Abstract
NAC transcription factors are a kind of plant specific transcription factor widely distributed in plants, and they play an important role in the process of plant growth and development. According to the transcriptome data, a transcription factor with typical NAC characteristics was isolated from Larix olgensis (common name “Dahurian larch”), that we named LoNAC5. The length of the coding sequence (CDS) was 1164 bp, encoding 387 amino acids. The LoNAC5 protein harbors a NAM (NAC family) domain at the 14–139 aa region of its N-terminus and an activation domain at the 324–364 aa region of the C-terminus. Phylogenetic tree analysis revealed that LoNAC5 belonged to the ATNAC3 subgroup. Cis-acting element analysis showed that there were multiple plant stress-resistance-related elements on the promoter of LoNAC5, including hormone and light responsiveness elements. LoNAC5 was localized in the nucleus by injection transformation of tobacco leaves. Results suggested that the LoNAC5 protein is active as a homodimer and that it binds to the GATGTG motif. The results of RT-qPCR showed that LoNAC5 is a highly expressed gene in L. olgensis, and the expression level is highest in 180-day needles. LoNAC5 responded to various hormone treatments and was induced by drought and salt stress. The yeast phenotype test showed that overexpression of LoNAC5 could make yeast grow better under drought and salt stress. It was speculated that LoNAC5 might act in L. olgensis as a positive regulator of drought and salt tolerance.
1. Introduction
The families of the plant-specific transcription factors (TFs) are defined by their characteristic DNA-binding domains [1]. The NAC family of TFs (including NAM, ATAF, and CUC) is a plant-specific transcription factor family, which has a characteristic domain of about 150 amino acids [2,3]. This domain binds specific DNA sequences and is the basis for the classification of NAC family members [4]. Previous studies have found that the conserved domain of NAC protein is generally located in the N-terminus, which can be generally divided into five motifs: A–E sub-domains [5,6,7]. This study found that sub-domains A, C, and D are highly conserved. Among them, C and D contain nuclear localization signals, which is presumed to be related to the nuclear localization in transcription factors and the recognition of specific DNA sequences. In contrast, B and E are varied, and are only conserved in some NAC subgroups, which is presumed to be associated with different functions of NAC TFs [6,7]. The C-terminus of NAC protein usually has simple amino acids with high repeatability (Thr, Ser, Pro, Glu, or other acidic amino acid residues), which makes it have diverse transcriptional activation domains in the C-terminus [8,9].
The NAC transcription factor family is very large. More than 8000 NAC family transcription factors have been found in plants, and 117 and 140 NAC family members have been found in Arabidopsis thaliana (L.) Heynh [10] and Oryza sativa L. [11] alone. According to the results of sequence clustering analysis, some researchers divided rice NAC family members into three subfamilies: NAM, ATAF, and OsNAC3 [12]. Alternatively, the NAC family was divided into five groups based on phylogenetic analyses in rice: OsNAC7, NAC1, NAM/CUC, GRAB2, and NAC2 [13]. Results of evolutionary studies of NAC family genes in rice and Arabidopsis showed that these NAC family proteins were classified into 2 groups and 18 subgroups by sequence similarity [14]. In Populus spp. 163 full-length NAC genes were identified that were divided into 18 different subfamilies according to the results of a phylogenetic tree [15]. Gene structure, motif composition, sequence conservation, and function tended to be more similar within than among NAC family subgroups [16,17].
NAC transcription factors have a variety of biological functions, which are not only involved in plant growth and morphogenesis [18,19], but also in response to various stresses [20,21,22]. NAM is one of the first discovered NAC family genes, and mutation of this gene in Petunia embryos will lead to abnormal development of shoot apical meristem [7]. CUCs from Arabidopsis thaliana were found to be associated with ovule number [23] and shoot meristem formation [18,24]. NAM and CUC genes in maize have also been shown to be related to cotyledon formation [25]. VNDs have been shown to switch plant metaxylem and protoxylem vessel formation [26,27,28,29]. The overexpression of AtNAC1 [30] or GmNAC20 [20] can promote lateral root formation in plants. In addition, researchers have found that NAC family genes can not only regulate plant tolerance to abiotic stresses such as drought [31,32,33], salt [34,35], cold [36,37,38], and high temperature [39], but also participate in plant response processes to biotic stresses such as viruses [40,41,42,43], pathogens [44,45,46], and pests [47,48].
With technological advancements, plant genome sequencing has progressed significantly, leading to a comprehensive analysis of the NAC family genes in most plants [15,49,50]. In Cryptomeria fortunei Hooibrenk, 33 CfNACs that may be involved in lignin biosynthesis, xylem development, and SCW formation have been identified [51]. In addition, three genes belonging to the NAC family in Larix olgensis were also successfully identified and named LoNAC1–3 [52,53]. However, due to the extensive genome and intricate genetic background of larch, research on NAC genes in this family remains scarce [52]. In this study, the NAC family gene LoNAC5 and its promoter were identified and isolated from L. olgensis based on transcriptome data, and the sequence information was analyzed using bioinformatics methods. The expression characteristics of LoNAC5 were studied through subcellular localization assay and yeast assay. Furthermore, we examined the expression pattern of LoNAC5 and characterized its role under drought and salt stress conditions in yeast. This study provides a theoretical and molecular foundation for further exploration of LoNAC5’s function.
2. Results
2.1. LoNAC5 Belongs to the AtNAC3 Subgroup
Using LoNAC5 amplification primers, DNA fragments of approximately 1000 bp and 3000 bp in length were isolated from the cDNA and gDNA of L. olgensis, respectively. Sequencing results revealed that the total length of the LoNAC5 sequence was 3205 bp, consisting of three exons and two introns. The length of LoNAC5 coding region is 1164 bp, encoding 387 amino acids. The theoretical molecular weight of LoNAC5 protein is 43,740.05 Da, with a predicted theoretical PI of 6.12. The corresponding protein consists of 43 positively charged and 47 negatively charged residues, an aliphatic index of 66.36, an instability index of 49.01, and average hydrophobicity of −0.681. These results indicate that the LoNAC5 protein is unstable and likely to be short-lived.
The sequence information of the L. olgensis NAC and Arabidopsis NAC family protein was obtained from TAIR (The Arabidopsis Information Resource) and NCBI (National Center for Biotechnology Information), and the phylogenetic tree was constructed together for homology analysis [14]. The results demonstrated that the 15 LoNACs were phylogenetically classified into six distinct subgroups (Figure 1), suggesting functional diversification within the NAC family in larch. Notably, LoNAC5 clustered within the AtNAC3 subgroup, forming a highly supported clade with Arabidopsis stress-responsive NAC members ANAC019, ANAC055, and ANAC072. This evolutionary conservation implies that LoNAC5 may share functional conservation with its Arabidopsis homologs ANAC019, ANAC055, and ANAC072.
