Effective Agrobacterium-Mediated Transformation System for Eureka Lemon Using Whole Cotyledonary Node
Abstract
1. Introduction
2. Results
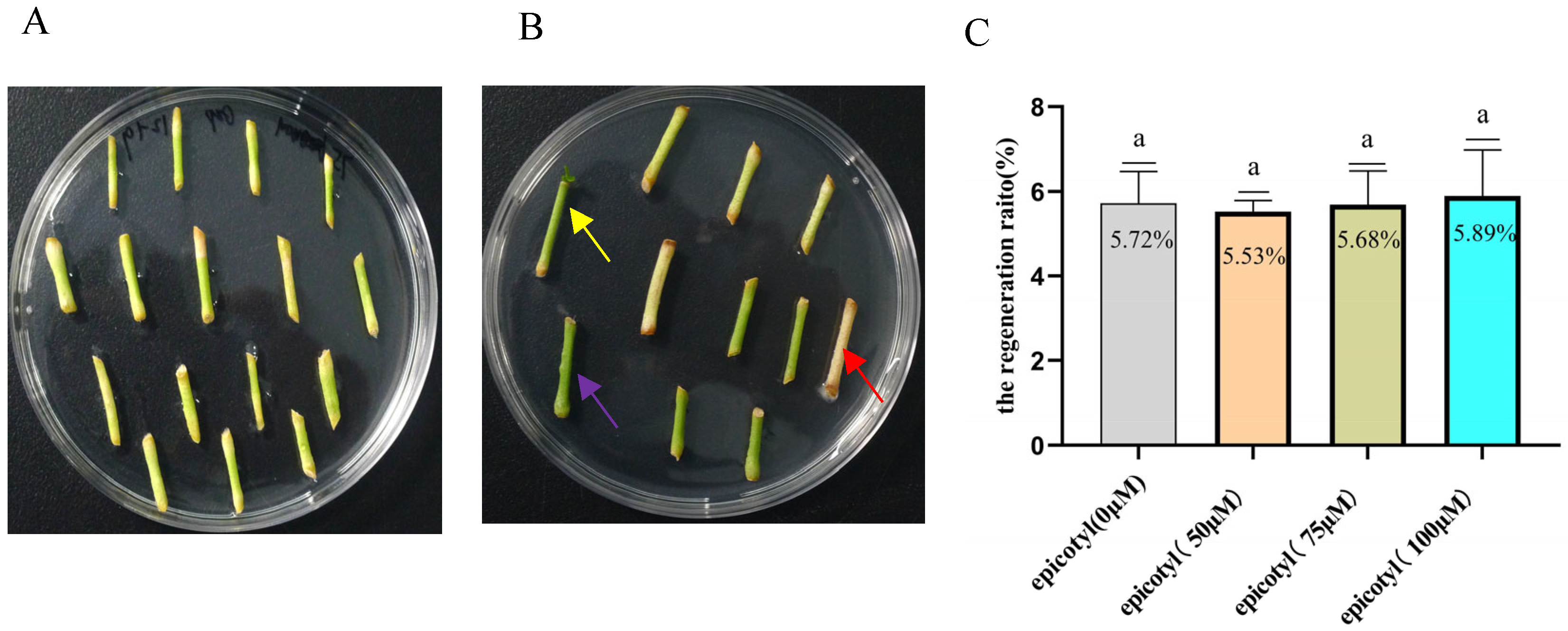
2.1. Epicotyls of Eureka Lemon Are Highly Recalcitrant to Agrobacterium-Mediated Transformation
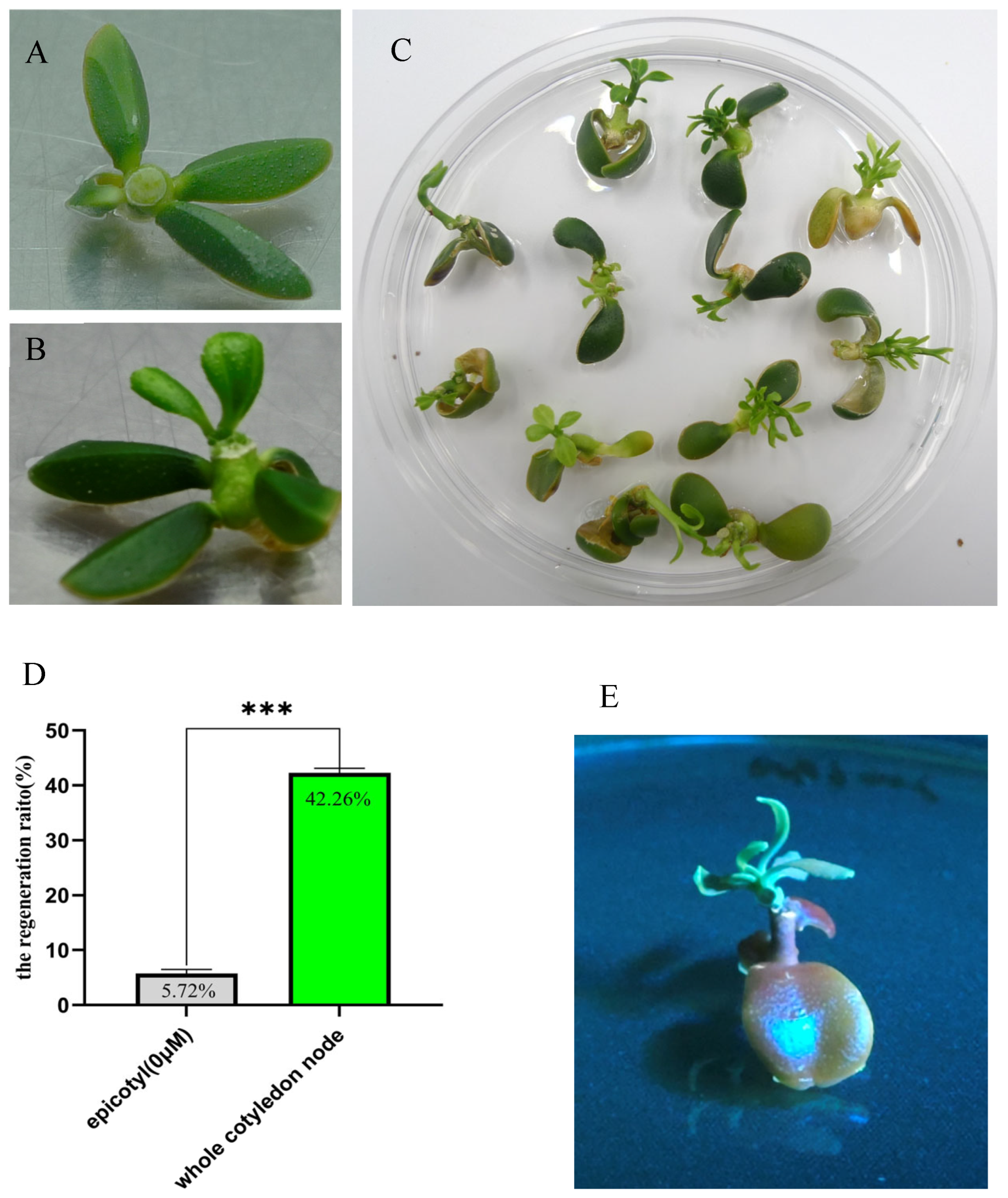
2.2. Whole Cotyledonary Node Explants from Eureka Lemon Exhibited Higher Transformation Efficiency than Epicotyl Explant
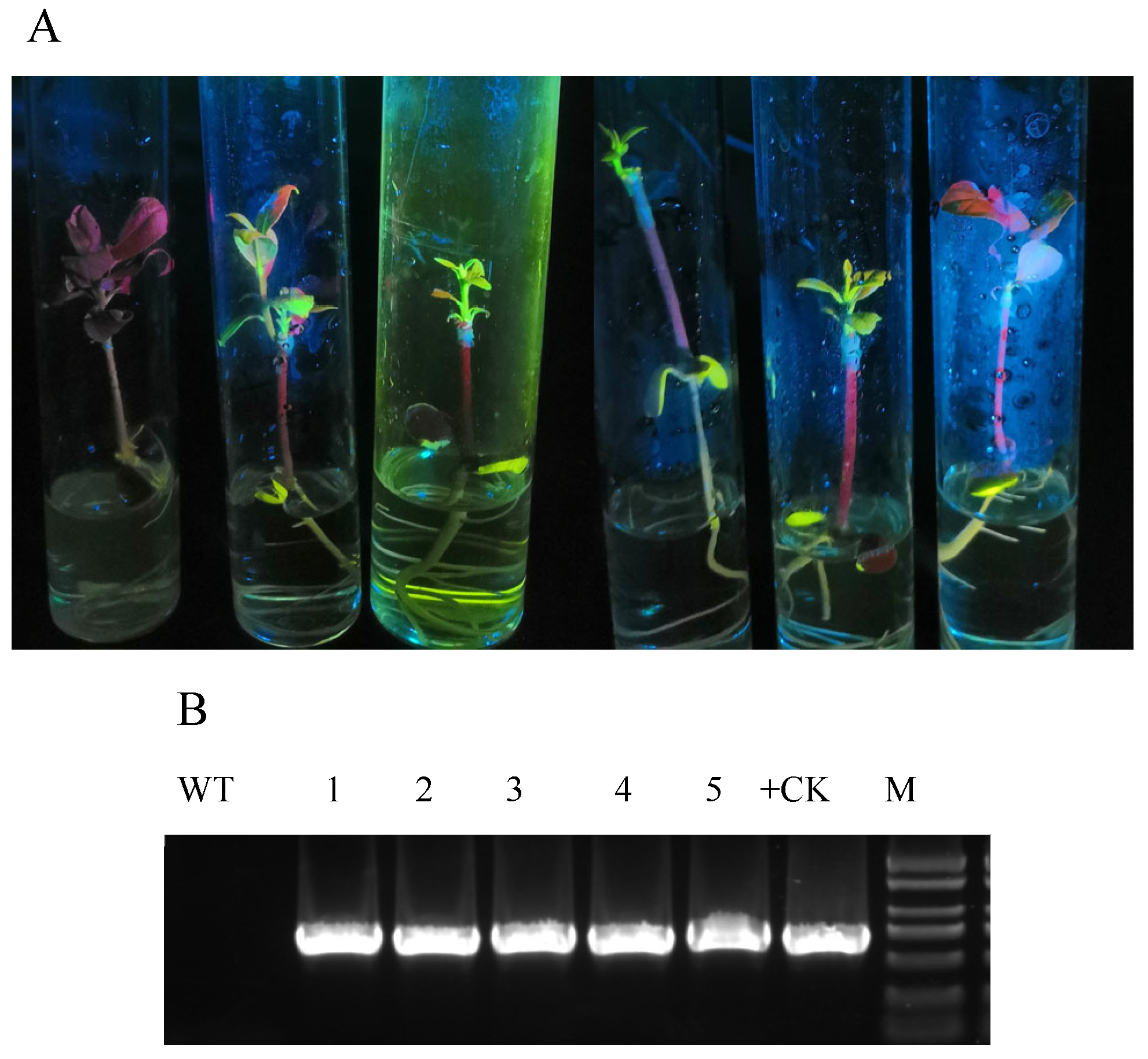
2.3. PCR Identification of Transgenic Plants
3. Discussion
4. Materials and Methods
4.1. Vector and Medium
4.2. Plant Material and Explant Preparations
4.3. Agrobacterium Preparation
4.4. Co-Cultivation and Shoot Regeneration
4.5. Micrografting and Identification of Transgenic Plants by Polymerase Chain Reaction (PCR)
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT Database Results. 2023. Available online: http://www.fao.org/faostat/en/#data (accessed on 2 June 2023).
- Coletta-Filho, H.D.; Castillo, A.I.; Laranjeira, F.F.; Andrade, E.D.D.; Silva, N.T.; Souza, A.A.D.; Bossi, M.E.; Almeida, R.P.P.; Lopes, J.R.S. Citrus variegated chlorosis: An overview of 30 years of research and disease management. Trop. Plant Pathol. 2020, 45, 175–191. [Google Scholar] [CrossRef]
- Shahbaz, E.; Ali, M.; Shafiq, M.; Atiq, M.; Hussain, M.; Balal, R.M.; Sarkhosh, A.; Alferez, F.; Sadiq, S.; Shahid, M.A. Citrus canker pathogen, its mechanism of infection, eradication, and impacts. Plants 2022, 12, 123. [Google Scholar] [CrossRef]
- Jaouad, M.; Moinina, A.; Ezrari, S.; Lahlali, R. Key pests and diseases of citrus trees with emphasis on root rot diseases: An overview. Mor. J. Agric. Sci. 2020, 1, 149–160. [Google Scholar]
- Li, X.; Ruan, H.Q.; Zhou, C.Q.; Meng, X.C.; Chen, W.L. Controlling citrus huanglongbing: Green sustainable development route is the future. Front. Plant Sci. 2021, 12, 760481. [Google Scholar] [CrossRef]
