Identification of the NTL Gene Family in Beta vulgaris L. and Functional Role of BvNTL2 in Drought Resistance
Abstract
1. Introduction
2. Results
2.1. Identification and Prediction of the Physicochemical Properties of BvNTLs Family Members
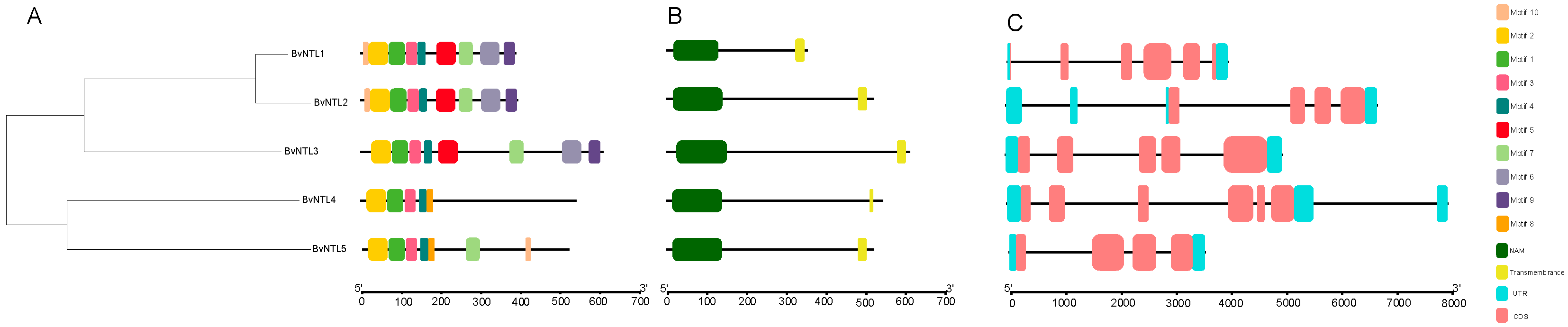
2.2. BvNTLs Family Gene and Protein Structure Analysis
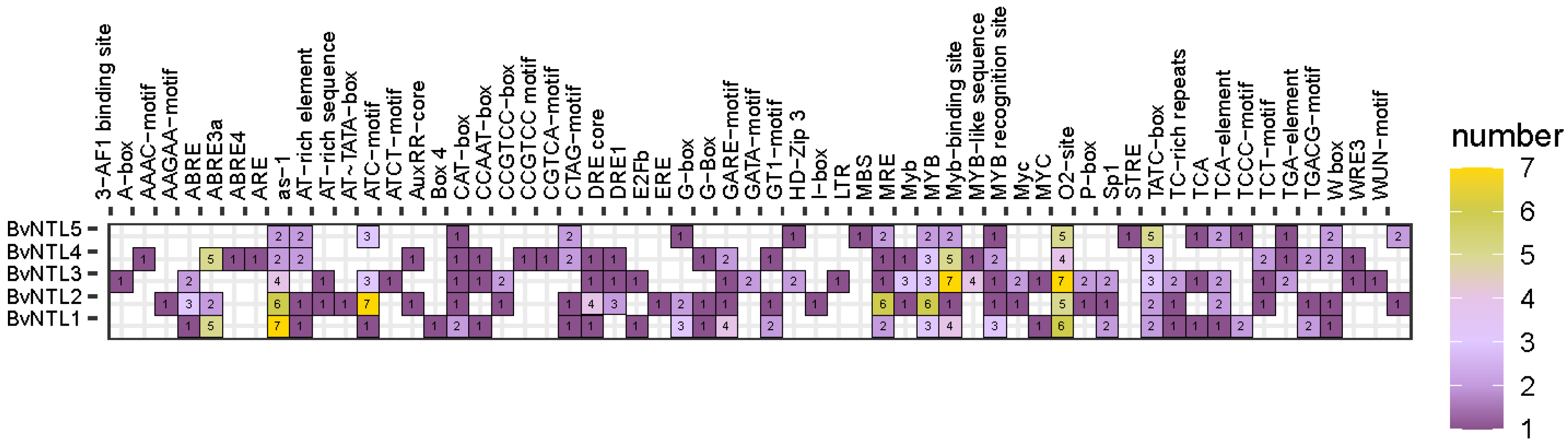
2.3. Prediction of Cis-Acting Elements in the Promoter Region of the BvNTLs Family
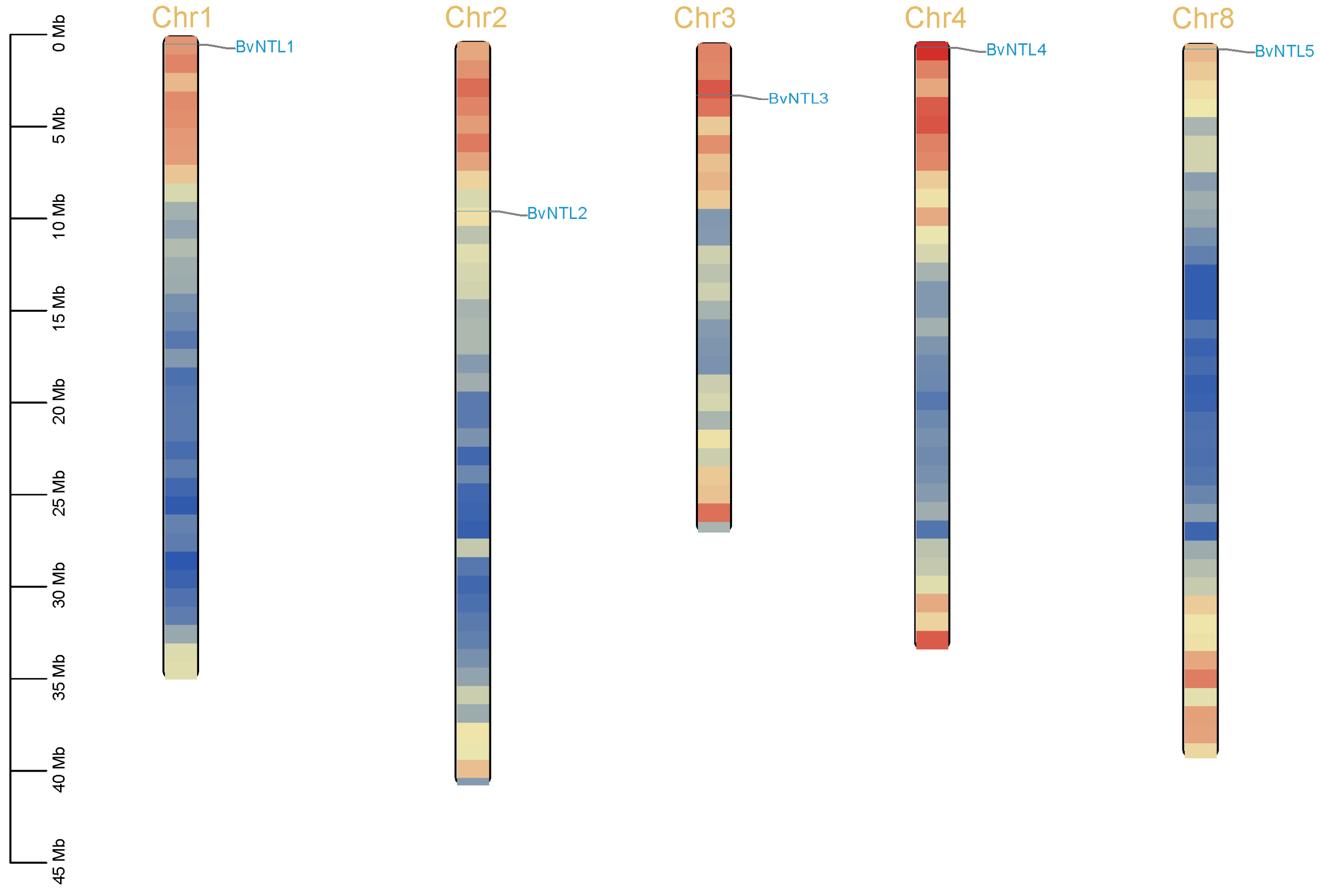
2.4. Collinearity Analysis of NTLs in Multiple Species
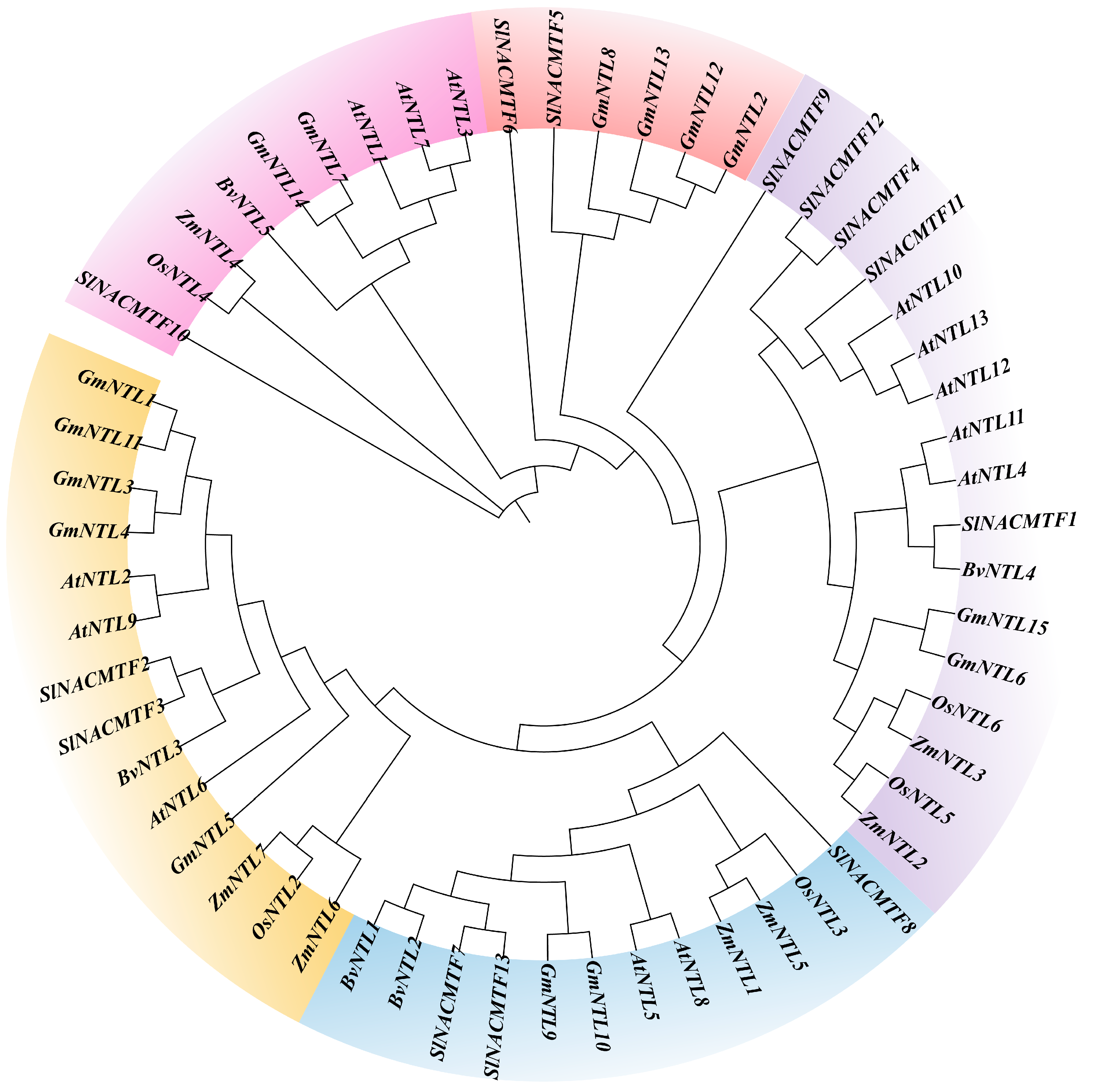
2.5. Phylogenetic Tree of the BvNTLs Family
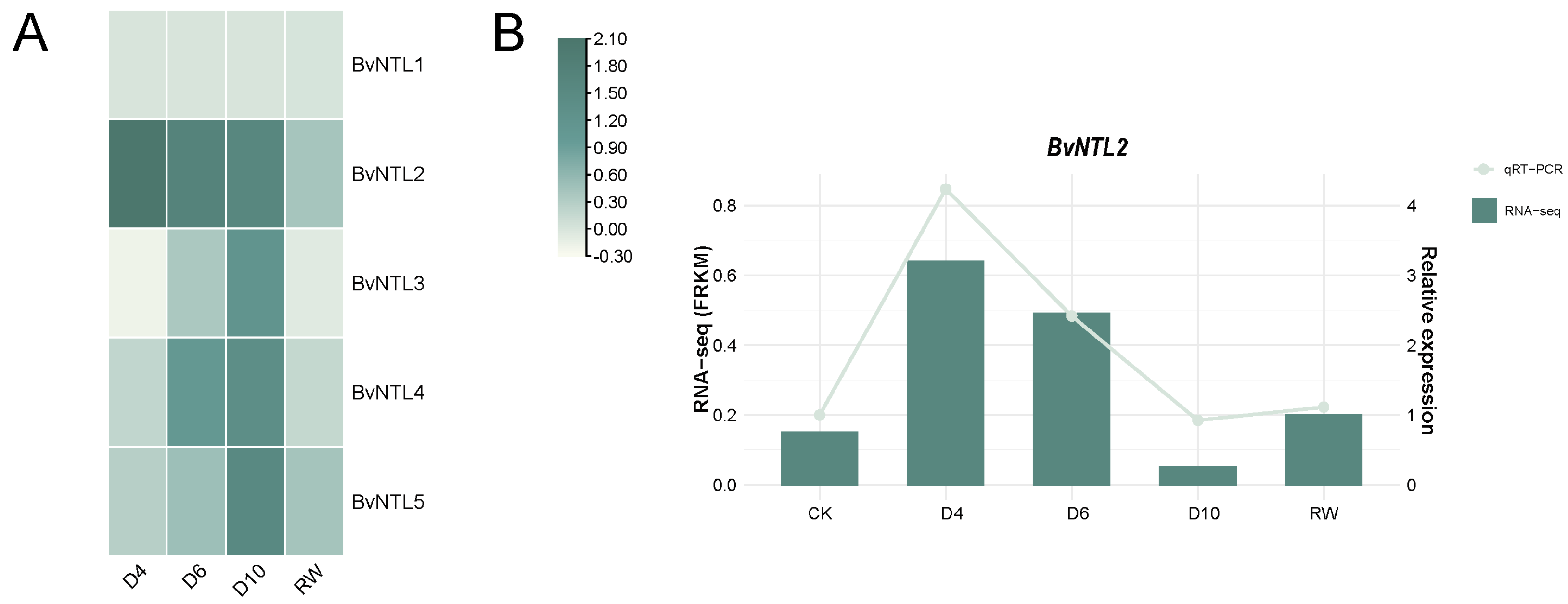
2.6. Expression Analysis of BvNTLs Members in Sugar Beet Leaves
2.7. Functional Analysis of BvNTL2
2.7.1. Subcellular Localization Analysis of BvNTL2
2.7.2. Drought Resistance Identification of BvNTL2 Overexpression
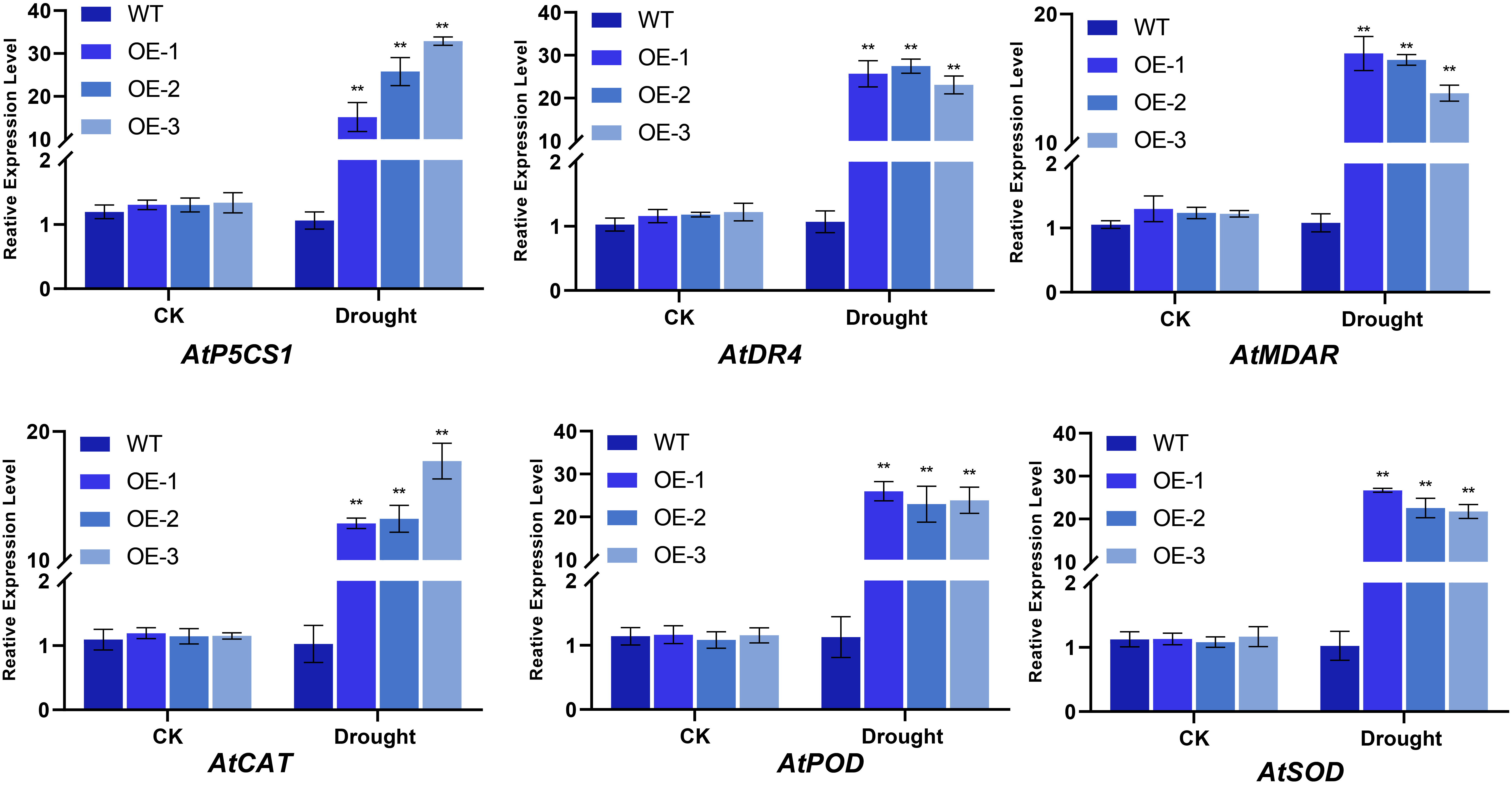
2.7.3. Impact of BvNTL2 Overexpression on Physiological Traits Associated with Stress Tolerance
2.7.4. Impact of BvNTL2 Overexpression on the Expression of Stress-Responsive Genes
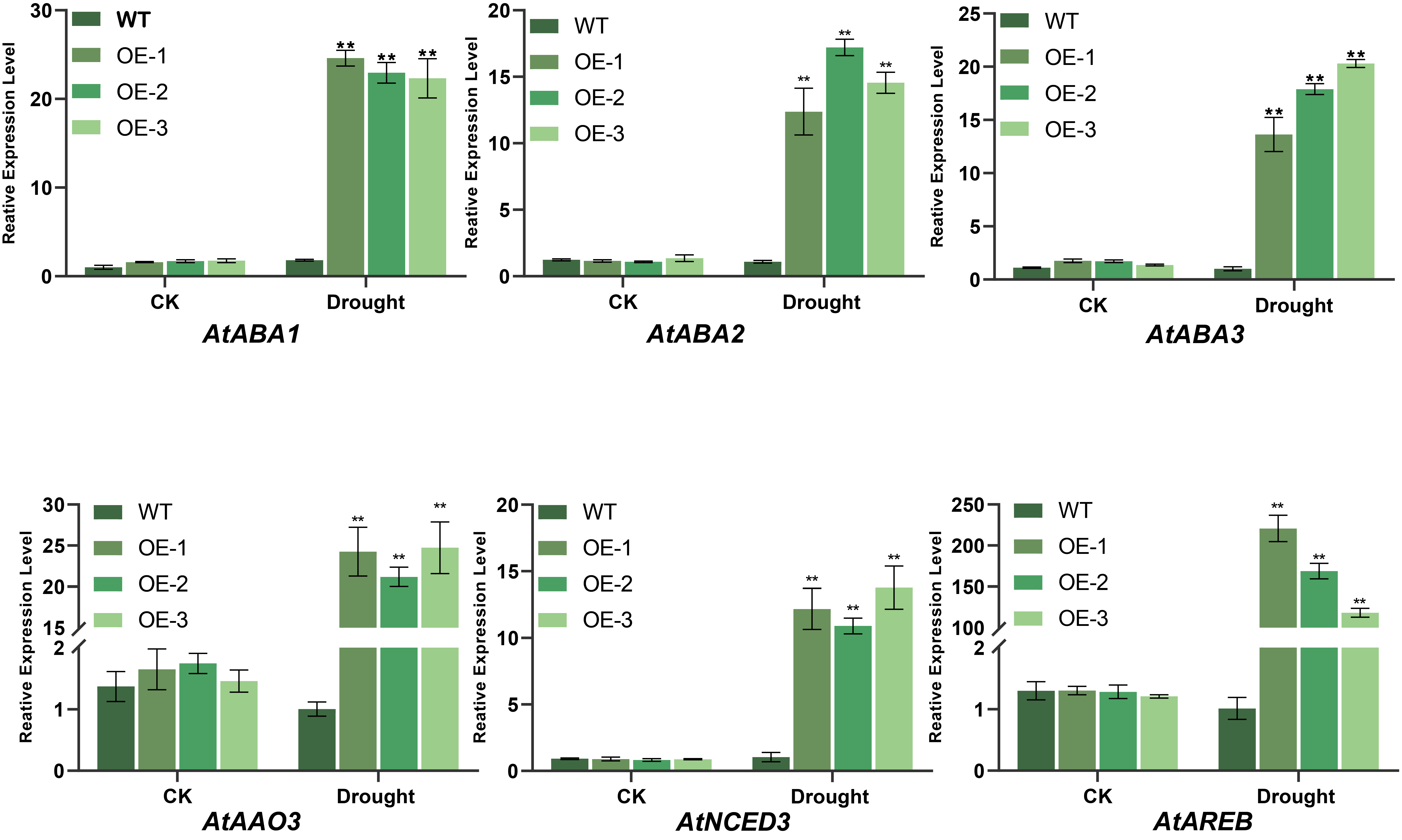
Impact of BvNTL2 Overexpression on the Expression of ABA Biosynthesis-Related Genes
3. Discussion
3.1. The Relationship Between the NTL Gene Family and Drought Resistance in Sugar Beet
