Identification and Abiotic Stress Expression Profiling of Malic Enzyme-Associated Genes in Maize (Zea mays L.)
Abstract
1. Introduction
2. Results
2.1. Identification of the Maize ME Gene Family
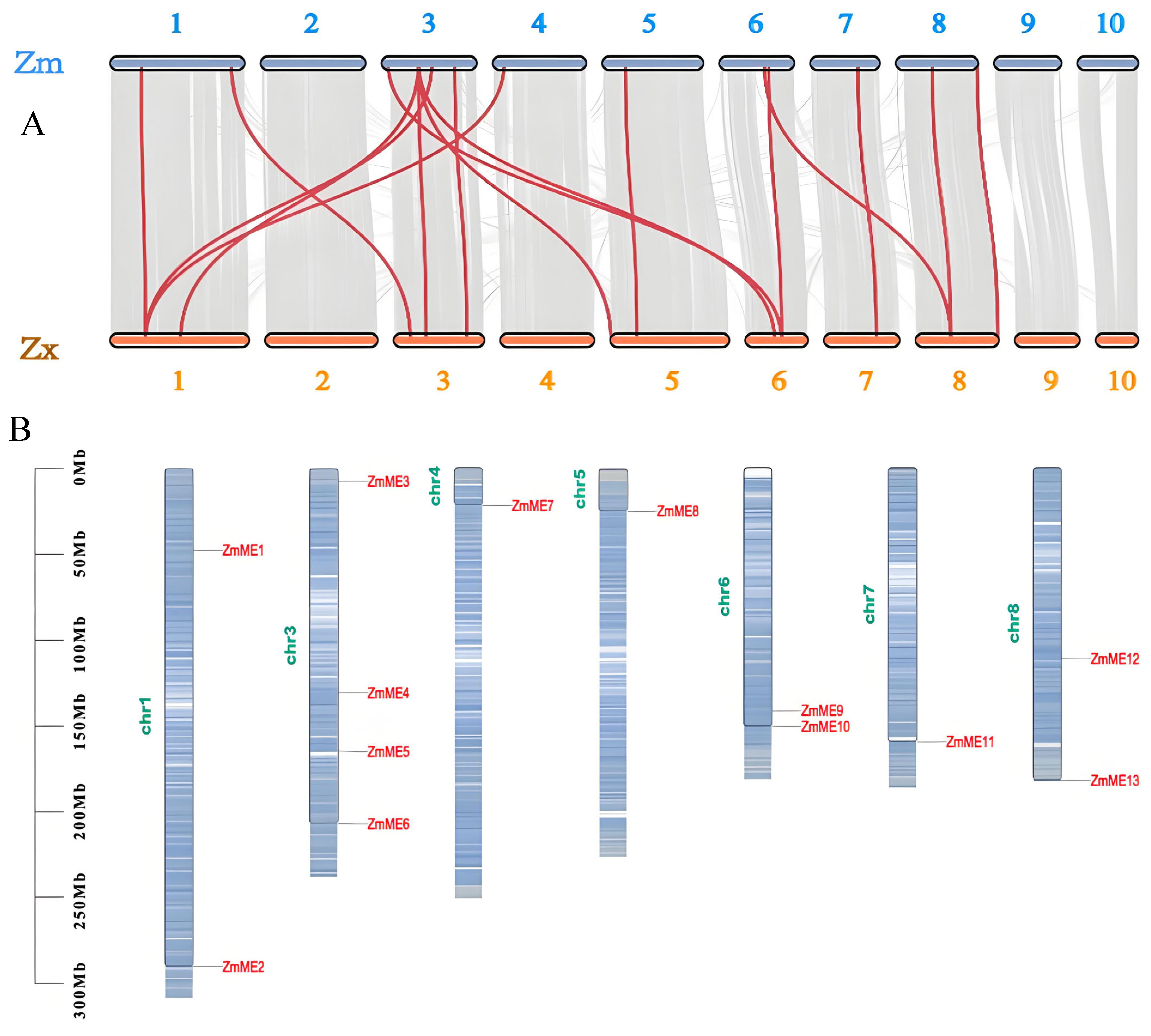
2.2. Chromosomal Localization and Collinearity Analysis
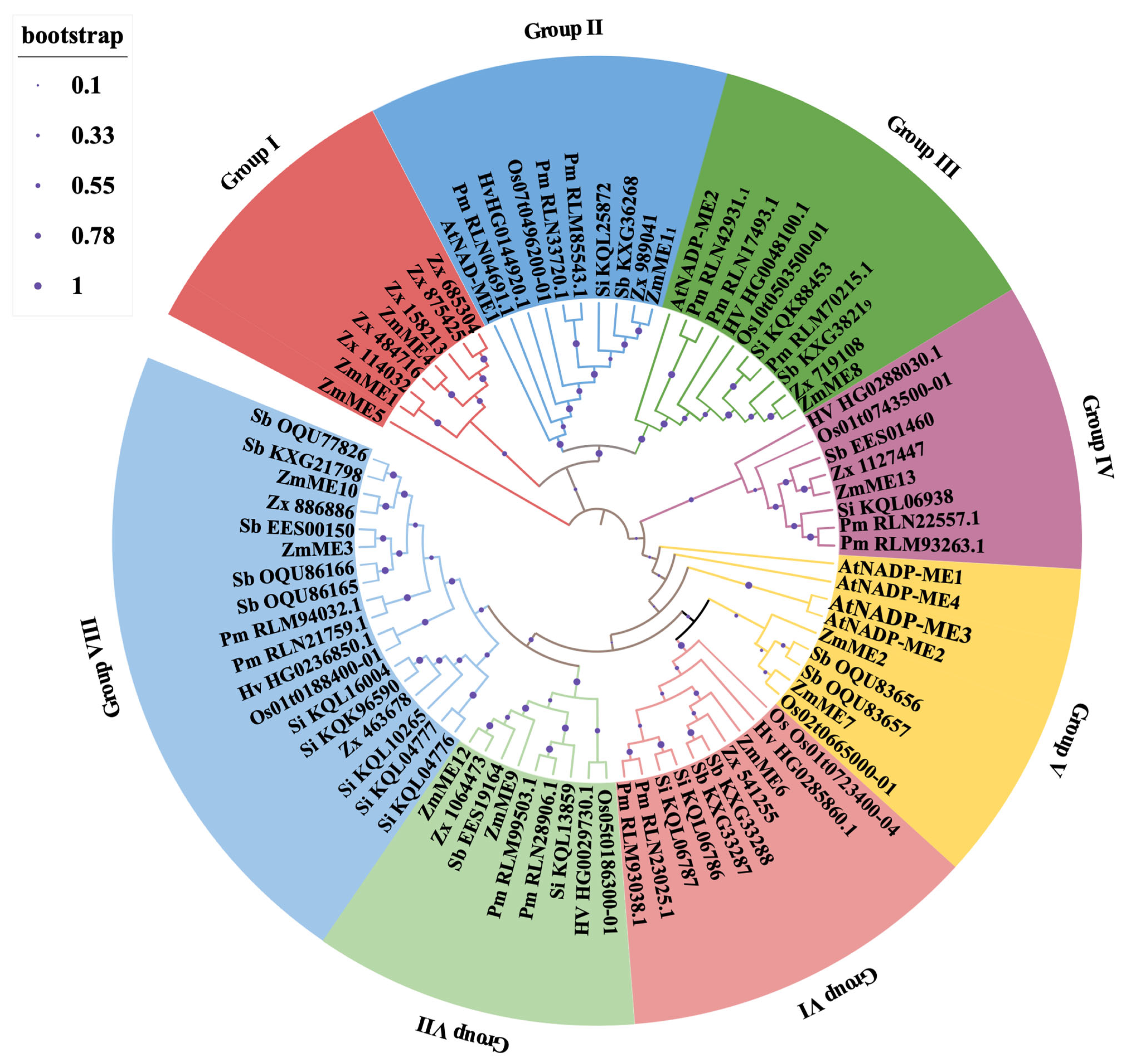
2.3. Phylogenetic Analysis of Gene Families
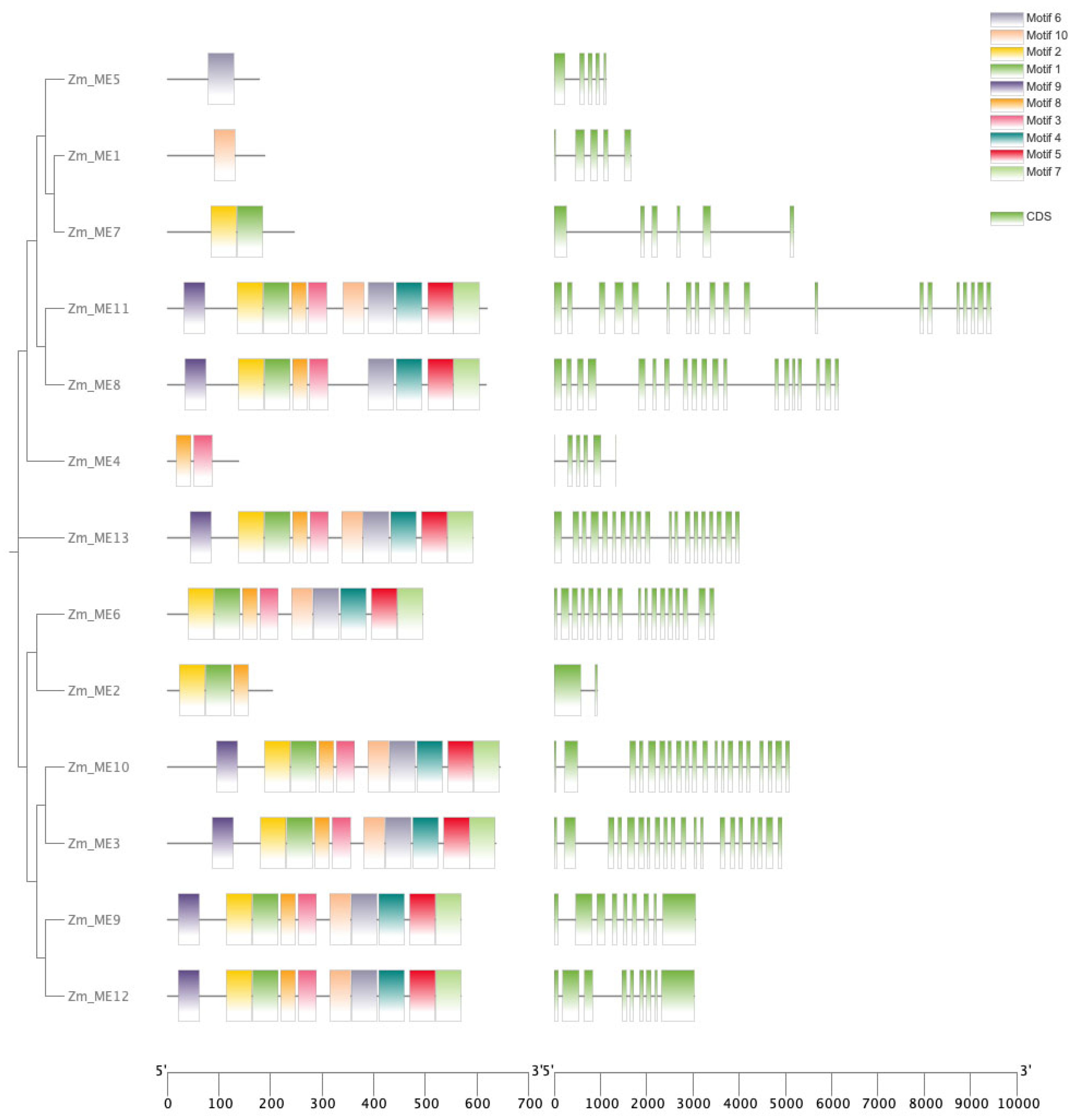
2.4. Conserved Motifs, Structural Domains and Gene Structure Analysis of Gene Family Members
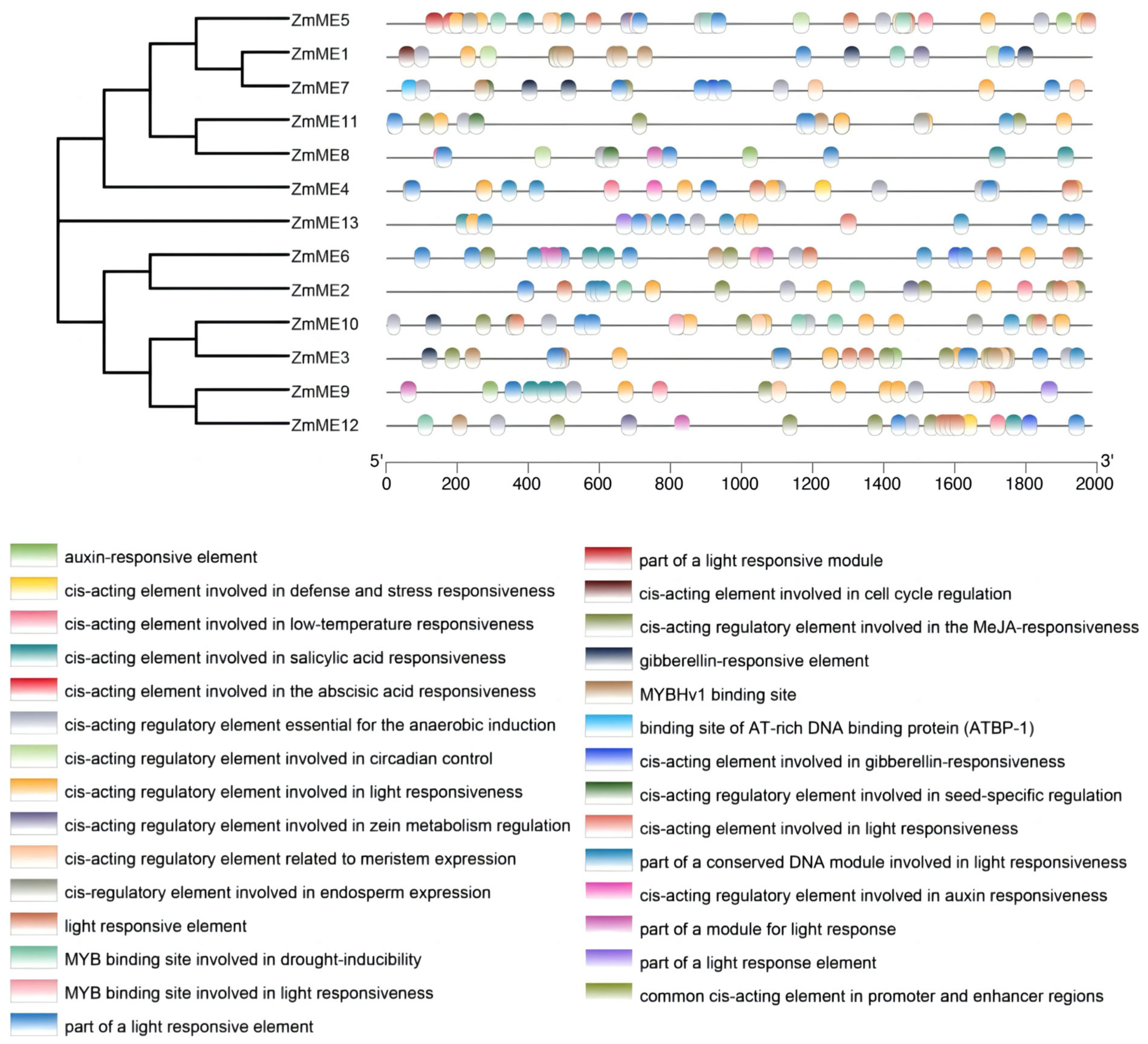
2.5. Analysis of Promoter Cis-Acting Elements of Maize ME Family Members
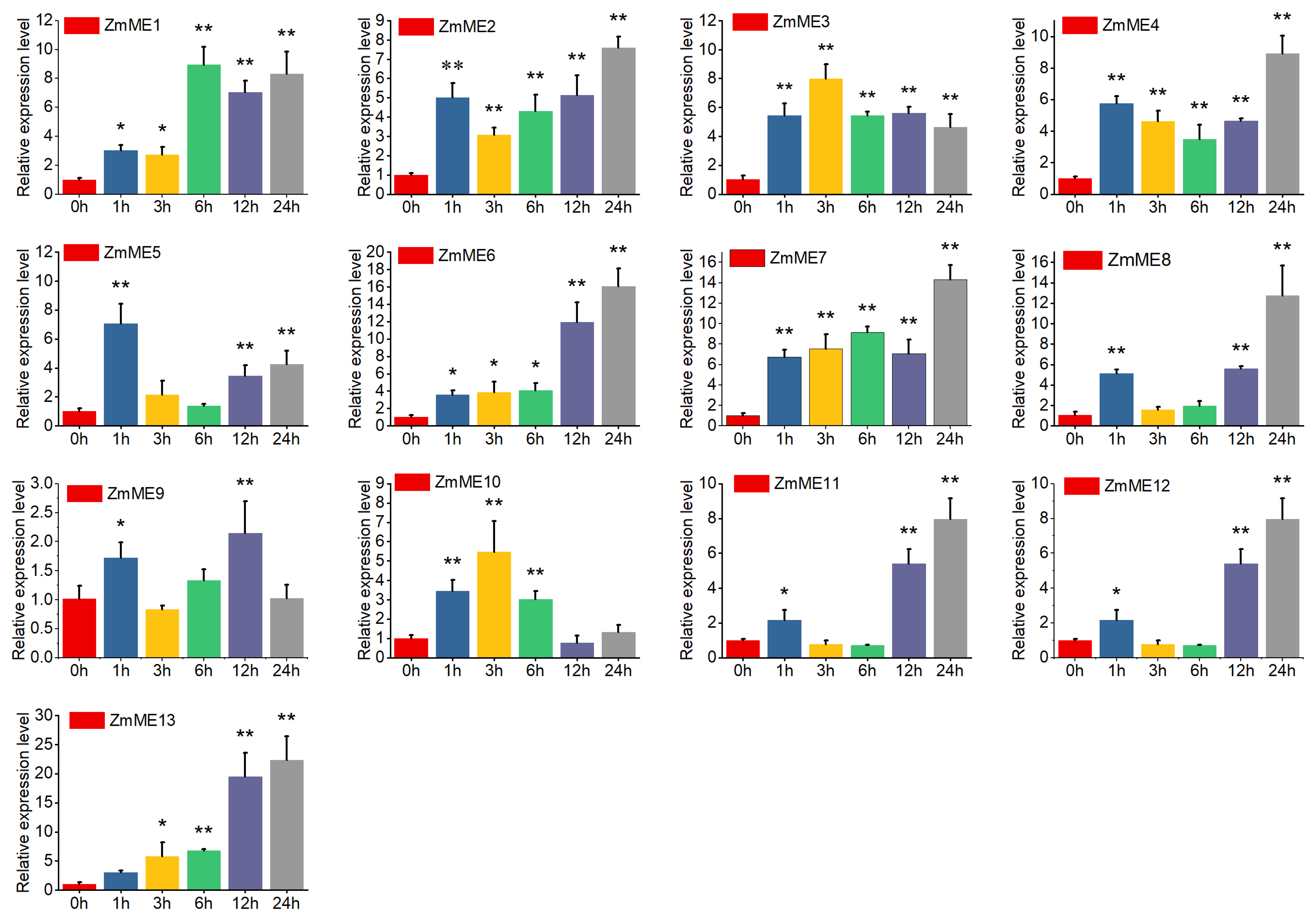
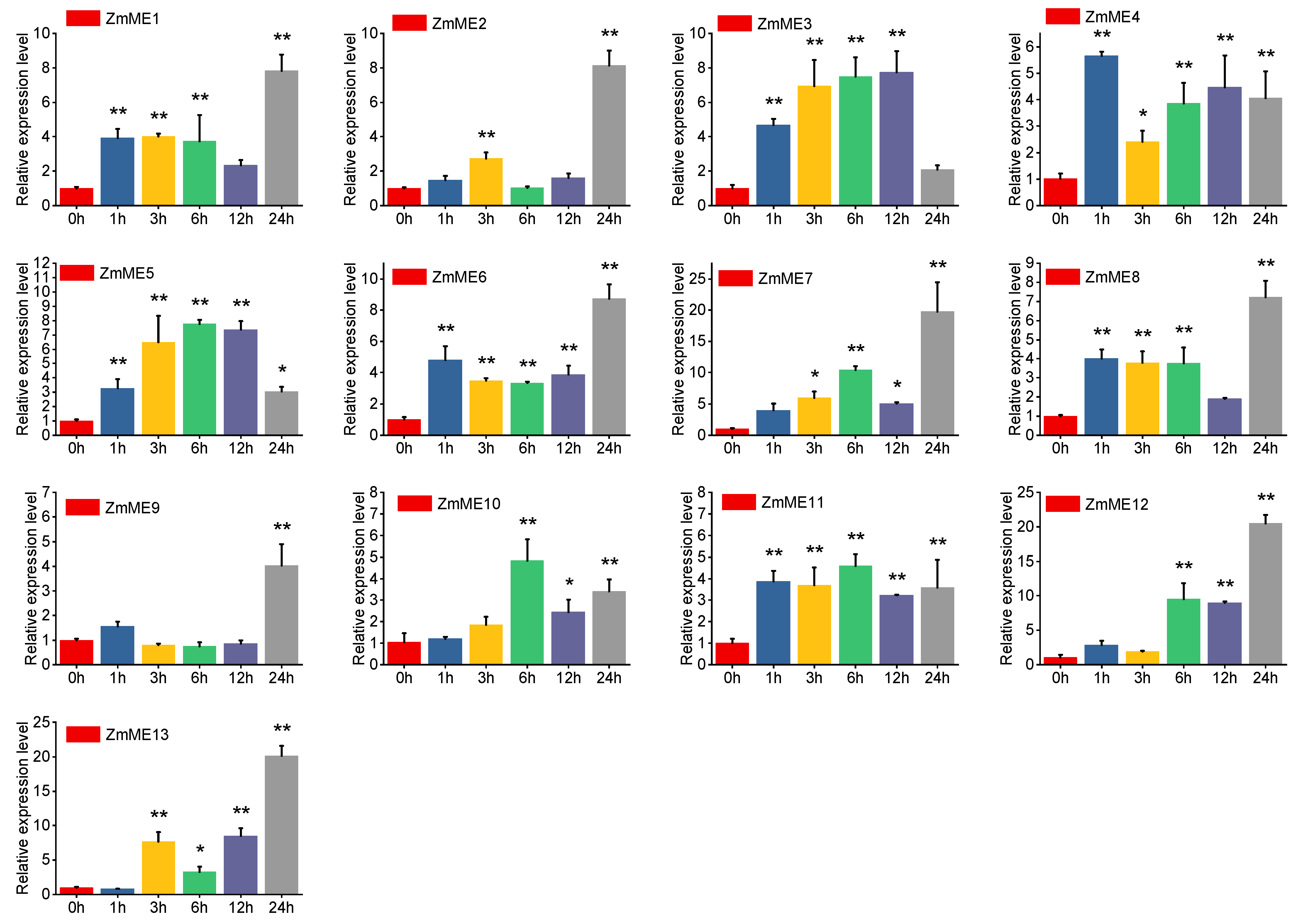
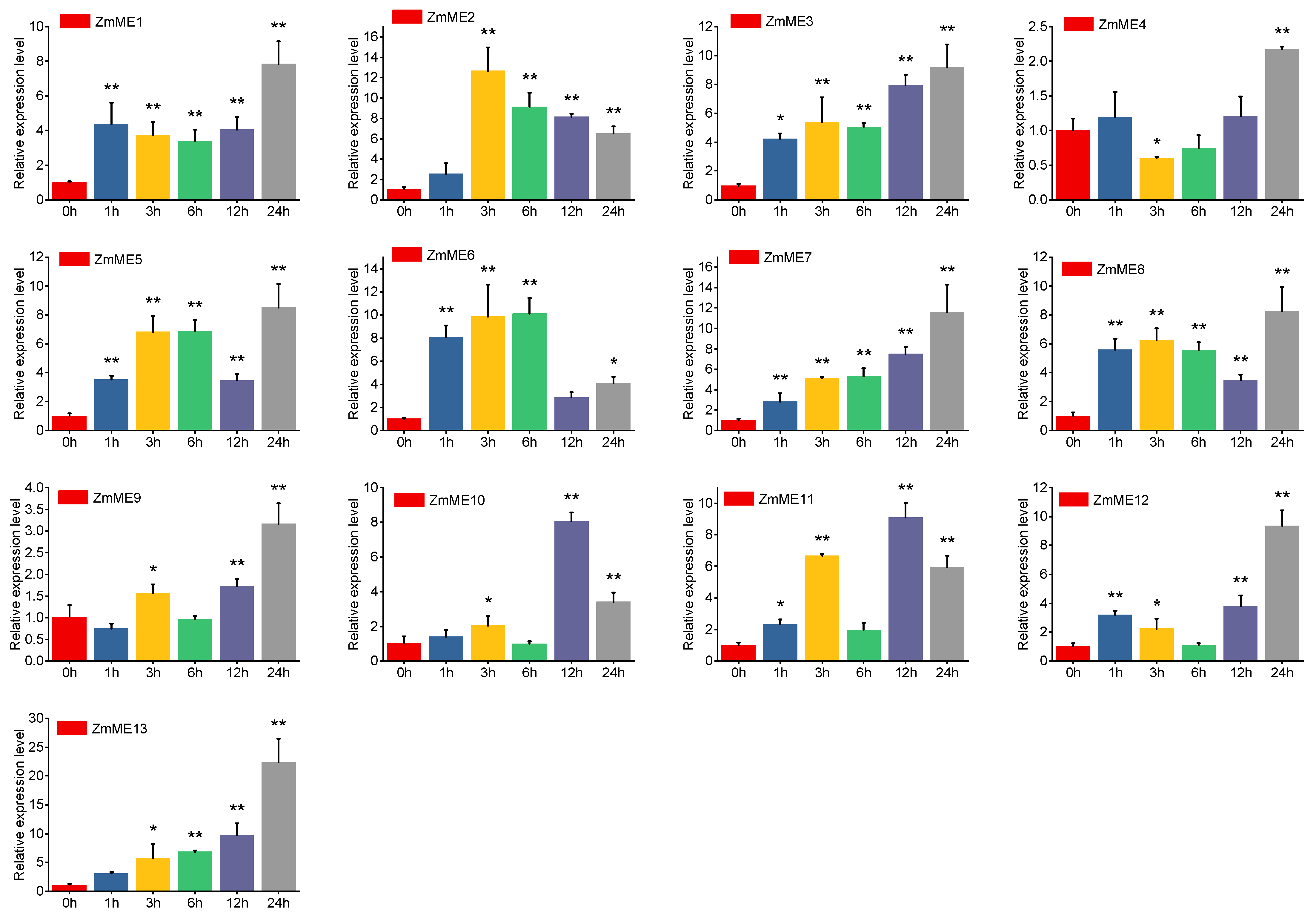
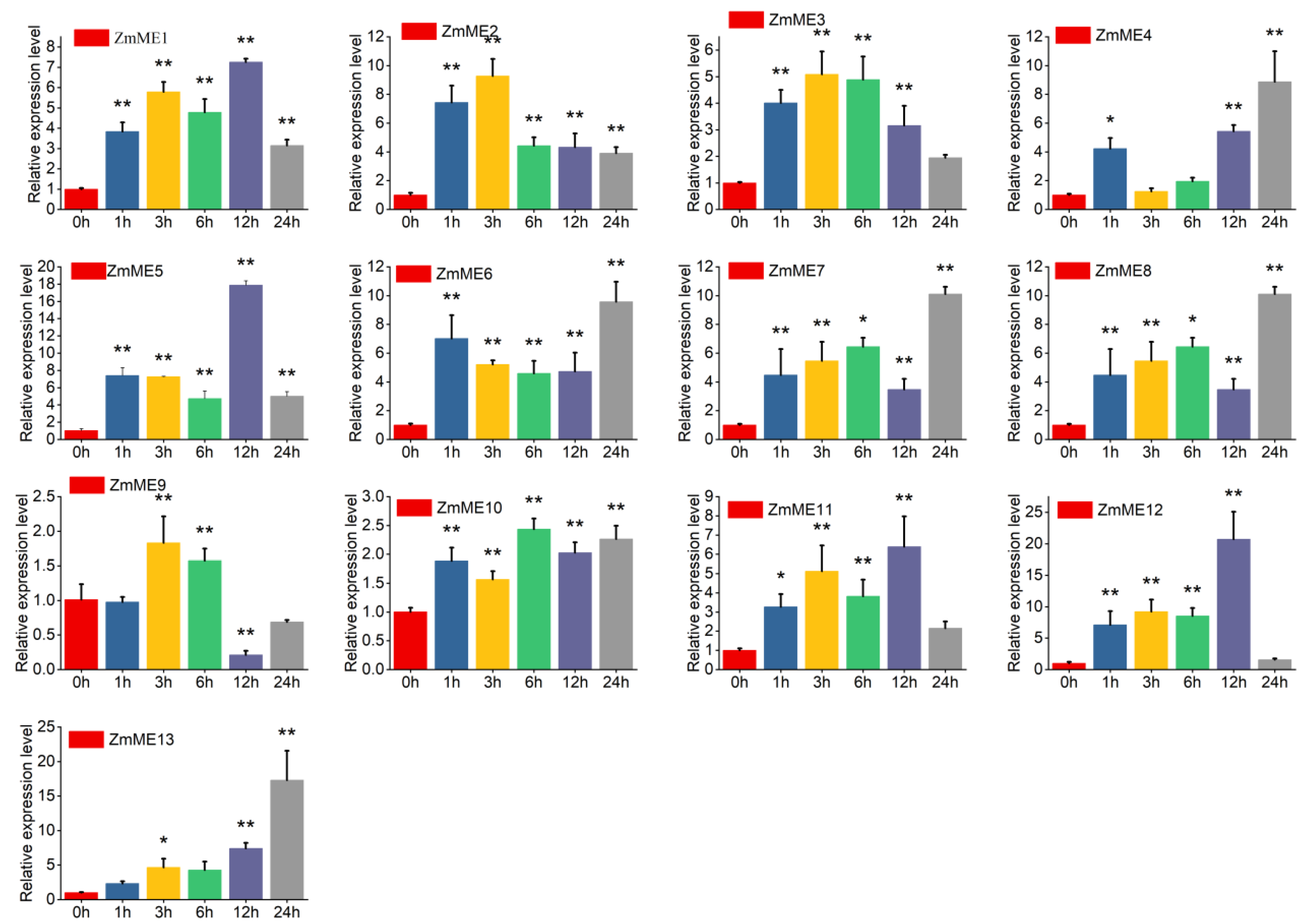
2.6. Expression Analysis of Maize Malic Enzyme Gene Under Abiotic Stresses
2.6.1. Drought Stress Treatments
2.6.2. High-Temperature Stress at 42 °C
2.6.3. 4 °C Low-Temperature Stress Treatment
2.6.4. 20% PEG Stress Treatment
2.6.5. 250 mmol·L−1 NaCl Stress Treatments
3. Discussion
4. Materials and Methods
4.1. Experimental Methods
4.1.1. Identification of ZmME Gene Family Members
4.1.2. Chromosomal Localization and Collinearity Analysis
4.1.3. Phylogenetic Analysis of Gene Families
4.1.4. Conserved Motifs, Structural Domains and Gene Structure Analysis of Gene Family Members
4.1.5. Gene Family Promoter Cis-Acting Element Analysis
4.1.6. ZmME Gene Family Expression Analysis
4.1.7. Maize ZmME Gene qRT-PCR Validation
4.1.8. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Kopecka, R.; Kameniarova, M.; Cerny, M.; Brzobohaty, B.; Novak, J. Abiotic stress in crop production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, J.M.; Hoisington, D.A.; Deutsch, J.A.; Jiang, C.; Gonzalez-de-Leon, D. Identification of quantitative trait loci under drought conditions in tropical maize. 1. Flowering parameters and the anthesis-silking interval. Theor. Appl. Genet. 1996, 92, 905–914. [Google Scholar] [CrossRef]
