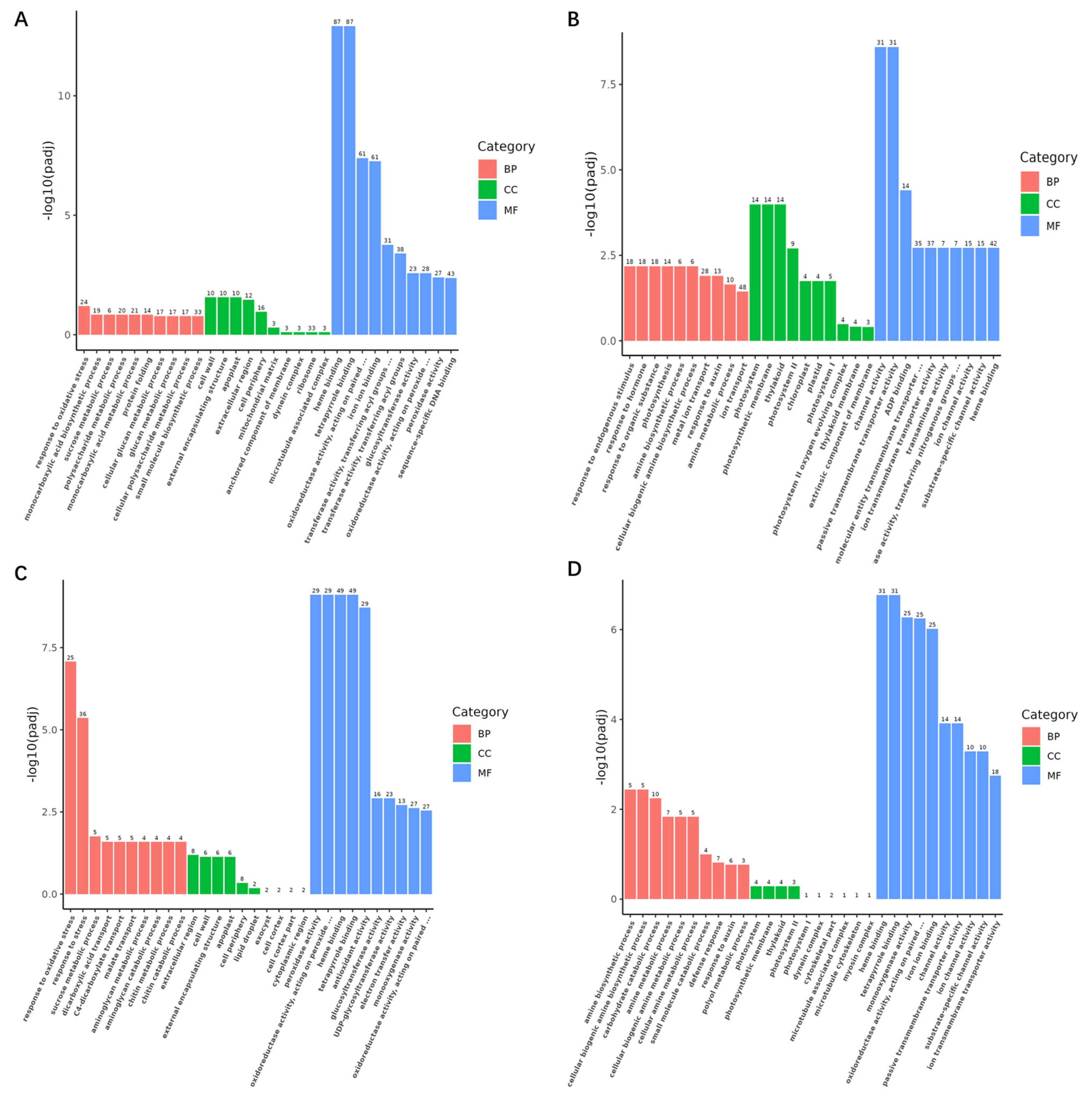
2.4.2. GO Enrichment Analysis
2.4.3. KEGG Enrichment Analysis
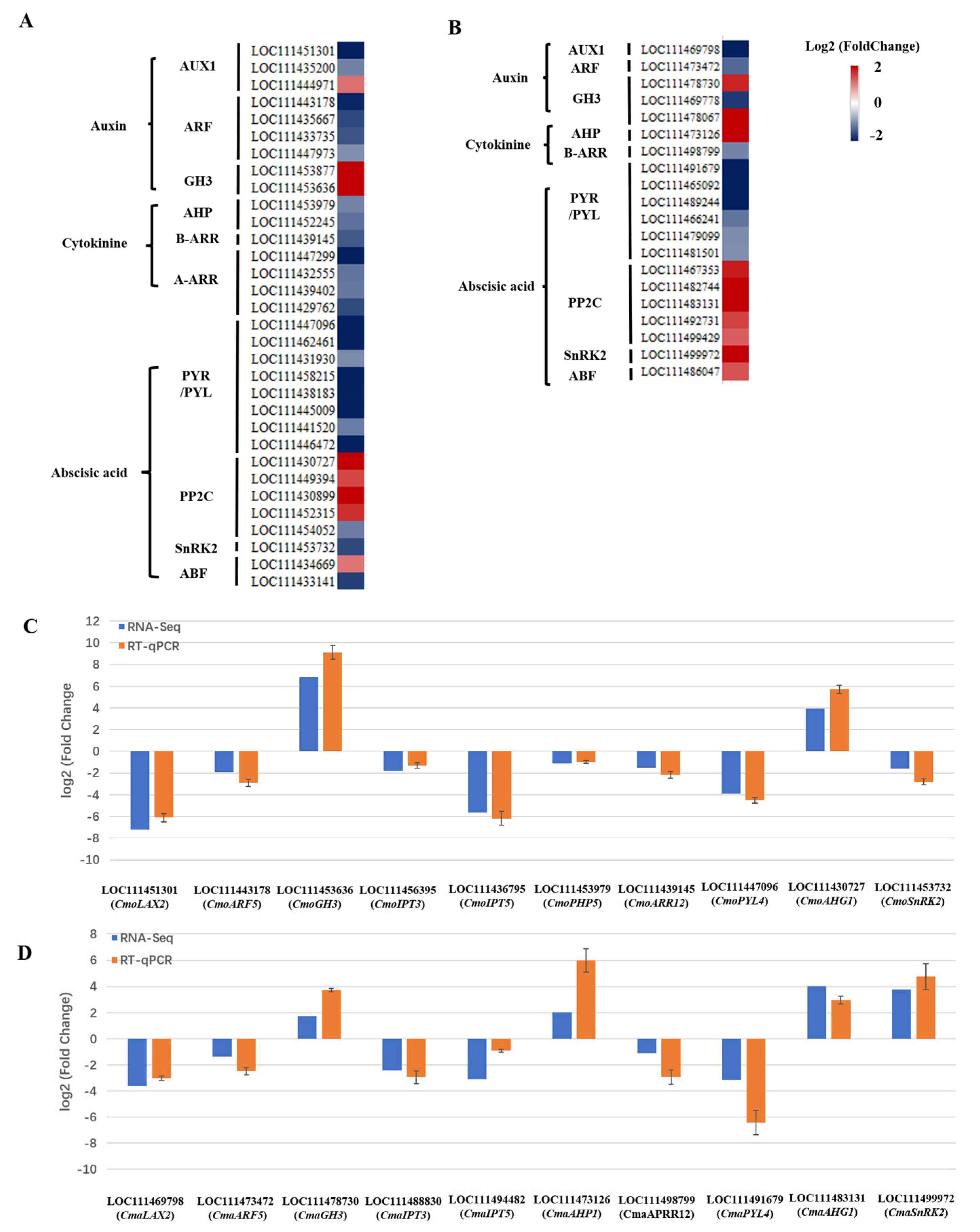
2.4.4. Identification of DEGs Involved Plant Hormone Signaling Transduction
3. Discussion
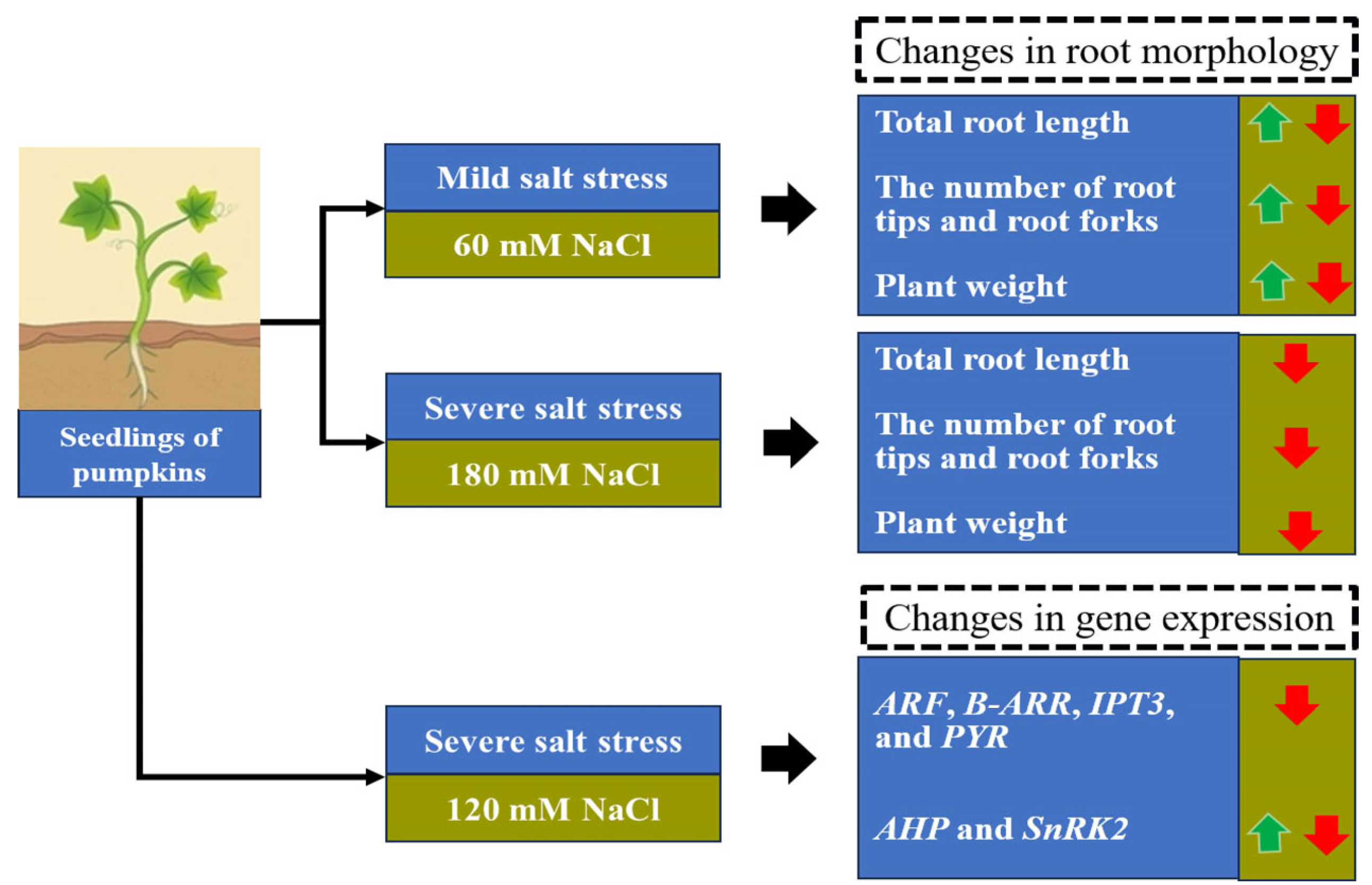
3.1. Changes in the Root Architecture Under Mild and Severe Salt Stress Conditions
3.1.1. Total Root Length
3.1.2. Lateral Root Initiation
3.1.3. Salt-Tolerant Pumpkin
3.2. Changes in Gene Expression in the Roots of C. Moschata and C. Maxima
3.2.1. The Number and Function of DEGs
3.2.2. Auxin and Cytokinin Signaling Pathway
3.2.3. Abscisic Acid Signaling Pathway
3.2.4. Sucrose Non-Fermenting-1-Related Protein Kinase 2 (SnRK2)
4. Materials and Methods
4.1. Plant Materials
4.2. Experiment 1 (Effect of NaCl on Root Morphology of Pumpkins at the Germination Stage)
4.3. Experiment 2 (Effect of NaCl on Root Morphology of Pumpkins at Seedling Stage)
4.4. Measurement of Root Morphological Parameter
4.5. Experiment3 (RNA-Seq Analysis of Roots of Cmo-3 and Cma-2 Under NaCl Treatment)
4.5.1. Experiment Design
4.5.2. RNA Extraction, Library Construction and Sequencing
4.5.3. RNA-Seq Analysis
4.5.4. RT-qPCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A-ARR | type-A response regulator |
| ABA | Abscisic acid |
| ABF | abscisic acid-responsive transcription factors |
| AHP | histidine-containing phosphotransmitter |
| AUX1 | auxin resistant 1 |
| B-ARR | type-B response regulator |
| C. maxima | Cucurbita maxima |
| C. moschata | Cucurbita moschata |
| CK | Cytokinin |
| DEGs | differential expression genes |
| GO | Gene Ontology |
| GH3 | gretchen hagen 3 |
| IPT3 | isopentenyl transferase 3 |
| IPT5 | isopentenyl transferase 5 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PYR | pyrabactin resistance |
| PYL | PYR-LIKE |
| PP2C | 2C type protein phosphatases |
| RNA-seq | RNA sequencing |
| SnRK2 | sucrose non-fermenting-1-related protein kinase 2 |
References