3.2. Subcellular Localization and Functional Analysis of BvNTL2 in Drought Tolerance
4. Materials and Methods
4.1. Identification and Prediction of the Physicochemical Properties of BvNTLs Family Members
4.2. Gene Structure and Conserved Motif Analysis of BvNTLs
4.3. Comparative Synteny and Phylogenetic Tree Analysis of BvNTLs
4.4. Cis-Regulatory Element Analysis of BvNTLs Promoters
4.5. Generation of Transgenic Arabidopsis Plants
4.6. Plant Materials and Growth Conditions
4.7. Quantitative Real-Time PCR Analysis
4.8. Physiological Index Analysis of Transgenic Arabidopsis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The No Apical Meristem Gene of Petunia Is Required for Pattern Formation in Embryos and Flowers and Is Expressed at Meristem and Primordia Boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.-H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef]
- Chen, Y.-N.; Slabaugh, E.; Brandizzi, F. Membrane-tethered transcription factors in Arabidopsis thaliana: Novel regulators in stress response and development. Curr. Opin. Plant Biol. 2008, 11, 695–701. [Google Scholar] [CrossRef]
- Kim, M.J.; Park, M.-J.; Seo, P.J.; Song, J.-S.; Kim, H.-J.; Park, C.-M. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2. 8 in the drought-stress response. Biochem. J. 2012, 448, 353–363. [Google Scholar] [CrossRef]