- Liang, Y.; Kang, K.; Gan, L.; Ning, S.; Xiong, J.; Song, S.; Xi, L.; Lai, S.; Yin, Y.; Gu, J.; et al. Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in brassica napus and arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol. J. 2019, 17, 2123–2142. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Roberts, M.J.; Schlenker, W.; Braun, N.; Little, B.B.; Rejesus, R.M.; Hammer, G.L. Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science 2014, 344, 516–519. [Google Scholar] [CrossRef]
- Li, Y.; Guan, K.; Schnitkey, G.D.; DeLucia, E.; Peng, B. Excessive rainfall leads to maize yield loss of a comparable magnitude to extreme drought in the United States. Glob. Change Biol. 2019, 25, 2325–2337. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, B.M. Effects of climatechange and drought tolerance onmaize growth. Plants 2023, 12, 3548. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant 2018, 162, 2–12. [Google Scholar] [CrossRef]
- Nykiel, M.; Gietler, M.; Fidler, J.; Prabucka, B.; Rybarczyk-Płońska, A.; Graska, J.; Boguszewska-Mańkowska, D.; Muszyńska, E.; Morkunas, I.; Labudda, M. Signal transduction incereal plants struggling with environmental stresses: From perception to Response. Plants 2022, 11, 1009. [Google Scholar] [CrossRef]
- Voll, L.M.; Zell, M.B.; Engelsdorf, T.; Saur, A.; Wheeler, M.G.; Drincovich, M.F.; Weber, A.P.; Maurino, V.G. Loss of cytosolic NADP-malic enzyme 2 in arabidopsis thaliana is associated with enhanced susceptibility tocolletotrichum higginsianum. New Phytol. 2012, 195, 189–202. [Google Scholar] [CrossRef]