- Waheed, A.; Zhuo, L.; Wang, M.; Hailiang, X.; Tong, Z.; Wang, C.; Aili, A. Integrative mechanisms of plant salt tolerance: Biological pathways, phytohormonal regulation, and technological innovations. Plant Stress 2024, 14, 100652. [Google Scholar] [CrossRef]
- Yun, P.; Kaya, C.; Shabala, S. Hormonal and epigenetic regulation of root responses to salinity stress. Crop J. 2024, 12, 1309–1320. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Katagiri, S.; Umezawa, T. Growth Promotion or Osmotic Stress Response: How SNF1-Related Protein Kinase 2 (SnRK2) Kinases Are Activated and Manage Intracellular Signaling in Plants. Plants 2021, 10, 1443. [Google Scholar] [CrossRef]
- Li, L.; Chen, G.; Sun, Q.; Wang, Q.; Wang, S.; Wang, H.; Ni, Z.; Jiang, C.; Li, L.; Li, T. Evaluation of Salt Resistance of Six Apple Rootstocks. Int. J. Mol. Sci. 2024, 25, 12568. [Google Scholar] [CrossRef]
- Li, P.; Yang, X.; Wang, H.; Pan, T.; Wang, Y.; Xu, Y.; Xu, C.; Yang, Z. Genetic control of root plasticity in response to salt stress in maize. Theor. Appl. Genet. 2021, 134, 1475–1492. [Google Scholar] [CrossRef]
- Rahnama, A.; Munns, R.; Poustini, K.; Watt, M. A screening method to identify genetic variation in root growth response to a salinity gradient. J. Exp. Bot. 2011, 62, 69–77. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Dissanayake, B.M.; Staudinger, C.; Munns, R.; Taylor, N.L.; Millar, A.H. Distinct salinity-induced changes in wheat metabolic machinery in different root tissue types. J. Proteom. 2022, 256, 104502. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Qiu, Z.; Zeng, B.; Zhang, Y.; Wang, X.; Chen, J.; Zhong, C.; Deng, R.; Fan, C. Transcriptome and structure analysis in root of Casuarina equisetifolia under NaCl treatment. PeerJ 2021, 9, e12133. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; de Zeeuw, T.; Duijts, K.; Kawa, D.; Lamers, J.; Munzert, K.S.; Li, H.; Zou, Y.; Meyer, A.J.; et al. Root branching under high salinity requires auxin-independent modulation of LATERAL ORGAN BOUNDARY DOMAIN 16 function. Plant Cell 2023, 36, 899–918. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carrasco, K.; Antognoni, F.; Coulibaly, A.K.; Lizardi, S.; Covarrubias, A.; Martínez, E.A.; Molina-Montenegro, M.A.; Biondi, S.; Zurita-Silva, A. Variation in salinity tolerance of four lowland genotypes of quinoa (Chenopodium quinoa Willd.) as assessed by growth, physiological traits, and sodium transporter gene expression. Plant Physiol. Biochem. 2011, 49, 1333–1341. [Google Scholar] [CrossRef]
- Aggarwal, G.; Edhigalla, P.; Walia, P.; Jindal, S.; Sandal, S.S. A method for screening salt stress tolerance in Indian mustard (Brassica juncea) (L.) Czern & Coss at seedling stage. Sci. Rep. 2024, 14, 12705. [Google Scholar] [CrossRef]
- Ramadan, E.; Freeg, H.A.; Shalaby, N.; Rizk, M.S.; Ma, J.; Du, W.; Ibrahim, O.M.; Alwutayd, K.M.; AbdElgawad, H.; Jo, I.-H.; et al. Response of nine triticale genotypes to different salt concentrations at the germination and early seedling stages. PeerJ 2023, 11, e16256. [Google Scholar] [CrossRef]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef]
- Nyabera, L.A.; Nzuki, I.W.; Runo, S.M.; Amwayi, P.W. Assessment of genetic diversity of pumpkins (Cucurbita spp.) from western Kenya using SSR molecular markers. Mol. Biol. Rep. 2021, 48, 2253–2260. [Google Scholar] [CrossRef]
- Hernandez, C.O.; Labate, J.; Reitsma, K.; Fabrizio, J.; Bao, K.; Fei, Z.; Grumet, R.; Mazourek, M. Characterization of the USDA Cucurbita pepo, C. moschata, and C. maxima germplasm collections. Front. Plant Sci. 2023, 14, 1130814. [Google Scholar] [CrossRef]
- Lee, J.-M.; Kubota, C.; Tsao, S.J.; Bie, Z.; Echevarria, P.H.; Morra, L.; Oda, M. Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci. Hortic. 2010, 127, 93–105. [Google Scholar] [CrossRef]
- Alonso, S.; Gautam, K.; Iglesias-Moya, J.; Martínez, C.; Jamilena, M. Crosstalk between Ethylene, Jasmonate and ABA in Response to Salt Stress during Germination and Early Plant Growth in Cucurbita pepo. Int. J. Mol. Sci. 2024, 25, 8728. [Google Scholar] [CrossRef]
- Nejad-Alimoradi, F.; Nasibi, F.; Kalantari, K.M. 24-epibrassinolide pre-treatment alleviates the salt-induced deleterious effects in medicinal pumpkin (Cucurbita pepo) by enhancement of GABA content and enzymatic antioxidants. S. Afr. J. Bot. 2019, 124, 111–117. [Google Scholar] [CrossRef]