- Seo, P.J. Recent advances in plant membrane-bound transcription factor research: Emphasis on intracellular movement. J. Integr. Plant Biol. 2014, 56, 334–342. [Google Scholar] [CrossRef]
- Seo, P.J.; Kim, S.-G.; Park, C.-M. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008, 13, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-G.; Lee, S.; Seo, P.J.; Kim, S.-K.; Kim, J.-K.; Park, C.-M. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 2010, 95, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yu, Y.; Liu, Z.; Li, S.; Wang, Z.; Xiang, F. Membrane-bound NAC transcription factors in maize and their contribution to the oxidative stress response. Plant Sci. 2016, 250, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, N.; Ji, D.; Xue, Z.; Yu, Y.; Jiang, Y.; Liu, J.; Liu, Z.; Xiang, F. Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol. 2016, 172, 1804–1820. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Das, R.; Mandal, A.; Kundu, P. Functional characterization of tomato membrane-bound NAC transcription factors. Plant Mol. Biol. 2017, 93, 511–532. [Google Scholar] [CrossRef]
- Shu, L.; Li, L.; Jiang, Y.-Q.; Yan, J. Advances in membrane-tethered NAC transcription factors in plants. Plant Sci. 2024, 342, 112034. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, S.-G.; Park, J.-E.; Park, H.-Y.; Lim, M.-H.; Chua, N.-H.; Park, C.-M. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, A.K.; Yoon, H.K.; Park, C.M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008, 55, 77–88. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.-S.; Kim, S.-G.; Jung, J.-H.; Woo, J.-C.; Park, C.-M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011, 156, 537–549. [Google Scholar] [CrossRef]
- Sun, H.; Xie, Y.; Yang, W.; Lv, Q.; Chen, L.; Li, J.; Meng, Y.; Li, L.; Li, X. Membrane-bound transcription factor TaNTL1 positively regulates drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2022, 182, 182–193. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Xiang, J.; Teng, R.; Zhang, D.; Teng, N. A lily membrane-associated NAC transcription factor, LlNAC014, is involved in thermotolerance via activation of the DREB2-HSFA3 module. J. Exp. Bot. 2023, 74, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef] [PubMed]
- Bui, L.T.; Shukla, V.; Giorgi, F.M.; Trivellini, A.; Perata, P.; Licausi, F.; Giuntoli, B. Differential submergence tolerance between juvenile and adult Arabidopsis plants involves the ANAC017 transcription factor. Plant J. 2020, 104, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef]