- Maurino, V.G.; Saigo, M.; Andreo, C.S.; Drincovich, M.F. Non-photosynthetic ‘malic enzyme’ from maize: A constituvely expressed enzyme that responds to plant defence inducers. Plant Mol. Biol. 2001, 45, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.P.; Chen, H.M.; Hong, C.T.; Jiang, D.; Zheng, B.S. Exogenous malic acid alleviates cadmium toxicity in miscanthus sacchariflorus through enhancing photosynthetic capacity and restraining ROS accumulation. Ecotoxicol. Environ. Saf. 2017, 141, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lu, S.X.; Cao, X.J.; Ma, Z.H.; Chen, B.H.; Mao, J. Physiological and transcriptome analyses of the effects of excessive water deficit on malic acid accumulation in apple. Tree Physiol. 2023, 43, 851–866. [Google Scholar] [CrossRef]
- Katayama, N.; Iwazumi, K.; Suzuki, H.; Osanai, T.; Ito, S. Malic enzyme, not malatedehydrogenase, mainly oxidizes malate that originates from the tricarboxylic acid cycle in cyanobacteria. mBio 2022, 13, e0218722. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Zell, M.B.; Maurino, V.G. Malate decarboxylases: Evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 2011, 62, 3061–3069. [Google Scholar] [CrossRef] [PubMed]
- Doubnerová Hýsková, V.; Miedzińska, L.; Dobrá, J.; Vankova, R.; Ryšlavá, H. Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J. Plant Physiol. 2014, 171, 19–25. [Google Scholar] [CrossRef]
- Sun, X.; Han, G.L.; Meng, Z.; Lin, L.; Sui, N. Roles of malic enzymes in plant development and stress responses. Plant Signal Behav. 2019, 14, e1644596. [Google Scholar] [CrossRef]
- Chen, Q.Q.; Wang, B.P.; Ding, H.Y.; Zhang, J.; Li, S.C. Review: The role of NADP-malic enzyme in plants under stress. Plant Sci. 2019, 281, 206–212. [Google Scholar] [CrossRef]
- Cushman, J.C. Characterization and expression of a NADP-malic enzyme cDNA induced by salt stress from the facultative crassulacean acid metabolism plant, Mesembryanthemum crystallinum. Eur. J. Biochem. 1992, 208, 259–266. [Google Scholar] [CrossRef]