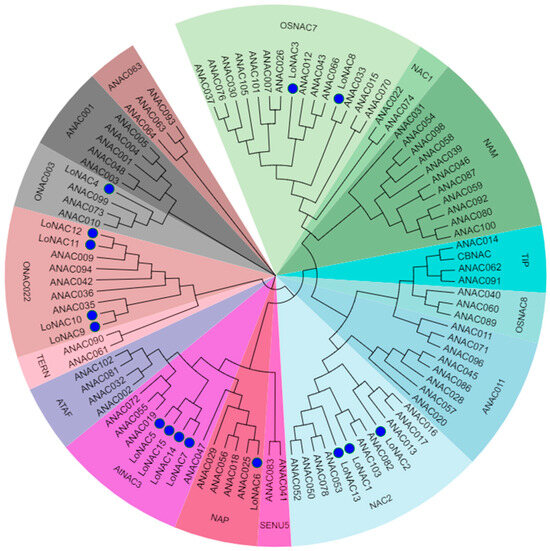
Figure 1.
Subgroup classification analysis of LoNAC5.
Conserved domain (CD)-search analysis identified a conserved NAM domain spanning amino acid residues 14 to 139 in the LoNAC5 protein. Motif analysis showed that the N-terminal domain can be divided into five motifs (Figure 2A), consistent with the structural features of the NAC family. Secondary structure predictions indicated that the encoded protein consisted of 27.13% alpha helices, 13.44% extended chains, and 59.43% random coils (Figure 2B). The SwissMode homology modeling method was used to predict the tertiary structure of LoNAC5 (Figure 2C), and the compared model was a NAC transcription factor (protein: A9NZG8.1, organism: Picea sitchensis/Sitka spruce/Pinus sitchensis) with 94.04% sequence identity. The LoNAC5 protein does not contain signal peptides or transmembrane domains, and its subcellular localization is predicted in the nucleus.
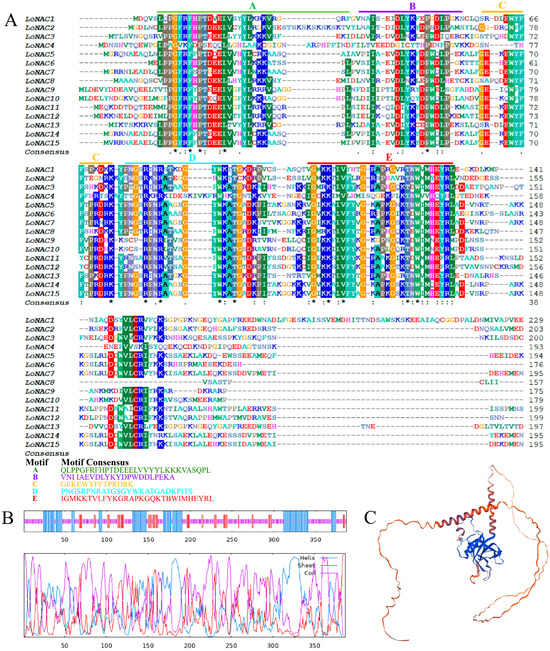
Figure 2.
Bioinformatics analysis of LoNAC5 protein. (A) The motif analysis of LoNAC5, “*” represents a conserved amino acid; “:” a conservative replacement; “.” a non-conservative replacement; (B) secondary structure prediction of LoNAC5; (C) tertiary structure prediction of LoNAC5.
2.2. LoNAC5 Conforms to the Characteristics of NAC Transcription Factors
To determine whether LoNAC5 has transcriptional activation activity, full-length LoNAC5 was fused to the GAL4 DNA-binding domain (pGBKT7-LoNAC5) and introduced into the Y2HGold yeast strain. The empty vector of pGBKT7 was used as the negative control. The yeast transformed with full-length LoNAC5 fused to GAL4-BD grew on the selective media without Trp or without Trp, HIS and Ade, indicating that LoNAC5 exhibits transcriptional activation activity. To further analyze the location of the activation domain, we divided LoNAC5 into different fragments and inserted them separately into pGBKT7, and then repeated the above operations of transferring into yeast and observing growth status (Figure 3A). The results showed that the activation domain was located at the C-terminus and between F3–R4.
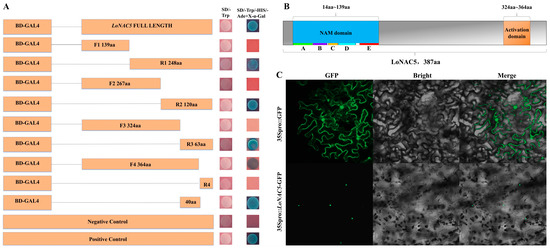
Figure 3.
Expression characteristics of LoNAC5 protein. (A) Activation domain analysis of LoNAC5; (B) structure of LoNAC5 protein; (C) the results of subcellular localization observed by laser confocal microscopy.
Based on the analysis of conserved and transcriptional activation domains, the structure of the LoNAC5 protein is depicted in Figure 3B. A conserved NAM domain is located between residues 14 and 139 at the N-terminus. This domain can be further divided into five sub-domains. Additionally, a transcriptional activation domain is located between 324 aa and 363 aa at the C-terminus. These findings suggest that LoNAC5 exhibits the same structural characteristics as most NAC transcription factors described previously.
LoNAC5 was fused to GFP and was successfully expressed in leaves of Nicotiana benthamiana. Observation by laser confocal microscopy revealed that green fluorescence signals could be detected throughout the tobacco epidermal cells in the control group, while in the experimental group (VB191104-LoNAC5-GFP), green fluorescence could only be observed in the nucleus of the tobacco epidermal cells (Figure 3C). The results showed that the subcellular localization of LoNAC5 was in the cell nucleus, which was consistent with the results predicted by WOLF PSORT.