- Peng, Y.; Cao, H.; Cui, L.; Wang, Y.; Wei, L.; Geng, S.; Yang, L.; Huang, Y.; Bie, Z. CmoNAC1 in pumpkin rootstocks improves salt tolerance of grafted cucumbers by binding to the promoters of CmoRBOHD1, CmoNCED6, CmoAKT1;2 and CmoHKT1;1 to regulate H2O2, ABA signaling and K+/Na+ homeostasis. Hortic. Res. 2023, 10, uhad157. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cao, H.; Cheng, J.; He, X.; Sohail, H.; Niu, M.; Huang, Y.; Bie, Z. Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber. Int. J. Mol. Sci. 2018, 19, 2648. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liu, L.; Chen, Z.; Huang, Y.; Yang, L.; Wang, P.; Xue, S.; Bie, Z. CmCNIH1 improves salt tolerance by influencing the trafficking of CmHKT1;1 in pumpkin. Plant J. 2023, 114, 1353–1368. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, R.-J.; Han, T.-T.; Cai, W.; Fu, Z.-W.; Lu, Y.-T. Salt Stress Reduces Root Meristem Size by Nitric Oxide-Mediated Modulation of Auxin Accumulation and Signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and Environmental Regulation of Root Development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010, 2, a001537. [Google Scholar] [CrossRef]
- Lu, C.; Chen, M.-X.; Liu, R.; Zhang, L.; Hou, X.; Liu, S.; Ding, X.; Jiang, Y.; Xu, J.; Zhang, J.; et al. Abscisic Acid Regulates Auxin Distribution to Mediate Maize Lateral Root Development Under Salt Stress. Front. Plant Sci. 2019, 10, 716. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T.; et al. Analysis of Cytokinin Mutants and Regulation of Cytokinin Metabolic Genes Reveals Important Regulatory Roles of Cytokinins in Drought, Salt and Abscisic Acid Responses, and Abscisic Acid Biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Nishiyama, R.; Tran, C.D.; Kusano, M.; Nakabayashi, R.; Okazaki, Y.; Matsuda, F.; Chávez Montes, R.A.; Mostofa, M.G.; Li, W.; et al. Defective cytokinin signaling reprograms lipid and flavonoid gene-to-metabolite networks to mitigate high salinity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2105021118. [Google Scholar] [CrossRef]