- Holtum, J.A.; Winter, K. Activity of enzymes of carbon metabolism during the induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum L. Planta 1982, 155, 8–16. [Google Scholar] [CrossRef]
- Li, X.F.; Zhang, X.X.; Takio, N.; Liu, C.K. Expression characteristics of Rice (Oryza sativa L.) malic enzyme (OsNADP-ME3) gene under environmental stress. Genom. Appl. Biol. 2012, 31, 327–332. [Google Scholar]
- Cheng, Y.X.; Long, M. A cytosolic NADP-malic enzyme gene from rice (Oryza sativa L.) confers salt tolerance in transgenic Arabidopsis. Biotechnol. Lett. 2007, 29, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.B.; Liu, Z.H.; Zhang, Z.B.; Chen, Q.J.; Chu, L.Y.; Brestic, M. Biological roles of crop NADP malic enzymes and molecular mechanisms involved in abiotic stress. Afr. J. Biotechnol. 2011, 10, 4947–4953. [Google Scholar]
- Wen, Z.B.; Zhang, M.L. Possible involvement of phosphoenolpyruvate carboxylase and NAD-malic enzyme in response to drought stress. A case study: A succulent nature of the C4-NAD-ME type desert plant, Salsola lanata (Chenopodiaceae). Funct. Plant Biol. 2017, 44, 1219–1228. [Google Scholar] [CrossRef]
- Abbas, K.; Li, J.R.; Gong, B.B.; Lu, Y.S.; Wu, X.L.; Lü, G.Y.; Gao, H.B. Drought stress tolerance in vegetables: The functional role of structural features, key gene pathways, and exogenous hormones. Int. J. Mol. Sci. 2023, 24, 13876. [Google Scholar] [CrossRef]
- Xu, W.; Yan, Q. Advances in the research of cold resistance in sugarcane. Sugarcane 2003, 10, 8–12. [Google Scholar]
- Liu, Z.H.; Zhang, Z.B.; Chu, L.Y.; Shao, H.B. The corresponding relationship between roles of NADP-malic enzymes and abiotic stress in plants. Emir. J. Food Arg. 2010, 22, 239. [Google Scholar]
- Zou, Y.; Zhang, Z.X.; Zeng, Y.J.; Hu, H.Y.; Hao, Y.J.; Huang, S.; Li, B. Common methods for phylogenetic tree construction and their implementation in R. Bioengineering 2024, 11, 480. [Google Scholar] [CrossRef]