- Zheng, X.-y.; Zhou, M.; Yoo, H.; Pruneda-Paz, J.L.; Spivey, N.W.; Kay, S.A.; Dong, X. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. USA 2015, 112, 9166–9173. [Google Scholar] [CrossRef]
- McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Pritchard, L.; Gomez, S.; Morales, J.; Whisson, S.C.; Beynon, J.L.; Birch, P.R. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 2013, 9, e1003670. [Google Scholar] [CrossRef]
- Sun, Y.; Li, C.; Li, Z.; Jian, C.; Li, N.; Zhang, S.; Li, G. Construction of Gene Expression Profiles of the NAC Transcription Factor Family in Sugar Beet Under Abiotic Stress. Sugar Tech 2024, 26, 1232–1242. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, S.-G.; Kim, Y.-S.; Seo, P.J.; Bae, M.; Yoon, H.-K.; Park, C.-M. Exploring membrane-associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res. 2007, 35, 203–213. [Google Scholar] [CrossRef]
- Guo, Y.L. Gene family evolution in green plants with emphasis on the origination and evolution of a Arabidopsis thaliana genes. Plant J. 2013, 73, 941–951. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef]
- Kiraga, J.; Mackiewicz, P.; Mackiewicz, D.; Kowalczuk, M.; Biecek, P.; Polak, N.; Smolarczyk, K.; Dudek, M.R.; Cebrat, S. The relationships between the isoelectric point and: Length of proteins, taxonomy and ecology of organisms. BMC Genom. 