- Mason, M.G.; Jha, D.; Salt, D.E.; Tester, M.; Hill, K.; Kieber, J.J.; Eric Schaller, G. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 2010, 64, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Koevoets, I.T.; Mol, S.; Hoefsloot, H.; Feron, R.; Tester, M.A.; Keurentjes, J.J.B.; Korte, A.; Haring, M.A.; de Boer, G.-J.; et al. Genetic Components of Root Architecture Remodeling in Response to Salt Stress. Plant Cell 2017, 29, 3198–3213. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, Y.; Testerink, C. Root dynamic growth strategies in response to salinity. Plant Cell Environ. 2022, 45, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Hoefsloot, H.C.J.; Mol, S.; Feron, R.; de Boer, G.-J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis Root Architecture Dynamics with root-fit Reveals Diversity in Responses to Salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef]
- Tang, K.; An, C.; Li, L.; Sun, T.; Song, J.; Zhao, J. Effects of drought and salt stress on the root phenotype of wheat seedlings and underlying gene expression analysis. Front. Plant Sci. 2024, 15, 1475500. [Google Scholar] [CrossRef]
- Teerarak, M.; Bhinija, K.; Thitavasanta, S.; Laosinwattana, C. The impact of sodium chloride on root growth, cell division, and interphase silver-stained nucleolar organizer regions (AgNORs) in root tip cells of Allium cepa L. Sci. Hortic. 2009, 121, 228–232. [Google Scholar] [CrossRef]
- Duan, L.; Sebastian, J.; Dinneny, J.R. Salt-Stress Regulation of Root System Growth and Architecture in Arabidopsis Seedlings. In Plant Cell Expansion: Methods and Protocols; Estevez, J.M., Ed.; Springer: New York, NY, USA, 2015; pp. 105–122. [Google Scholar]
- Wang, F.; Iki, Y.; Tanoi, K.; Naito, K. Phenotypic responses in the root of salt-tolerant accessions of Vigna marina and Vigna luteola under salt stress. Genet. Resour. Crop Evol. 2024, 71, 2631–2640. [Google Scholar] [CrossRef]