- Frenkel, R. Regulation and physiological functions of malic enzymes. Curr. Top. Cell Regul. 1975, 9, 157–181. [Google Scholar] [CrossRef]
- Nei, M. Phylogenetic analysis in molecular evolutionary genetics. Annu. Rev. Genet. 1996, 30, 371–403. [Google Scholar] [CrossRef]
- Peng, X.J.; Zhao, Y.Z.; Li, X.M.; Wu, M.; Chai, W.B.; Sheng, L.; Wang, Y.; Dong, Q.; Jiang, H.Y.; Cheng, B.J. Genomewide identification, classification and analysis of NAC type gene family in maize. J. Genet. 2015, 94, 377–390. [Google Scholar] [CrossRef]
- Xu, J.H.; Messing, J. Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proc. Natl. Acad. Sci. USA 2008, 105, 14330–14335. [Google Scholar] [CrossRef]
- Li, H.Z.; Xiao, J.; Chen, J.H.; Shen, X.; Luo, J.; Guo, F.G.; Wang, S.F.; Xu, L.Y.; Guo, X.; Wang, S.; et al. Identification of the cassava NADP-ME gene family and its response and regulation in photosynthesis. Front. Plant Sci. 2025, 16, 1525193. [Google Scholar] [CrossRef] [PubMed]
- Taboada, J.; González-Gordo, S.; Muñoz-Vargas, M.A.; Palma, J.M.; Corpas, F.J. NADP-dependent malic enzyme genes in sweet pepper fruits: Involvement in ripening and modulation by nitric oxide (NO). Plants 2023, 12, 2353. [Google Scholar] [CrossRef]
- Fu, Z.Y.; Zhang, Z.B.; Hu, X.J.; Shao, H.B.; Ping, X. Cloning, identification, expression analysis and phylogenetic relevance of two NADP-dependent malic enzyme genes from hexaploid wheat. Comptes Rendus Biol. 2009, 332, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.C.; Tronconi, M.A.; Drincovich, M.F.; Andreo, C.S.; Flügge, U.I.; Maurino, V.G. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 2005, 139, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.X.; Guo, C.C.; Shan, H.Y.; Kong, H.Z. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Chow, C.N.; Chiang-Hsieh, Y.F.; Chien, C.H.; Zheng, H.Q.; Lee, T.Y.; Wu, N.Y.; Tseng, K.C.; Hou, P.F.; Chang, W.C. Delineation of condition specific Cis- and Trans-acting elements in plant promoters under various Endo- and exogenous stimuli. BMC Genom. 