- Liu, C.H.; Liu, H.; Hurst, J.; Timko, M.P.; Zhou, C.Y. Recent advances on citrus yellow vein clearing virus in citrus. Hortic. Plant J. 2020, 6, 216–222. [Google Scholar] [CrossRef]
- Sharma, S. A review on citrus tristeza virus (CTV) and its management approaches. Turk. J. Agric. For. 2023, 11, 803–810. [Google Scholar] [CrossRef]
- Wang, L.J.; Chen, S.C.; Peng, A.H.; Xie, Z.; He, Y.R.; Zou, X.P. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wancheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 2019, 13, 501–510. [Google Scholar] [CrossRef]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a modified plant thionin enhances disease resistance to citrus canker and huanglongbing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef]
- Soler, N.; Plomer, M.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R. Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol. J. 2012, 10, 597–608. [Google Scholar] [CrossRef]
- Poles, L.; Licciardello, G.; Distefano, G.; Nicolosi, E.; Gentile, A.; La Malfa, S. Recent advances of in vitro culture for the application of new breeding techniques in citrus. Plants 2020, 9, 938. [Google Scholar] [CrossRef]
- Conti, G.; Xoconostle-Cázares, B.; Marcelino-Pérez, G.; Hopp, H.E.; Reyes, C.A. Citrus genetic transformation: An overview of the current strategies and insights on the new emerging technologies. Front. Plant Sci. 2021, 12, 768197. [Google Scholar] [CrossRef]
- Omar, A.A.; Song, W.Y.; Grosser, J.W. Introduction of Xa21, a Xanthomonas-resistance gene from rice, into ‘Hamlin’ sweet orange [Citrus sinensis (L.) Osbeck] using protoplast-GFP co-transformation or single plasmid transformation. J. Hortic. Sci. Biotech. 2007, 82, 914–923. [Google Scholar] [CrossRef]
- Dutt, M.; Grosser, J.W. An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of citrus. Plant Cell Rep. 2010, 29, 1251–1260. [Google Scholar] [CrossRef]