- Li, F.; Liu, B.; Zhang, H.; Zhang, J.; Cai, J.; Cui, J. Integrative multi-omics analysis of chilling stress in pumpkin (Cucurbita moschata). BMC Genom. 2024, 25, 1042. [Google Scholar] [CrossRef]
- Li, F.; Lu, X.; Duan, P.; Liang, Y.; Cui, J. Integrating transcriptome and metabolome analyses of the response to cold stress in pumpkin (Cucurbita maxima). PLoS ONE 2021, 16, e0249108. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, C.; Zou, B.; Wang, C.; Xu, W.; Cui, C.; Qu, S. Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression. Int. J. Mol. Sci. 2019, 20, 3185. [Google Scholar] [CrossRef]
- Xue, S.; Huang, H.; Xu, Y.; Liu, L.; Meng, Q.; Zhu, J.; Zhou, M.; Du, H.; Yao, C.; Jin, Q.; et al. Transcriptomic analysis reveals the molecular basis of photoperiod-regulated sex differentiation in tropical pumpkins (Cucurbita moschata Duch.). BMC Plant Biol. 2024, 24, 90. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L.; Chen, B.H.; Chen, X.J.; Guo, Y.Y.; Yang, H.L.; Li, X.Z.; Wang, G.Y. Transcriptome profiling of pumpkin (Cucurbita moschata Duch.) leaves infected with powdery mildew. PLoS ONE 2018, 13, e0190175. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Peng, L.; Zhou, X. SnRK2s: Kinases or Substrates? Plants 2025, 14, 1171. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mo, X.; Shou, H.; Wu, P. Cytokinin-Mediated Cell Cycling Arrest of Pericycle Founder Cells in Lateral Root Initiation of Arabidopsis. Plant Cell Physiol. 2006, 47, 1112–1123. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Mateo-Bonmatí, E.; Šimura, J.; Pěnčík, A.; Novák, O.; Staswick, P.; Ljung, K. Inactivation of the entire Arabidopsis group II GH3s confers tolerance to salinity and water deficit. New Phytol. 2022, 235, 263–275. [Google Scholar] [CrossRef]
- Dang, T.V.T.; Cho, H.S.; Lee, S.; Hwang, I. Salt stress-accelerated proteasomal degradation of LBD11 suppresses ROS-mediated meristem development and root growth in Arabidopsis. Plant Commun. 2025, 6, 101241. [Google Scholar] [CrossRef]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, P.M.Y.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA Signaling Promotes Lateral Root Quiescence during Salt Stress in Arabidopsis Seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.-H.; Hu, S.; Li, X.-B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef]