2018, 19, 85. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Girin, T.; Lejay, L.; Wirth, J.; Widiez, T.; Palenchar, P.M.; Nazoa, P.; Touraine, B.; Gojon, A.; Lepetit, M. Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant Cell Environ. 2007, 30, 1366–1380. [Google Scholar] [CrossRef]
- Li, R.X.; Zhu, F.D.; Duan, D. Function analysis and stress-mediated cis-element identification in the promoter region of VqMYB15. Plant Signal Behav. 2020, 15, 1773664. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Cao, H.L.; Lin, H.Z.; Hu, J.; Ye, Y.J.; Li, J.M.; Hao, Z.L.; Hao, X.Y.; Sun, Y.; Yang, Y.J.; et al. Expression patterns of alpha-amylase and beta-amylase genes provide insights into the molecular mechanisms underlying the responses of tea plants (Camellia sinensis) to stress and postharvest processing treatments. Planta 2019, 250, 281–298. [Google Scholar] [CrossRef]
- Jwa, N.S.; Agrawal, G.K.; Tamogami, S.; Yonekura, M.; Han, O.; Iwahashi, H.; Rakwal, R. Role of defense/stress-related marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiol. Biochem. 2006, 44, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.J.; Ren, Q.Y.; Chen, Y.L.; Xu, G.L.; Qian, Y.X. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021, 21, 427. [Google Scholar] [CrossRef]
- Sun, S.B.; Shen, Q.R.; Wan, J.M.; Liu, Z.P. Induced expression of the gene for NADP-malic enzyme in leaves of Aloe vera L. under salt stress. Acta Biochim. Biophys. Sin. 2003, 35, 423–429. [Google Scholar]
- Crecelius, F.; Streb, P.; Feierabend, J. Malate metabolism and reactions of oxidoreduction in cold-hardened winter rye (Secale cereale L.) leaves. J. Exp. Bot. 2003, 54, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Babayev, H.; Mehvaliyeva, U.; Aliyeva, M.; Feyziyev, Y.; Guliyev, N. The study of NAD-malic enzyme in Amaranthus cruentus L. under drought. Plant Physiol. Biochem. 2014, 81, 84–89. [Google Scholar] [CrossRef]