- de Oliveira, M.L.P.; Moore, G.; Thomson, J.G.; Stover, E. Agrobacterium-mediated transformation of Mexican lime (Citrus aurantifolia Swingle) using optimized systems for epicotyls and cotyledons. Adv. Biosci. Biotechnol. 2015, 6, 657–668. [Google Scholar] [CrossRef][Green Version]
- Khan, E.U.; Fu, X.Z.; Liu, J.H. Agrobacterium-mediated genetic transformation and regeneration of transgenic plants using leaf segments as explants in Valencia sweet orange. Plant Cell Tiss Org. 2012, 109, 383–390. [Google Scholar] [CrossRef]
- Singh, S.; Rajam, M.V. Highly efficient and rapid plant regeneration in citrus sinensis. J. Plant Biochem. Biotechnol. 2010, 19, 195–202. [Google Scholar] [CrossRef]
- Gentile, A.; Deng, Z.; La Malfa, S.; Distefano, G.; Domina, F.; Vitale, A.; Polizzi, G.; Lorito, M.; Tribulato, E. Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 2007, 126, 146–151. [Google Scholar] [CrossRef]
- Mahmoud, L.M.; Killiny, N.; Dutt, M. Melatonin supplementation enhances browning suppression and improves transformation efficiency and regeneration of transgenic rough lemon plants (Citrus × jambhiri). PLoS ONE 2024, 19, e0294318. [Google Scholar] [CrossRef]
- Ge, X.Y.; Xu, J.T.; Yang, Z.E.; Yang, X.F.; Wang, Y.; Chen, Y.L.; Wang, P.; Li, F.G. Efficient genotype-independent cotton genetic transformation and genome editing. J. Integr. Plant Biol. 2023, 65, 907–917. [Google Scholar] [CrossRef]
- Liu, H.Q.; Zhao, J.Y.; Chen, F.F.; Wu, Z.M.; Tan, J.Y.; Nguyen, H.N.; Cheng, Z.H.; Weng, Y.Q. Improving Agrobacterium tumefaciens—Mediated genetic transformation for gene function studies and mutagenesis in cucumber (Cucumis sativus L.). Genes 2023, 14, 601. [Google Scholar] [CrossRef]