2007, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Keren, H.; Lev-Maor, G.; Ast, G. Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 2010, 11, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Hatemi, P.K.; Gillespie, N.A.; Eaves, L.J.; Maher, B.S.; Webb, B.T.; Heath, A.C.; Medland, S.E.; Smyth, D.C.; Beeby, H.N.; Gordon, S.D.; et al. A genome-wide analysis of liberal and conservative political attitudes. J. Politics 2011, 73, 271–285. [Google Scholar] [CrossRef]
- Amtmann, A.; Bohnert, H.J.; Bressan, R.A. Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 2005, 138, 127–130. [Google Scholar] [CrossRef]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010, 61, 661–671. [Google Scholar] [CrossRef]
- Yan, J.; Li, Y.; Zhao, P.; Mu, B.; Chen, Q.; Li, X.; Cui, X.; Wang, Z.; Li, J.; Li, S.; et al. Membrane-bound transcriptional activator NTL1 from rapeseed positively modulates leaf senescence through targeting genes involved in reactive oxygen species production and programmed cell death. J. Agric. Food Chem. 2021, 69, 4968–4980. [Google Scholar] [CrossRef]
- Lu, J.; Wu, T.; Zhang, B.; Liu, S.; Song, W.; Qiao, J.; Ruan, H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021, 19, 60. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate–stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef]