2.3. LoNAC5 Can Respond to Drought and Salt Stress
The expression pattern of LoNAC5 in L. olgensis was analyzed by reverse transcription quantitative PCR (RT-qPCR). The experiment found that fluorescent signals could be detected in the roots, stems, and leaves of larch at three different stages, indicating that LoNAC5 was expressed in various parts at each stage. Through the analysis of RT-qPCR results, it can be seen that LoNAC5 has the highest relative expression level in the leaves of 180-day L. olgensis (Figure 4A). The expression level of LoNAC5 changed under different abiotic stresses. Under the treatment of 28% PEG 6000, the expression level of LoNAC5 increased with the prolongation of treatment time. The expression level reached the maximum in seedlings after 96 h, which was 35.26 times that of the untreated seedlings. Under the treatment of 0.2 M NaCl solution, the expression level of LoNAC5 was down-regulated in seedlings treated for 12 h and 24 h, whilst it was up-regulated in seedlings treated for 48 h and 96 h. The relative expression level was the lowest in seedlings treated for 12 h, with a 14.37-fold down-regulation compared to the untreated seedlings. The expression level reached the maximum after 48 h of treatment, with a relative expression level of 15.17 times that of the control. Under the treatment of NaHCO3, the change in the expression level of LoNAC5 was relatively small. The relative expression level of LoNAC5 was 2.12 times the control at 96 h. LoNAC5 expression was down-regulated in seedlings treated for 12 h, 24 h, and 48 h. The expression level was the lowest in seedlings treated for 48 h, with a 3-fold down-regulation compared to the untreated seedlings. Under the treatment of 50 mg/L MeJA at four time points, the expression level of LoNAC5 was down-regulated. The greatest down-regulation occurred in seedlings treated with methyl jasmonate for 96 h, with a 17.28-fold decrease compared to the untreated seedlings. Under the treatment of IAA, the relative expression level was the highest at 96 h, with a 10.66-fold increase compared to the untreated seedlings. At 48 h, the relative expression level was down-regulated by 15.75 times the control. Under the treatment of ABA, the expression level was down-regulated at 12 h and 24 h, and up-regulated at 48 h and 96 h. The relative expression level of LoNAC5 in seedlings at 96 h was up-regulated by 14.82 times the control. Under the treatments of MeJA, IAA, and ABA, |log2RT| > 3 at multiple time points, indicating more significant changes in the expression level (Figure 4B).
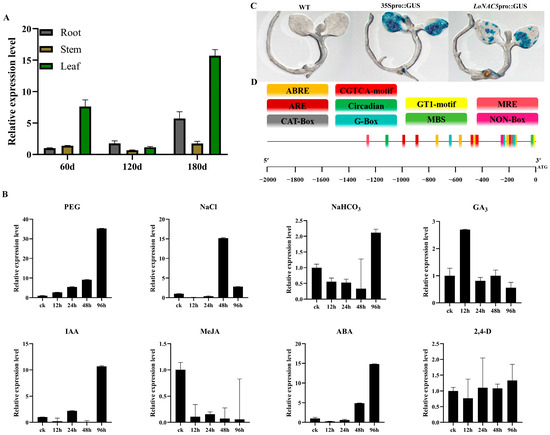
Figure 4.
Expression analysis of LoNAC5 and LoNAC5 promoter. (A) Expression analysis of LoNAC5 in different growth stages of L. olgensis; (B) expression analysis of LoNAC5 under different treatment; (C) analysis of promoter activity through GUS staining; (D) the cis-acting elements in LoNAC5 promoter sequence.
Based on genomic data of L. kaempferi (taxid: 54,800), we predicted the sequence information of the LoNAC5 promoter, which corresponds to the fragment ranging from position 1,076,477 to 1,078,477 of the contig LKA1p01161 (GenBank accession: BSBM01001161.1). The plasmids of the constructed recombinant vector (LoNAC5pro::GUS) and empty vector (35Spro::GUS) were expressed separately in tobacco (Figure 4C). GUS stain was not detected in the negative control tobacco (WT). In the experimental tobacco (LoNAC5pro::GUS), leaves were a major part of GUS staining, and the intensity and area of GUS staining were lower than those in the positive control tobacco (35Spro::GUS). These results indicate that the LoNAC5 promoter has a certain initiating activity and is strongest in plant leaves.
The analysis of cis-acting elements revealed the presence of multiple elements with different LoNAC5 promoter functions as follows: (1) Plant hormone responsive elements: five cis-acting elements are involved in abscisic acid response (ABRE) and methyl jasmonate response (CGTCA motif); (2) Elements involved in plant stress response: two cis-regulatory elements involved in anaerobic induction (ARE) and one MYB binding site involved in drought induction (MBS); (3) Elements related to photosynthesis: four light responsive elements (G-box and GT1 motif) and one MYB binding site participate in light response (MRE). In addition, there are cis-regulatory elements related to circadian rhythm control (Circadian), meristematic expression (CAT box), and meristematic specific activation (NON box) in the LoNAC5 promoter. Nearly all cis-acting elements are distributed from the starting codon (ATG) to the position 1300 bp upstream, with a significant clustering of these elements around 200 bp and 450 bp upstream of the starting codon (Figure 4D).
2.4. Functional Characterization of LoNAC5 in Yeast
The result of RT-qPCR revealed that LoNAC5 could respond to drought and salt stresses in larch. To validate its role in drought and salt tolerance, LoNAC5 was functionally characterized in yeast [54]. The negative control yeast (pYES2-NTB) could grow on the media of SG-U + 0 mM PEG 3350, SG-U + 30 mM PEG 3350, SG-U + 60 mM PEG 3350, and SG-U + 90 mM PEG 3350, but could not grow under the conditions of SG-U + 120 mM PEG 3350 and SG-U + 135 mM PEG 3350. However, growth of yeast carrying pYES2-NTB-LoNAC5 was better than the negative control and could grow under the condition of SG-U + 120 mM PEG 3350. Similarly, the negative control yeast could grow on the culture medium plates of SG-U + 0 M NaCl, SG-U + 0.5 M NaCl, and SG-U + 1.0 M NaCl, but could not grow on the plates of SG-U + 1.3 M NaCl, SG-U + 1.5 M NaCl, and SG-U + 2.0 M NaCl. In contrast, the yeast carrying pYES2-NTB-LoNAC5 not only could grow on the culture medium of SG-U + 1.3 M NaCl, but also its growth state on the SG-U + 1.0 M NaCl plate was better than that of the negative control (Figure 5). In conclusion, the growth state of the yeast carrying pYES2-NTB-LoNAC5 was better than the yeast carrying pYES2-NTB, speculating that LoNAC5 is a positive regulatory factor for salt and drought resistance.
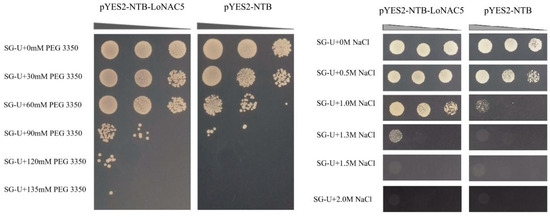
Figure 5.
Influence of LoNAC5 expression in the INVSC1 yeast strain under PEG and salt stress.