- Duan, S.Y.; Xin, R.J.; Guan, S.X.; Li, X.T.; Riwen Fei, R.W.; Cheng, W.; Pan, Q.; Sun, X.M. Optimization of callus induction and proliferation of Paeonia lactiflora Pall. and Agrobacterium-mediated genetic transformation. Front. Plant Sci. 2022, 13, 996690. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.H.; Wu, T.L. Rapid and efficient regeneration in soybean [Glycine max (L.) Merrill] from whole cotyledonary node explants. Acta Physiol. Plant. 2008, 30, 209–216. [Google Scholar] [CrossRef]
- Dai, H.Y.; Li, W.R.; Mao, W.J.; Zhang, L.; Han, G.F.; Zhao, K.; Liu, Y.X.; Zhang, Z.H. Development of an efficient regeneration and Agrobacterium-mediated transformation system in crab apple (Malus micromalus) using cotyledons as explants. Vitr. Cell. Dev. Biol. 2014, 50, 1–8. [Google Scholar] [CrossRef]
- Moharana, A.; Das, A.; Subudhi, E.; Naik, S.K.; Bari, D.P. High frequency shoot proliferation from cotyledonary node of Lawsonia inermis L. and validation of their molecular finger printing. J. Crop Sci. Biotechnol. 2017, 20, 405–416. [Google Scholar] [CrossRef]
- Singh, S.; Tarannum, Z.; Kokane, S.; Ghosh, D.K.; Sharma, A.K.; Chauhan, H. Efficient transformation and regeneration of transgenic plants in commercial cultivars of Citrus aurantifolia and Citrus sinensis. Transgenic Res. 2023, 32, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Tang, D.; Liu, Z.G.; Chen, J.J.; Cheng, B.P.; Kumar, R.; Yer, H.Y.; Li, Y. An improved procedure for Agrobacterium-mediated transformation of ‘Carrizo’ Citrange. Plants 2022, 11, 1457. [Google Scholar] [CrossRef]
- Ghorbel, R.; Domínguez, A.; Navarro, L.; Peña, L. High efficiency genetic transformation of sour orange (Citrus aurantium) and production of transgenic trees containing the coat protein gene of citrus tristeza virus. Tree Physiol. 2000, 20, 1183–1189. [Google Scholar] [CrossRef]