- Nadeau, J.; Sack, F. Stomatal development in Arabidopsis. Arab. Book 2002, 2002, e0066. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A. The role of abscisic acid in drought stress: How ABA helps plants to cope with drought stress. In Drought Stress Tolerance in Plants, Vol 2: Molecular and Genetic Perspectives; Springer: Cham, Switzerland, 2016; pp. 123–151. [Google Scholar]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ødum, M.T.; Teufel, F.; Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Winther, O.; Nielsen, H. DeepLoc 2.1: Multi-label membrane protein type prediction using protein language models. Nucleic Acids Res. 2024, 52, W215–W220. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- González, L.; González-Vilar, M. Determination of Relative Water Content. In Handbook of Plant Ecophysiology Techniques; Reigosa Roger, M.J., Ed.; Springer: Dordrecht, The Netherlands, 2001; pp. 207–212. [Google Scholar]
- Jambunathan, N. Determination and Detection of Reactive Oxygen Species (ROS), Lipid Peroxidation, and Electrolyte Leakage in Plants. In Plant Stress Tolerance: Methods and Protocols; Sunkar, R., Ed.; Humana Press: Totowa, NJ, Canada, 2010; pp. 291–297. [Google Scholar]
- Liang, Y.K.; Xie, X.; Lindsay, S.E.; Wang, Y.B.; Masle, J.; Williamson, L.; Leyser, O.; Hetherington, A.M. Cell wall composition contributes to the control of transpiration efficiency in Arabidopsis thaliana. Plant J. 2010, 64, 679–686. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P. Ultraviolet-B-and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 1996, 110, 125–136. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Z.; Xu, Y.; Sun, Y.; Li, N.; Zhang, S.; Li, G. Identification of the NTL Gene Family in Beta vulgaris L. and Functional Role of BvNTL2 in Drought Resistance. Plants 2025, 14, 1528. https://doi.org/10.3390/plants14101528
Fan Z, Xu Y, Sun Y, Li N, Zhang S, Li G. Identification of the NTL Gene Family in Beta vulgaris L. and Functional Role of BvNTL2 in Drought Resistance. Plants. 2025; 14(10):1528. https://doi.org/10.3390/plants14101528
Chicago/Turabian StyleFan, Ziqi, Yanni Xu, Yaqing Sun, Ningning Li, Shaoying Zhang, and Guolong Li. 2025. "Identification of the NTL Gene Family in Beta vulgaris L. and Functional Role of BvNTL2 in Drought Resistance" Plants 14, no. 10: 1528. https://doi.org/10.3390/plants14101528
APA StyleFan, Z., Xu, Y., Sun, Y., Li, N., Zhang, S., & Li, G. (2025). Identification of the NTL Gene Family in Beta vulgaris L. and Functional Role of BvNTL2 in Drought Resistance. Plants, 14(10), 1528. https://doi.org/10.3390/plants14101528