- Farris, J.S.; Albert, V.A.; Källersjö, M.; Lipscomb, D.; Kluge, A.G. Parsimony jackknifing outperforms neighbor-joining. Cladistics 1996, 12, 99–124. [Google Scholar] [CrossRef]

| Gene Name | Chromosomes | Start | End | Gene Length | Number of Amino Adds | Molecular Weight | Theoretical Pl | CDS (bp) | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|---|
| Zm ME1 | chr1 | 47,534,292 | 47,535,943 | 1652 | 189 | 21,681.92 | 9.2 | 567 | −0.324 |
| Zm ME2 | chr1 | 290,259,636 | 290,260,669 | 1034 | 204 | 23,001.29 | 5.22 | 612 | −0.361 |
| Zm ME3 | chr3 | 7,184,482 | 7,190,135 | 5654 | 636 | 69,818.85 | 6.2 | 1908 | −0.168 |
| Zm ME4 | chr3 | 130,704,894 | 130,717,659 | 12766 | 138 | 15,622.11 | 5.56 | 414 | 0.291 |
| Zm ME5 | chr3 | 164,884,990 | 164,886,965 | 1976 | 178 | 19,890.73 | 8.87 | 534 | −0.33 |
| Zm ME6 | chr3 | 206,946,472 | 206,951,192 | 4721 | 480 | 52,749.47 | 5.93 | 1440 | −0.081 |
| Zm ME7 | chr4 | 2,155,524 | 21,560,705 | 5182 | 240 | 27,447.67 | 7.07 | 720 | −0.248 |
| Zm ME8 | chr5 | 24,840,267 | 24,847,307 | 7041 | 618 | 67,809.04 | 8 | 1854 | −0.121 |
| Zm ME9 | chr6 | 141,031,530 | 141,035,585 | 4056 | 570 | 62,893.99 | 5.69 | 1710 | −0.147 |
| Zm ME10 | chr6 | 150,321,630 | 150,375,807 | 54178 | 644 | 70,694.85 | 6.35 | 1932 | −0.144 |
| Zm ME11 | chr7 | 159,213,544 | 159,223,682 | 10139 | 619 | 69,133.99 | 6.09 | 1857 | −0.251 |
| Zm ME12 | chr8 | 110,806,909 | 110,810,557 | 3649 | 570 | 62,703.59 | 5.74 | 1710 | −0.154 |
| Zm ME13 | chr8 | 181,572,829 | 181,578,195 | 5367 | 593 | 64,958.31 | 5.82 | 1779 | −0.092 |
| Primer Name | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| ZmME1 | ACCTCTTCCTCTCGAGGCTT | ACTGAAGGCATTAGGCCACC |
| ZmME2 | GCTTGCTTGCCGATCACATT | GGGGTCAGAAGCTTCCAGTT |