2.5. LoNAC5 Can Form Homodimers and Bind to CATGTG Motif
The yeast carrying PGBKT7-LoNAC5-F3 can grow on the QDO (selective media without Trp, Leu, HIS, and Ade) with 40 μL/L X-α-Gal and appears blue when co-transformed with pGADT7-LoNAC5 (Figure 6A), indicating that LoNAC5 can form homodimers. NAC transcription factors mainly bind to the NACRS with core sequences or “CATGTG” to regulate gene expression [55]. The NAC proteins ANAC019/055/072 in the same subgroup as LoNAC5 bound specifically to the CATGTG motif [6]. The Y187 yeast strain carrying pGADT7-LoNAC5 can grow on the TDO (selective media without Trp, Leu, and HIS) when co-transformed with pHIS2-CATGTG (Figure 6B), indicating that LoNAC5 can bind to the CATGTG motif.
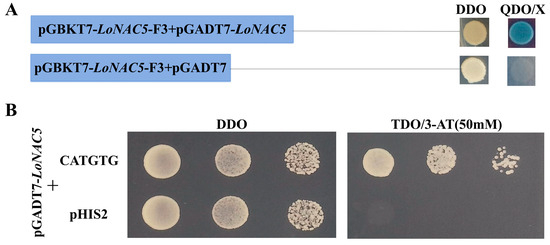
Figure 6.
Expression characteristics of LoNAC5 in yeast. (A) Homodimer analysis of LoNAC5; (B) binding of CATGTG motif to LoNAC5.
3. Discussion
Larch is an important timber tree species in northeast China. However, research on NAC transcription factors in larch remains scarce and warrants further exploration. NAC transcription factors typically exhibit a high degree of conservation at the N-terminus. This conserved domain, usually composed of approximately 150 amino acids, can frequently be partitioned into five distinct sub-domains. In contrast, their C-termini are diverse, and sometimes a transcriptional activation domain can be found therein. The OsNAC19 protein harbors a plant-specific NAC domain a in the N-terminus, and the activation domain has been detected within 181–240 aa of its C-terminal region [56]. NAC transcription factors such as VND7 [27], TaNAC8 [57], and CarNAC5 [58] exhibit highly similar structural characteristics, which are also consistent with the structure of LoNAC5: a NAM domain is present from 14 to 139 aa at the N-terminus and a transcriptional activation domain is located from 324 to 364 aa at the C-terminus (Figure 3B). Studies have revealed that NAC transcription factors have conserved intron–exon structure [59], and some NACs in rice and sorghum have been found to have two introns [60,61]. In this experiment, it was found that LoNAC5 also has two introns, and the coding region is 1164 bp. The LoNAC5-GFP fusion protein was localized in the nucleus, exhibiting typical transcription factor characteristics [58]. Collectively, these results suggest that LoNAC5, isolated from the cDNA of L. olgensis in this experiment, is a canonical transcription factor of the NAC family.
Drought and salinity, two major forms of abiotic stress, severely disrupt the growth, development, and reproduction processes of plants. As a consequence, this not only undermines the yield and quality of plants but also incurs significant economic losses [53]. NAC transcription factors exhibit a wide array of biological functions. The AtNAC3 subgroup, to which the LoNAC5 protein belongs, has been designated as the SNAC group (stress-responsive NAC) [62]. Previous research has shown that this subgroup is particularly crucial in the plant’s response to abiotic stress [21,62,63]. Through homologous analysis by constructing a phylogenetic tree, it was found that LoNAC5 and Arabidopsis ANAC019/055/072 belong to the same subgroup (Figure 1). Overexpressing any one of ANAC019/055/072 is sufficient to augment the abiotic stress tolerance capabilities of Arabidopsis thaliana [64]. Additionally, PwNAC2, a NAC transcription factor identified through NCBI sequence alignment and highly homologous to LoNAC5, has been shown to enhance the drought and salt tolerance of Arabidopsis thaliana when overexpressed [65]. The acquisition of transgenic plants through the in vitro regeneration system of larch is characterized by a protracted timeline. Concurrently, the cell embryogenesis system manifests pronounced instability, accompanied by a notably elevated incidence of somatic embryo malformations [66].
Through bioinformatics analysis, the transcription factor LoNAC5, which is related to plant abiotic stress, has been preliminarily identified. The results of RT-qPCR indicated that the expression of LoNAC5 is induced by drought and salt stress. Remarkably, yeast strains carrying LoNAC5 exhibited significantly enhanced tolerance to both drought and salt stress. These findings strongly suggest that LoNAC5 may act as a positive regulator under drought and salt stress, providing a certain theoretical basis for subsequent research. The role of LoNAC5 in the drought and salt resistance of larch is worthy of further investigation.
ABA coordinates the plant stress response and regulates complex metabolic and physiological mechanisms that are vital for survival in a dynamically changing environment [67]. MeJA is a member of the jasmonic acid family, playing an important role in plant stress response [68]. Jasmonate- and ABA-mediated signaling pathways play crucial roles in activating plants’ defense responses against both biotic and abiotic stresses [69]. ANAC019/055/072 may play a dual role in regulating ABA response and jasmonate response [70]. Additionally, ANAC019/055/072 are crucial in the signal transduction of ABA-induced leaf senescence [63]. In this research, it was revealed that the promoter of LoNAC5 harbors multiple elements associated with plant stress resistance, along with MeJA-responsive elements and ABA-responsive elements. The results of RT-qPCR indicate that the expression of LoNAC5 is not only induced by drought and salt, but also significantly altered under the treatments of ABA and MeJA. In the larch seedlings treated with MeJA, the expression of LoNAC5 was down-regulated. In contrast, after treatment with ABA, the expression of LoNAC5 was up-regulated. The regulation of LoNAC5 by MeJA and ABA may trigger a cascade of events, potentially activating downstream genes involved in osmolyte synthesis, ion homeostasis, and antioxidant defense, which are crucial for plant adaptation to abiotic stresses. Moreover, LoNAC5 expression is robust in and specific to larch needles, indicating its potential functional significance in this specific tissue. Collectively, the evidence strongly suggests that LoNAC5 may also exert a dual regulatory function in modulating both MeJA and ABA responses. It is highly likely to participate in the regulation of plant tolerance to abiotic stresses via the Jasmonate- and ABA-mediated signaling. However, further in-depth investigations are warranted to fully elucidate the role of LoNAC5 in plant resistance to drought and salt stresses.
