Identification and Functional Validation of the PeDHN Gene Family in Moso Bamboo
Abstract
1. Introduction
2. Results
2.1. PeDHN Gene Identification and Basic Characterization
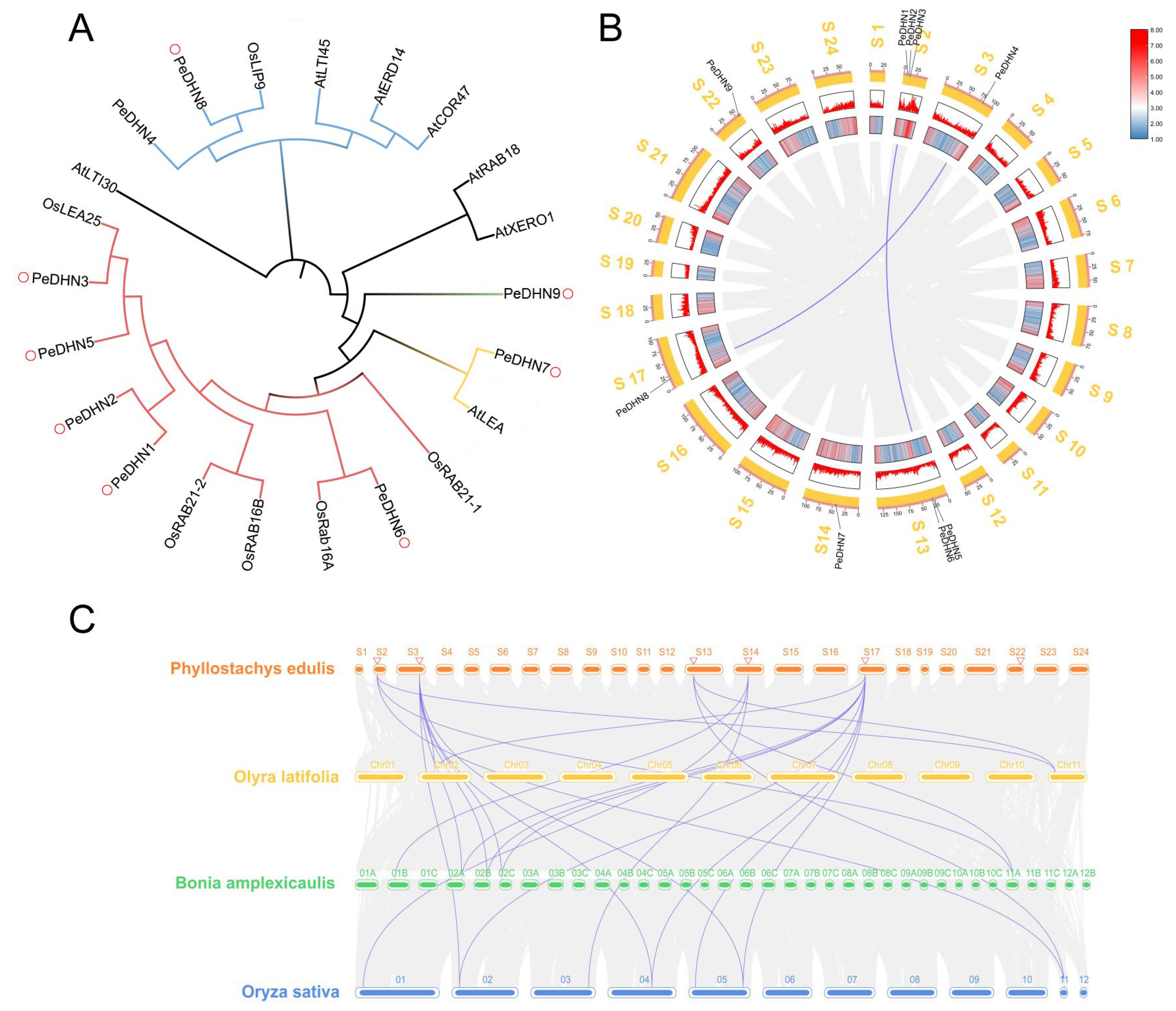
2.2. Phylogenetic Tree Construction and DHN Gene Synteny Analysis
2.3. Transcriptome Data Analysis of PeDHNs Under Various Treatment Conditions
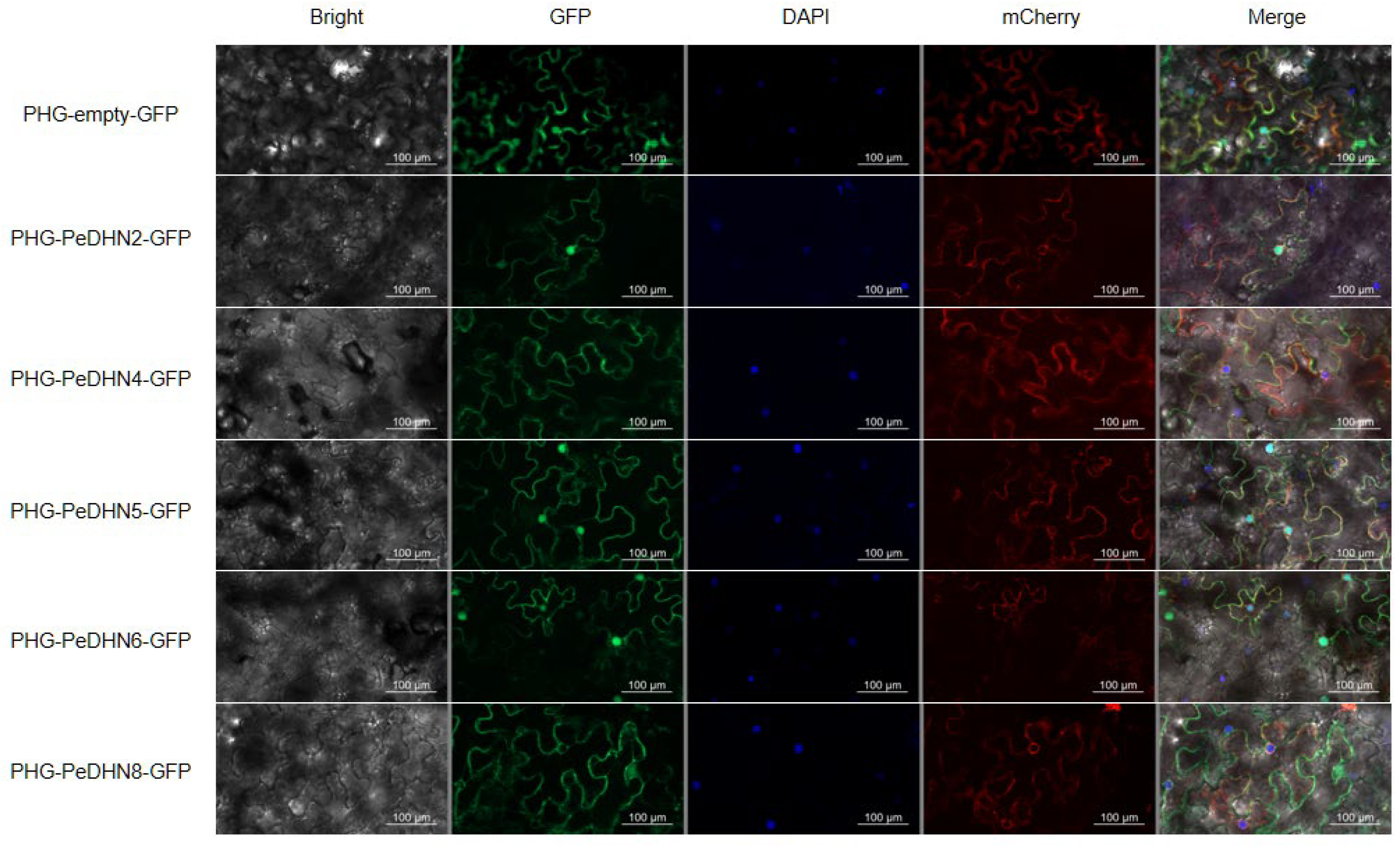