- Yang, G.; Yu, Z.; Gao, L.; Zheng, C. SnRK2s at the Crossroads of Growth and Stress Responses. Trends Plant Sci. 2019, 24, 672–676. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Li, C.; Ma, J.; Liu, J.; Zhu, X.; Li, J.; He, F.; Yang, C. Genome-Wide Identification and Expression Profile Analysis of the SnRK2 Gene Family in Nicotiana tabacum. Biochem. Genet. 2022, 60, 1511–1526. [Google Scholar] [CrossRef]
- Shao, Y.; Qin, Y.; Zou, Y.; Ma, F. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 2014, 552, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.; Luo, S.; Zhang, Z.; Liu, Z.; Qiao, Y.; Gao, X.; Yu, J.; Zhang, G. Identification and expression profile analysis of the SnRK2 gene family in cucumber. PeerJ 2022, 10, e13994. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, W.; Sun, J.; Liang, X.; Yang, X.; Wei, S.; Wang, X.; Zhou, Y.; Xiao, Q.; Yang, G.; et al. Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 2015, 237, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Chen, X.-j.; Wang, J.-h.; Zheng, J. Overexpression of a maize SNF-related protein kinase gene, ZmSnRK2.11, reduces salt and drought tolerance in Arabidopsis. J. Integr. Agric. 2015, 14, 1229–1241. [Google Scholar] [CrossRef]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef]
- Zhang, Y.; Wan, S.; Liu, X.; He, J.; Cheng, L.; Duan, M.; Liu, H.; Wang, W.; Yu, Y. Overexpression of CsSnRK2.5 increases tolerance to drought stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 150, 162–170. [Google Scholar] [CrossRef]
- Nagy, I.; Veeckman, E.; Liu, C.; Bel, M.V.; Vandepoele, K.; Jensen, C.S.; Ruttink, T.; Asp, T. Chromosome-scale assembly and annotation of the perennial ryegrass genome. BMC Genom. 2022, 23, 505. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

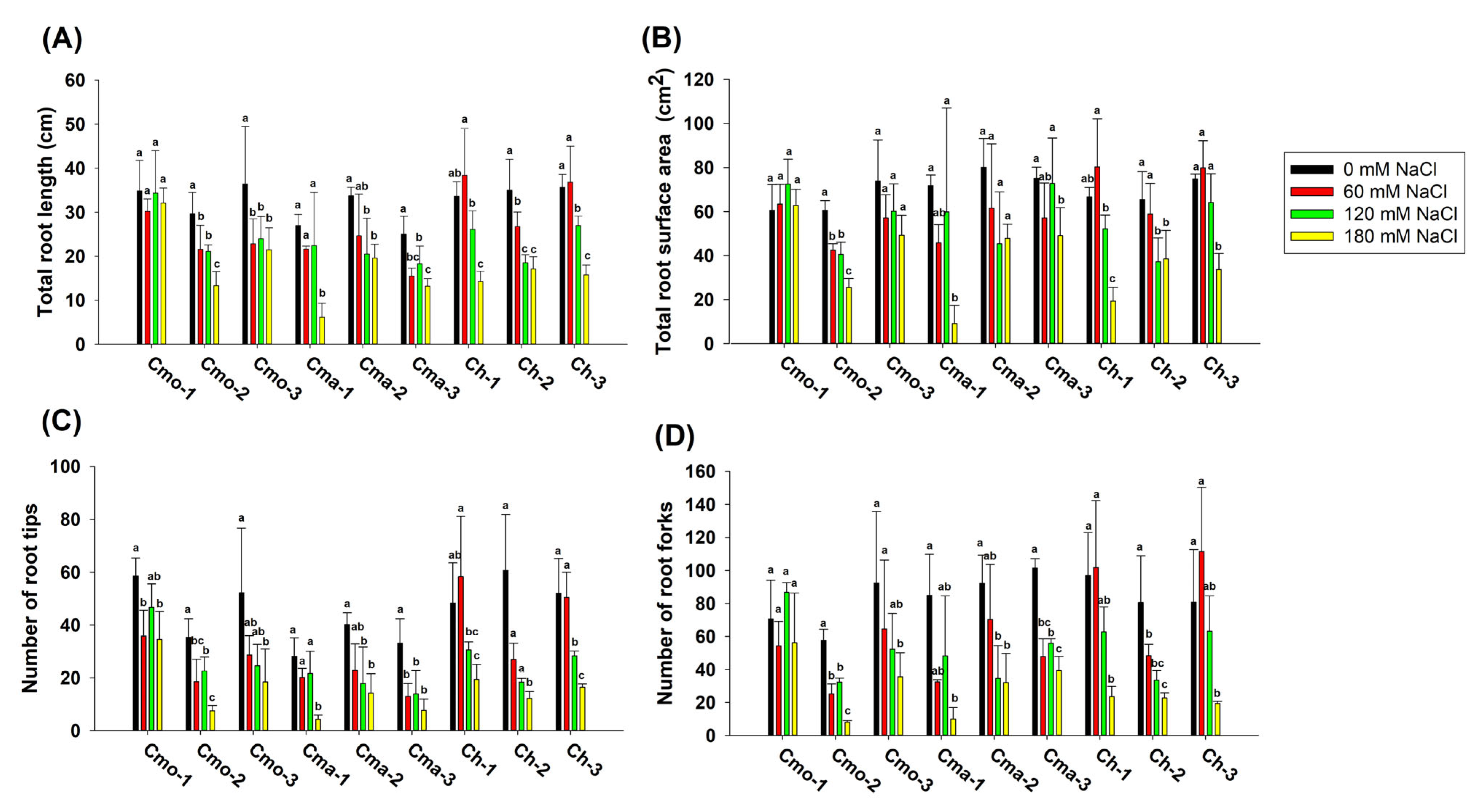