Transcription factors can bind to specific cis-acting elements on gene promoters to regulate gene expression [71]. CATGTG is an MYC-like sequence that plays a crucial role in the dehydration- and salt-induced expression of the Arabidopsis ERD1 [72]. This sequence has important functions in many promoters that respond to stresses such as drought, salt, and abscisic acid [73]. Previous analyses of stress-inducible genes in Arabidopsis have demonstrated that, under diverse stress conditions, predominantly drought and salinity, the sequence CATGTG is pivotal in regulating gene expression [74]. Researchers have previously used the yeast one-hybrid system to isolate ANAC019/055/072, which can bind to the CATGTG motif in the promoter region of erd1 [6]. This study found that LoNAC5 can also specifically bind to the CATGTG motif in yeast. It is speculated that LoNAC5 binds to the CATGTG motif to regulate downstream genes, thereby acting as a positive regulator of drought and salt stress tolerance. Plant transcription factors are classified according to their characteristic DNA-binding domains. Given the functional significance of LoNAC5’s binding to the CATGTG motif, we further explored its potential interactions with other LoNACs. Studies have shown that even genes within the same family can be divided into different subgroups based on sequence information. Genes within the same subgroup have more similar gene structures and often occupy the same position in the regulatory network and play similar roles [17,75]. This study demonstrated that LoNAC5 is capable of forming homodimers. Given the higher structural conservation among LoNAC5 and LoNAC7/14/15, which belong to the same subgroup, it is hypothesized that LoNAC7/14/15 are more likely to form heterodimers with LoNAC5 than homodimers. These heterodimers may collaborate to participate in the plant stress-resistance process.
In summary, several critical scientific questions demand meticulous attention and in-depth investigation. Firstly, the molecular mechanisms by which LoNAC5 modulates plant responses to abiotic stresses via Jasmonate- and ABA-mediated signaling cascades should be elucidated. Secondly, the potential formation of heterodimers between LoNAC5 and LoNAC7/14/15 (members of the stress-responsive NAC subgroup) and their subsequent roles in regulating the expression of downstream target genes should be explored. Addressing these scientific queries will not only facilitate the construction of a comprehensive molecular regulatory network in larch but also enable the identification and characterization of functionally significant genes. These efforts will ultimately provide invaluable genetic resources and a robust theoretical framework for the genetic improvement of larch, thereby enhancing its adaptability to adverse environmental conditions and promoting sustainable forestry practices.
4. Materials and Methods
4.1. Isolation of LoNAC5 and LoNAC5 Promoter
Based on the transcriptome data and the L. kaempferi genome from NCBI (taxid: 54,800, GenBank assembly GCA_013171265.2 Nucleotide BLAST), cloning primers for LoNAC5 and its promoter were designed. LoNAC5 forward primers were (5′-3′): ATGGGGAGGCAGAATGCA; reverse primers were (5′-3′): CTAATAAGAAGACCTTTGTAAATACCC. The LoNAC5 promoter forward primers (5′-3′) were: CCCGGGATATGTGGTGGGTGGGTGG; reverse primers (5′-3′) were: CCATGGGTTGTTAAAATTGCTATCGATTTAAC. cDNA and gDNA from L. olgensis were used as templates for PCR amplification. After purifying the target DNA bands (E.Z.N.A.® Gel Extraction Kit, Omega Bio-Tek, Norcross, Georgia), the sample was dispatched to Sangon Biotech Co., Ltd. (Shanghai, China) for Sanger sequencing to acquire the sequence information of LoNAC5 and its promoter.
4.2. Web Tools and Software Used for Bioinformatics Analysis
A total of 15 NAC proteins of L. olgensis, including LoNAC5 (GenBank accession: MW206687–MW206691 and OQ941818–OQ941827), which were retrieved from NCBI, along with the NAC proteins of Arabidopsis thaliana obtained from the database (https://www.arabidopsis.org/browse/genefamily/NAC.jsp (accessed on 8 June 2024)), were employed to construct a neighbor-joining phylogenetic tree using the MEGA 5.0 software [76]. Subgroup classification was conducted by referring to the existing research findings, and the phylogenetic tree was refined and formatted using Evolview (https://www.evolgenius.info/evolview/ (accessed on 10 June 2024)). The NCBI CD search was utilized to predict and analyze the domains of these proteins, while MEME (https://meme-suite.org/meme/tools/meme (accessed on 10 June 2024)) was applied to analyze their motifs. GOR4 (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html (accessed on 10 November 2024)) and SwissModel (https://www.swissmodel.expasy.org/ (accessed on 10 November 2024)) were, respectively, used to predict the secondary structure and the tertiary structure of the LoNAC5 protein. The prediction of signal peptides and transmembrane domains was accomplished by SignalP 6.0 (https://services.healthtech.dtu.dk/services/SignalP-6.0/ (accessed on 10 November 2024)) and DeepTMHMM (https://dtu.biolib.com/DeepTMHMM (accessed on 10 November 2024)), respectively. Furthermore, the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 13 February 2024)) was exploited to analyze the cis-acting elements existing in the promoter region.
4.3. Analysis of LoNAC5 Promoter Activity
The LoNAC5 promoter was inserted between the XmaI and NcoI cleavage sites of the VB191103-1905rcy plant expression vector, resulting in the construction of a recombinant plasmid, designated as LoNAC5pro::GUS. Subsequently, Agrobacterium GV3101 cells were transformed with either the recombinant plasmid or the empty vector plasmid. The transformed Agrobacterium cells were then cultured in LB medium supplemented with 50 mg/L Kanamycin (Kan) and Rifampicin (Rif) at 200 rpm and 28 °C. The cultivation was continued until OD600 ≈ 0.8, at which point the cells were ready for subsequent experiments. Using one-week-old tobacco seedlings as experimental materials, they were immersed in Agrobacterium with LoNAC5pro::GUS or 35S::GUS resuspended in 1/2 MS medium (OD₆₀₀ = 0.5–0.6). After shaking for 3–4 h at 120 rpm and 25 °C, the seedlings were placed on 1/2 MS medium and incubated in the dark at 25 °C for 2 days. Then, they were put into the prepared GUS staining solution and incubated in the dark at 37 °C for 12 h. Finally, the stained tobacco seedlings were placed in 95% ethanol for decolorization.
4.4. Subcellular Localization of LoNAC5
The LoNAC5 was inserted between the BamHI and HindIII restriction sites of the plant expression vector VB191104-GFP to obtain the recombinant plasmid (VB191104-LoNAC5-GFP). Agrobacterium harboring the recombinant plasmid and empty vector plasmid were cultured in LB medium containing 50 mg/L of both Kan and Rif at 28 °C with a shaking speed of 200 rpm until OD600 ≈ 0.8. After that, the Agrobacterium was resuspended in the prepared 1/2 MS solution to OD600 = 0.5–0.6. The infection solution was then loaded into a syringe. Using the leaves of Nicotiana benthamiana seedlings that had grown for about one month as experimental materials, the infection solution was injected into the tobacco leaves through the lower epidermis. The injected tobacco plants were then incubated in the dark at 25 °C for 2–3 days. Slices were made from the injected tobacco leaves, and the GFP signals were detected using a laser-confocal fluorescence microscope.