- Luth, D.; Moore, G. Transgenic grapefruit plants obtained by Agrobacterium tumefaciens-mediated transformation. Plant Cell Tiss. Organ Cult 1999, 57, 219–222. [Google Scholar] [CrossRef]
- Xu, C.J.; Ru, Z.W.; Li, L.; Zeng, B.Y.; Huang, J.M.; Huang, W.; Hu, O. The Effects of Polyphenol Oxidase and Cycloheximide on the Early Stage of Browning in Phalaenopsis Explants. Hortic. Plant J. 2015, 1, 172–180. [Google Scholar]
- Dutt, M.; Vasconcellos, M.; Grosser, J.W. Effects of antioxidants on Agrobacterium-mediated transformation and accelerated production of transgenic plants of Mexican lime (Citrus aurantifolia Swingle). Plant Cell Tiss. Org. 2011, 107, 79–89. [Google Scholar] [CrossRef]
- Zou, X.P.; Song, E.L.; Peng, A.H.; He, Y.R.; Xu, L.Z.; Lei, T.G.; Yao, L.X.; Chen, S.C. Activation of three pathogen-inducible promoters in transgenic citrus (Citrus sinensis Osbeck) after Xanthomonas axonopodis pv. citri infection and wounding. Plant Cell Tiss. Organ Cult. 2014, 117, 85–98. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Chen, Y.; Wang, J.; Wang, C.; Zhou, Y. Effective Agrobacterium-Mediated Transformation System for Eureka Lemon Using Whole Cotyledonary Node. Plants 2025, 14, 1629. https://doi.org/10.3390/plants14111629
Zhao J, Chen Y, Wang J, Wang C, Zhou Y. Effective Agrobacterium-Mediated Transformation System for Eureka Lemon Using Whole Cotyledonary Node. Plants. 2025; 14(11):1629. https://doi.org/10.3390/plants14111629
Chicago/Turabian StyleZhao, Jinfa, Yuan Chen, Jiajun Wang, Chunqing Wang, and Yan Zhou. 2025. "Effective Agrobacterium-Mediated Transformation System for Eureka Lemon Using Whole Cotyledonary Node" Plants 14, no. 11: 1629. https://doi.org/10.3390/plants14111629
APA StyleZhao, J., Chen, Y., Wang, J., Wang, C., & Zhou, Y. (2025). Effective Agrobacterium-Mediated Transformation System for Eureka Lemon Using Whole Cotyledonary Node. Plants, 14(11), 1629. https://doi.org/10.3390/plants14111629