| ZmME3 | GGACCTACAGGAACACCCTG | GACCCTTGTTGTGGTGTGGA |
| ZmME4 | AACGTGTGACTGTCGAGGAG | GCATTGCAGGACCCTATCCA |
| ZmME5 | ATGGTGCAGGCTCTAGCAAA | GAGGCCACAATCAGTCCCTC |
| ZmME6 | CTGGAGCTATCCGCGTTCAT | GGGGCTGTACATGCAACTCT |
| ZmME7 | CCTATGGCGACAACTTCGGT | CGTCTCCCCCTCCTTAGTGT |
| ZmME8 | TGGGATTGGTTTAGGTGCCC | CTCAGCCAGGTCTTCAGCAA |
| ZmME9 | AGGAGCATCCAGGTCATCGT | ACGGCGGTCATGAACTCTTCAA |
| ZmME10 | TGCTTCCTCCTGCAGTTGTT | AGGACCTTCCCTTTGTCCCT |
| ZmME11 | ACAAGAGCACCGTCTTGAGG | ACTCCAATACCAGCACTGCC |
| ZmME12 | TTCCCGTTGGCAAGCTCTC | GTAGTTTTGCTTGACGGCGGC |
| ZmME13 | ATTTGGCAGTGGAAGCCCAT | CATAAGCTTTTGCGGCGACA |
| ZmActin-1 | TGAAACCTTCGAATGCCCAG | GATTGGAACCTGGTGGCTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Li, Y.; Ma, Z.; Wang, R.; Zhou, Y.; Zhou, W.; He, H.; Wang, X.; Lian, X.; Dong, X.; et al. Identification and Abiotic Stress Expression Profiling of Malic Enzyme-Associated Genes in Maize (Zea mays L.). Plants 2025, 14, 1603. https://doi.org/10.3390/plants14111603
Yan H, Li Y, Ma Z, Wang R, Zhou Y, Zhou W, He H, Wang X, Lian X, Dong X, et al. Identification and Abiotic Stress Expression Profiling of Malic Enzyme-Associated Genes in Maize (Zea mays L.). Plants. 2025; 14(11):1603. https://doi.org/10.3390/plants14111603
Chicago/Turabian StyleYan, Haishan, Yongsheng Li, Zengke Ma, Ruihong Wang, Yuqian Zhou, Wenqi Zhou, Haijun He, Xiaojuan Wang, Xiaorong Lian, Xiaoyun Dong, and et al. 2025. "Identification and Abiotic Stress Expression Profiling of Malic Enzyme-Associated Genes in Maize (Zea mays L.)" Plants 14, no. 11: 1603. https://doi.org/10.3390/plants14111603
APA StyleYan, H., Li, Y., Ma, Z., Wang, R., Zhou, Y., Zhou, W., He, H., Wang, X., Lian, X., Dong, X., & Yao, L. (2025). Identification and Abiotic Stress Expression Profiling of Malic Enzyme-Associated Genes in Maize (Zea mays L.). Plants, 14(11), 1603. https://doi.org/10.3390/plants14111603