4.5. Research on Mechanism of LoNAC5
The full-length and fragments of LoNAC5 were, respectively, inserted between the BamHI and EcoRI restriction sites of pGBKT7. After the recombinant plasmids that were confirmed to be correct by sequencing were transformed into Y2HGold yeast strain, they were spread on SD/-Trp medium and incubated upside-down at 30 °C for 1–2 days. Single colonies were picked and suspended in 10 μL of physiological saline. After the bacterial suspension was verified to be correct, the remaining bacterial suspension was spotted on SD/-Trp/-His/-Ade/X-α-Gal screening medium and incubated upside-down at 30 °C for 2–3 days to observe the growth of the yeast. After inserting LoNAC5 into the SmaI site of pGADT7 to obtain the recombinant plasmid, it was co-transformed with the pGBKT7-LoNAC5 fragment plasmid without self-activation activity into the Y2HGold yeast strain. Then, the growth states of the yeast were observed after spreading the yeast bacterial suspension on DDO and QDO/X media, respectively. The classic NAC recognition sequence (NACRS)-CATGTG was repeated three times and inserted between the EcoRI and SacI restriction sites of the pHIS2. After obtaining the recombinant plasmid, it was co-transformed with pGADT7-LoNAC5 into Y187 yeast strain. The yeast suspension was spread on DDO and TDO media, respectively. After incubating the plates upside-down at 30 °C for 3–5 days, the growth of the yeast was observed.
4.6. Functional Characterization of LoNAC5
The plasmids of pYES2-NTB-LoNAC5 (experimental group) and pYES2-NTB (control group) were, respectively, transformed into the INVSC1 yeast strain. Positive clones were picked and resuspended in 2 mL of sterile water. After dilution (100, 10−1, 10−2), the suspensions were spread on the following plates: SG-U + 0 mM PEG 3350, SG-U + 30 mM PEG 3350, SG-U + 60 mM PEG 3350, SG-U + 90 mM PEG 3350, SG-U + 120 mM PEG 3350, SG-U + 135 mM PEG 3350; SG-U + 0 M NaCl, SG-U + 0.5 M NaCl, SG-U + 1.0 M NaCl, SG-U + 1.3 M NaCl, SG-U + 1.5 M NaCl, and SG-U + 2.0 M NaCl (Merck KGaA, Darmstadt, Germany). The plates were then placed in an incubator at 30 °C. After 7 days, the plates were observed and photographed. The lithium acetate method was applied to the transformation of all the yeast strains mentioned above [27,77].
4.7. Analysis of LoNAC5 Expression in L. olgensis
L. olgensis at different growth stages (2-month-old, 4-month-old, and 6-month-old) were used as experimental materials. The roots, stems, and needles of the plants were separated and sampled. Separately, different abiotic stress treatments (28% w/v PEG 6000, 250 mmol·L−1 NaCl, 50 mmol·L−1 NaHCO3, from Merck KGaA) and hormone treatments (50 mg·L−1 IAA, GA3, MeJA, ABA and 2,4-D, from PhytoTech Labs, Lenexa, KS, USA) were applied to 2-month-old seedlings. Each treatment was applied once at 0 h, 48 h, 72 h, and 84 h, with water as the control. Sampling was carried out after 96 h of treatment [53]. Total RNA was extracted from the samples using the Universal Plant Total RNA Extraction Kit (BIOTEKE, Beijing, China) according to the manufacturer’s protocol. Subsequently, the extracted total RNA was reverse transcribed into cDNA using the ReverseScript RT reagent Kit (TaKaRa, Shiga, Japan). The synthesized cDNA was then employed as the template for subsequent experiments.
The primers for RT-qPCR were designed with Primer Premier. The forward primer was (5′-3′): CTGATGTCAATAGGACTGCAAAG, and the reverse primer was (5′-3′): ATTCCTGGCACCTTTTCATC. These primers were meticulously screened via gel electrophoresis detection. The LoTubulin primers, as reported in reference [52,53], were selected as the internal control. Using different cDNA templates of L. olgensis, samples were loaded following the 20 μL reaction system specified in the TransStart® Top Green qPCR SuperMix Top (TransGen Biotech, Beijing, China). Amplification was then performed using an ABI 7500 real-time PCR instrument. In RT-qPCR, the results were analyzed using the 2−ΔΔCt method [78].
5. Conclusions
In this study, the NAC family transcription factor LoNAC5 was isolated from L. olgensis, which comprises three exons and two introns. The length of the CDS region is 1164 bp, encoding 387 amino acids. In the LoNAC5 protein there is a NAM domain located at positions 14aa to 139aa of the N-terminus, and a transcriptional activation domain at positions 324aa to 364aa of the C-terminus. The subcellular localization of LoNAC5 is in the cell nucleus. Overall, LoNAC5 is a typical transcription factor of the NAC family. Since LoNAC5 belongs to the AtNAC3 subgroup, it can be classified as a stress-responsive NAC. Through a series of well-designed experiments, we have provided solid evidence that LoNAC5 is more strongly expressed in plant leaves, can participate in the responses to ABA and MeJA, and plays a positive regulatory role in the processes of drought and salt tolerance. The yeast experiment results showed that LoNAC5 can form a homodimer and can specifically bind to the CATGTG motif. These findings lay a solid foundation for subsequent exploration of the molecular mechanisms underlying LoNAC5-mediated regulatory pathways.