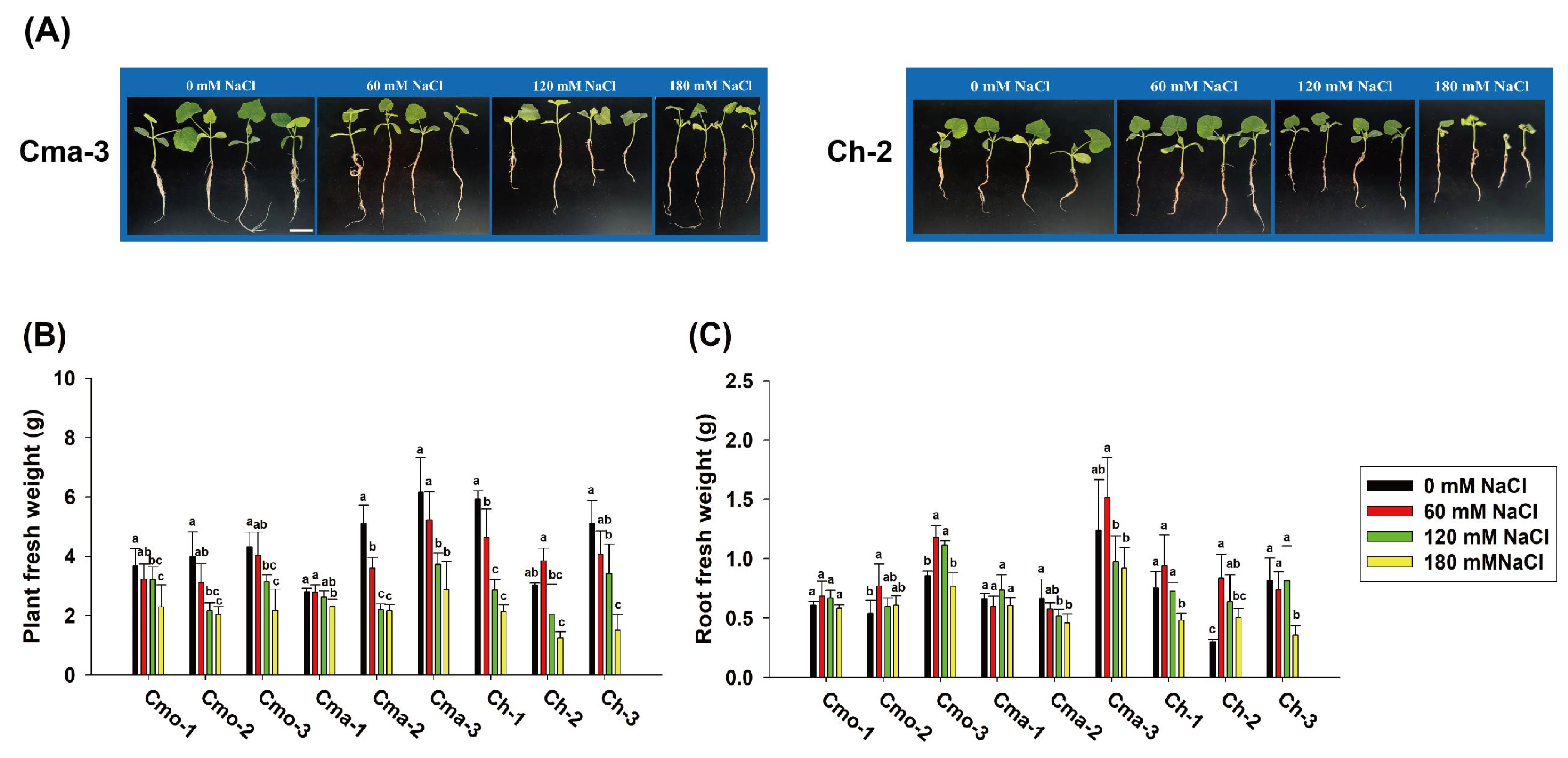


| Experimental Factors | Total Root Length | Total Root Surface Area | Number of Root Tips | Number of Root Forks | ||||
|---|---|---|---|---|---|---|---|---|
| F Value | p | F Value | p | F Value | p | F Value | p | |
| Cultivar | 9.07 | <0.0001 | 4.16 | 0.0004 | 9.54 | <0.0001 | 4.48 | 0.0002 |
| NaCl | 37.03 | <0.0001 | 23.79 | <0.0001 | 39.43 | <0.0001 | 25.94 | <0.0001 |
| Cultivar × NaCl | 1.91 | 0.0191 | 1.89 | 0.0205 | 1.48 | 0.103 | 1.44 | 0.121 |
| Experimental Factors | Total Root Length | Total Root Surface Area | Total Root Volume | Number of Root Tips | Number of Root Forks | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| F Value | p | F Value | p | F Value | p | F Value | p | F Value | p | |
| Cultivar | 51.69 | <0.0001 | 5.12 | <0.0001 | 2.64 | 0.0131 | 48.82 | <0.0001 | 10.58 | <0.0001 |
| NaCl | 38.38 | <0.0001 | 21.64 | <0.0001 | 0.45 | 0.7202 | 30.14 | <0.0001 | 19.19 | <0.0001 |
| Cultivar × NaCl | 5.08 | <0.0001 | 3.11 | 0.0001 | 1.44 | 0.1168 | 4.82 | <0.0001 | 5.37 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Ding, D.; Sun, Y.; Ma, R.; Yang, X.; Liu, J.; Zhang, G. Salt Stress Leads to Morphological and Transcriptional Changes in Roots of Pumpkins (Cucurbita spp.). Plants 2025, 14, 1674. https://doi.org/10.3390/plants14111674
Liu H, Ding D, Sun Y, Ma R, Yang X, Liu J, Zhang G. Salt Stress Leads to Morphological and Transcriptional Changes in Roots of Pumpkins (Cucurbita spp.). Plants. 2025; 14(11):1674. https://doi.org/10.3390/plants14111674
Chicago/Turabian StyleLiu, Hongjiu, Ding Ding, Yeshuo Sun, Ruiping Ma, Xiaoqing Yang, Jie Liu, and Guoxin Zhang. 2025. "Salt Stress Leads to Morphological and Transcriptional Changes in Roots of Pumpkins (Cucurbita spp.)" Plants 14, no. 11: 1674. https://doi.org/10.3390/plants14111674
APA StyleLiu, H., Ding, D., Sun, Y., Ma, R., Yang, X., Liu, J., & Zhang, G. (2025). Salt Stress Leads to Morphological and Transcriptional Changes in Roots of Pumpkins (Cucurbita spp.). Plants, 14(11), 1674. https://doi.org/10.3390/plants14111674