Author Contributions
Conceptualization, Q.C. and L.Z.; methodology, Q.C.; software, Q.C. and J.D.; validation, Q.C., J.D. and M.Y.; formal analysis, M.Y. and C.W.; investigation, T.Z.; resources, C.W. and Q.Z.; data curation, Q.C. and L.L.; writing—original draft preparation, Q.C.; writing—review and editing, L.Z. and H.Z.; visualization, Q.C.; supervision, Q.C., L.Z. and H.Z.; project administration, L.Z. and H.Z.; funding acquisition, H.Z. and L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Scientific and Technological Innovation 2030-Major Project of Agricultural Biological Breeding “Creation of Drought Resistant New Germplasm and Cultivation of New Varieties of Larch” (2023ZD040580204).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and can be made available on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, B.; Rizwan, H.M.; Sun, K.; Zeng, J.; Shi, M.; Guo, T.; Chen, F. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression analysis under fusarium kyushuense and drought stress conditions in passiflora edulis. Front. Plant Sci. 2022, 13, 972734. [Google Scholar] [CrossRef]
- Zafar, Z.; Fatima, S.; Bhatti, M.F. Comprehensive analyses of NAC transcription factor family in almond (prunus dulcis) and their differential gene expression during fruit development. Plants 2021, 10, 2200. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, J.P.; Yang, H.F. Identification and functional characterization of the NAC gene promoter from populus euphratica. Planta 2016, 244, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Taoka, K.; Yanagimoto, Y.; Daimon, Y.; Hibara, K.; Aida, M.; Tasaka, M. The NAC domain mediates functional specificity of cup-shaped cotyledon proteins. Plant J. 2004, 40, 462–473. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Moumeni, A.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Attia, K.; Kikuchi, S. Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (nils) generated from crossing aday selection (drought tolerant) and ir64. Mol. Genet. Genom. 2012, 287, 389–410. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; UeguchiTanaka, M.; Yoshida, K.T.; Nagato, Y.; Matsusoka, M.; Hirano, H.Y. Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 2000, 262, 1047–1051. [Google Scholar] [CrossRef]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zi, Y.; Yang, X.; Yang, X.; Zhu, L.; Cai, H.; Yin, T.; Liu, X.; Zhang, H. Genome-wide analysis of the CsAP2/ERF gene family of sweet orange (Citrus sinensis) and joint analysis of transcriptional metabolism under salt stress. Ann. Bot. 2025, mcaf 006, 39847492. [Google Scholar] [CrossRef]
- Fang, S.; Shang, X.; Yao, Y.; Li, W.; Guo, W. NST- and SND-subgroup NAC proteins coordinately act to regulate secondary cell wall formation in cotton. Plant Sci. 2020, 301, 110657. [Google Scholar] [CrossRef] [PubMed]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.; de Vries, S.C. The CUP-shaped cotyledon3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef]
- Sun, S.; Li, X.; Nie, N.; Chen, Y.; Gao, S.; Zhang, H.; He, S.; Liu, Q.; Zhai, H. Sweet potato NAC transcription factor NAC43 negatively regulates plant growth by causing leaf curling and reducing photosynthetic efficiency. Front. Plant Sci. 2023, 14, 1095977. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Pooam, M.; El-Ballat, E.M.; Jourdan, N.; Ali, H.M.; Hano, C.; Ahmad, M.; El-Esawi, M.A. SNAC3 transcription factor enhances arsenic stress tolerance and grain yield in rice (Oryza sativa L.) Through regulating physio-biochemical mechanisms, stress-responsive genes, and cryptochrome 1b. Plants 2023, 12, 2731. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, B.; Cheng, C.; Fu, B.; Qi, M.; Du, H.; Geng, S.; Zhang, X. Comparative transcriptomics analysis reveals the differences in transcription between resistant and susceptible pepper (Capsicum annuum L.) varieties in response to anthracnose. Plants 2024, 13, 527. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, M.; Manrique, S.; Cuesta, C.; Benkova, E.; Novak, O.; Colombo, L. CUP-shaped cotyledon1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. J. Exp. Bot. 2018, 69, 5169–5176. [Google Scholar] [CrossRef]
- Li, Y.; Xia, T.; Gao, F.; Li, Y. Control of plant branching by the CUC2/CUC3-DA1-UBP15 regulatory module. Plant Cell 2020, 32, 1919–1932. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Werr, W. Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol. Biol. 2005, 58, 669–685. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. Vascular-related NAC-domain7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-interacting2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef]
- Tan, T.T.; Endo, H.; Sano, R.; Kurata, T.; Yamaguchi, M.; Ohtani, M.; Demura, T. Transcription factors VND1-VND3 contribute to cotyledon xylem vessel formation. Plant Physiol. 2018, 176, 773–789. [Google Scholar] [CrossRef]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- Lu, M.; Ying, S.; Zhang, D.F.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. A maize stress-responsive NAC transcription factor, ZmsNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef]
- Jin, X.; Chai, Q.; Liu, C.; Niu, X.; Li, W.; Shang, X.; Gu, A.; Zhang, D.; Guo, W. Cotton GhNAC4 promotes drought tolerance by regulating secondary cell wall biosynthesis and ribosomal protein homeostasis. Plant J. 2024, 117, 1052–1068. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, L.Y.; Dai, J.X.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Shinde, H.; Dudhate, A.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Pearl millet stress-responsive NAC transcription factor PgNAC21 enhances salinity stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2019, 135, 546–553. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.; Wang, C.; Wang, Y.C.; Wang, L.Q. ThNAC12 from Tamarix hispida directly regulates ThPIP2.5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiol. Biochem. 2021, 163, 27–35. [Google Scholar] [CrossRef]
- Zhang, H.; Pei, Y.; Zhu, F.; He, Q.; Zhou, Y.; Ma, B.; Chen, X.; Guo, J.; Khan, A.; Jahangir, M.; et al. CaSNRK2.4-mediated phosphorylation of CaNAC035 regulates abscisic acid synthesis in pepper (Capsicum annuum L.) Responding to cold stress. Plant J. 2024, 117, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef]
- Yan, T.; Sun, M.; Su, R.; Wang, X.; Lu, X.; Xiao, Y.; Deng, H.; Liu, X.; Tang, W.; Zhang, G. Transcriptomic profiling of cold stress-induced differentially expressed genes in seedling stage of indica rice. Plants 2023, 12, 2675. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef]
- Ren, T.; Qu, F.; Morris, T.J. Hrt gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 2000, 12, 1917–1926. [Google Scholar] [CrossRef]
- Xie, Q.; Sanz-Burgos, A.P.; Guo, H.; Garcia, J.A.; Gutierrez, C. Grab proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 1999, 39, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Dogra, S.C.; Rasheed, M.S.; Healy, H.; Randles, J.W.; Rezaian, M.A. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 2005, 17, 311–325. [Google Scholar] [CrossRef]
- Wang, S.; Han, H.; Zhang, B.; Wang, L.; Wu, J.; Chen, Z.; Lin, K.; Hao, J.; Jia, R.; Zhang, Y. Identification of crucial genes and regulatory pathways in alfalfa against fusarium root rot. Plants 2023, 12, 3634. [Google Scholar] [CrossRef]
- Oh, S.K.; Lee, S.; Yu, S.H.; Choi, D. Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 2005, 222, 876–887. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeon, H.S.; Kim, H.G.; Park, O.K. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microrna164. New Phytol. 2017, 214, 343–360. [Google Scholar] [CrossRef]
- Jensen, M.K.; Rung, J.H.; Gregersen, P.L.; Gjetting, T.; Fuglsang, A.T.; Hansen, M.; Joehnk, N.; Lyngkjaer, M.F.; Col-linge, D.B. The HvNAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007, 65, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.P.; Kuo, C.H.; Lu, H.H.; Lo, H.S.; Yeh, K.W. The sweet potato NAC-domain transcription factor IbNAC1 is dynamically coordinated by the activator IbBHLH3 and the repressor IbBHLH4 to reprogram the defense mechanism against wounding. PLoS Genet. 2016, 12, e1006397. [Google Scholar] [CrossRef]
- Yoshii, M.; Shimizu, T.; Yamazaki, M.; Higashi, T.; Miyao, A.; Hirochika, H.; Omura, T. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to rice dwarf virus. Plant J. 2009, 57, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; Guignon, V.; Roux, N.; Rouard, M. Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol. Biol. 2014, 85, 63–80. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Xue, J.; Zhu, L. Expression and functional analysis of NAC transcription factors under five diverse growth stages reveal their regulatory roles during wood formation in chinese cedar (Cryptomeria Fortunei hooibrenk). Ind. Crops Prod. 2023, 197, 12. [Google Scholar] [CrossRef]
- Cao, Q.; An, P.; Zhang, S.; Wang, J.; Zhang, H.; Zhang, L. Preliminary analysis of two NAC transcription factor expression patterns in Larix olgensis. J. For. Res. 2022, 33, 601–609. [Google Scholar] [CrossRef]
- Cao, Q.; Hao, J.; Zhang, T.; Liu, L.; Xu, D.; Wang, C.; Zhao, Q.; Zhang, H.; Zhang, L. Isolation and functional analysis of the Larix olgensis LoNAC3 transcription factor gene. BMC Plant Biol. 2024, 24, 881. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Song, X.; Yue, W.; Liu, L.; Ge, F.; Wang, J. Identification and functional characterization of abiotic stress tolerance-related platz transcription factor family in barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2024, 25, 191. [Google Scholar] [CrossRef]
- He, L.; Xu, J.; Wang, Y.; Yang, K. Transcription factor ANAC074 binds to NRS1, NRS2, or MYBST1 element in addition to the NACRS to regulate gene expression. Int. J. Mol. Sci. 2018, 19, 3271. [Google Scholar] [CrossRef]
- Lin, R.; Zhao, W.; Meng, X.; Wang, M.; Peng, Y. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by magnaporthe grisea. Plant Sci. 2007, 172, 120–130. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Sun, Y.; Zhu, L.; Xu, L.; Chen, X.; Liu, B.; Yu, Y.; Wang, X.; Huang, L.; et al. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant Pathol. 2010, 74, 394–402. [Google Scholar] [CrossRef]
- Peng, H.; Cheng, H.; Yu, X.; Shi, Q.; Zhang, H.; Li, J.; Ma, H. Characterization of a chickpea (Cicer arietinum L.) NAC family gene, CarNAC5, which is both developmentally-and stress-regulated. Plant Physiol. Biochem. 2009, 47, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Cai, C.; Zhang, T.; Guo, W. Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 2009, 176, 352–359. [Google Scholar] [CrossRef]
- Ray, S.; Basnet, A.; Bhattacharya, S.; Banerjee, A.; Biswas, K. A comprehensive analysis of NAC gene family in Oryza sativa japonica: A structural and functional genomics approach. J. Biomol. Struct. Dyn. 2023, 41, 856–870. [Google Scholar] [CrossRef]
- Kadier, Y.; Zu, Y.; Dai, Q.; Song, G.; Lin, S.; Sun, Q.; Pan, J.; Lu, M. Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench]. Plant Growth Regul. 2017, 83, 301–312. [Google Scholar] [CrossRef]
- Li, X.; Li, N.; Wen, D.; Yu, J.; Hong, J.; Wu, M.; Cheng, L.; Meng, S. Identification of stress responsive NAC genes in Casuarina equisetifolia L. and its expression analysis under abiotic stresses. Agronomy 2024, 14, 535. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-as, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef]
- Han, X.; Feng, Z.; Xing, D.; Yang, Q.; Wang, R.; Qi, L.; Li, G. Two NAC transcription factors from Caragana intermedia altered salt tolerance of the transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 208. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, X.; Guo, Y.; Luo, C.; Zhang, L. Picea wilsonii transcription factor NAC2 enhanced plant tolerance to abiotic stress and participated in Rfcp1-regulated flowering time. Plant Mol. Biol. 2018, 98, 471–493. [Google Scholar] [CrossRef]
- Zhang, S.; Yan, S.; An, P.; Cao, Q.; Wang, C.; Wang, J.; Zhang, H.; Zhang, L. Embryogenic callus induction from immature zygotic embryos and genetic transformation of Larix kaempferi 3x Larix gmelinii 9. PLoS ONE 2021, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Partida, R.; Rosario, S.M.; Lozano-Juste, J. An update on crop aba receptors. Plants 2021, 10, 1087. [Google Scholar] [CrossRef]
- Mohamed, H.I.; Latif, H.H. Improvement of drought tolerance of soybean plants by using methyl jasmonate. Physiol. Mol. Biol. Plants 2017, 23, 545–556. [Google Scholar] [CrossRef]
- Bu, Q.; Jiang, H.; Li, C.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef]
- Jiang, H.; Li, H.; Bu, Q.; Li, C. The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating aba response and jasmonate response. Plant Signal. Behav. 2009, 4, 464–466. [Google Scholar] [CrossRef]
- Duval, I.; Lachance, D.; Giguere, I.; Bomal, C.; Morency, M.; Pelletier, G.; Boyle, B.; MacKay, J.J.; Seguin, A. Large-scale screening of transcription factor-promoter interactions in spruce reveals a transcriptional network involved in vascular development. J. Exp. Bot. 2014, 65, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Two different novel cis-acting elements of ERD1, a ClpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003, 33, 259–270. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Bhadouriya, S.L.; Suresh, A.; Gupta, H.; Mehrotra, S.; Gupta, D.; Mehrotra, R. In silico analysis of ccgac and catgtg cis-regulatory elements across genomes reveals their roles in gene regulation under stress. Curr. Genom. 2021, 22, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yuan, M.; Sun, B.; Zhang, D.; Zhang, J.; Li, C.; Shao, Y.; Liu, W.; Jiang, L. Evolutionary divergence and biased expression of NAC transcription factors in hexaploid bread wheat (Triticum aestivum L.). Plants 2021, 10, 382. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).