2.4. Subcellular Localization of DHN Gene Family Members
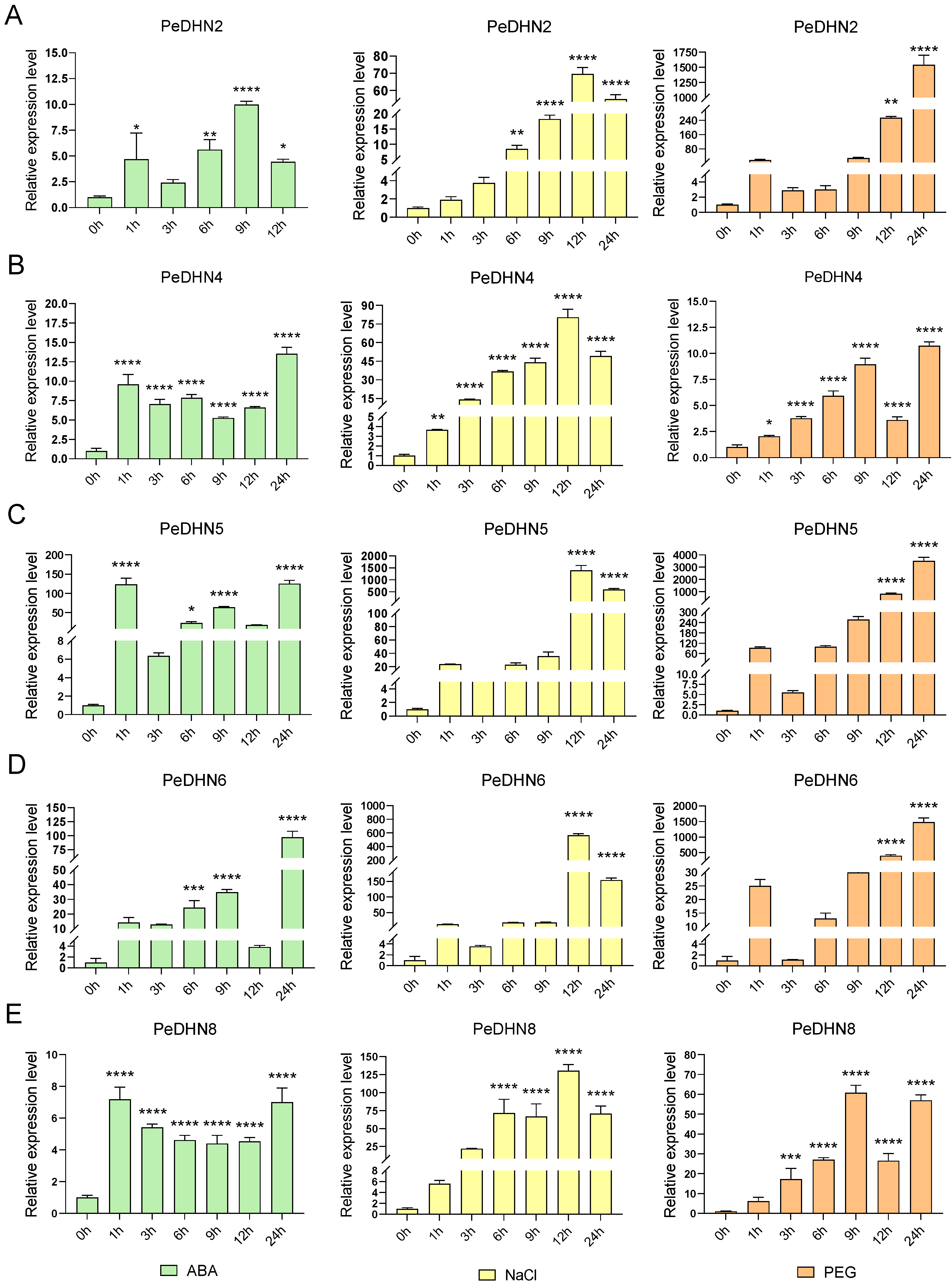
2.5. Expression Pattern Analysis of Five PeDHN Genes Under Various Stress Treatments
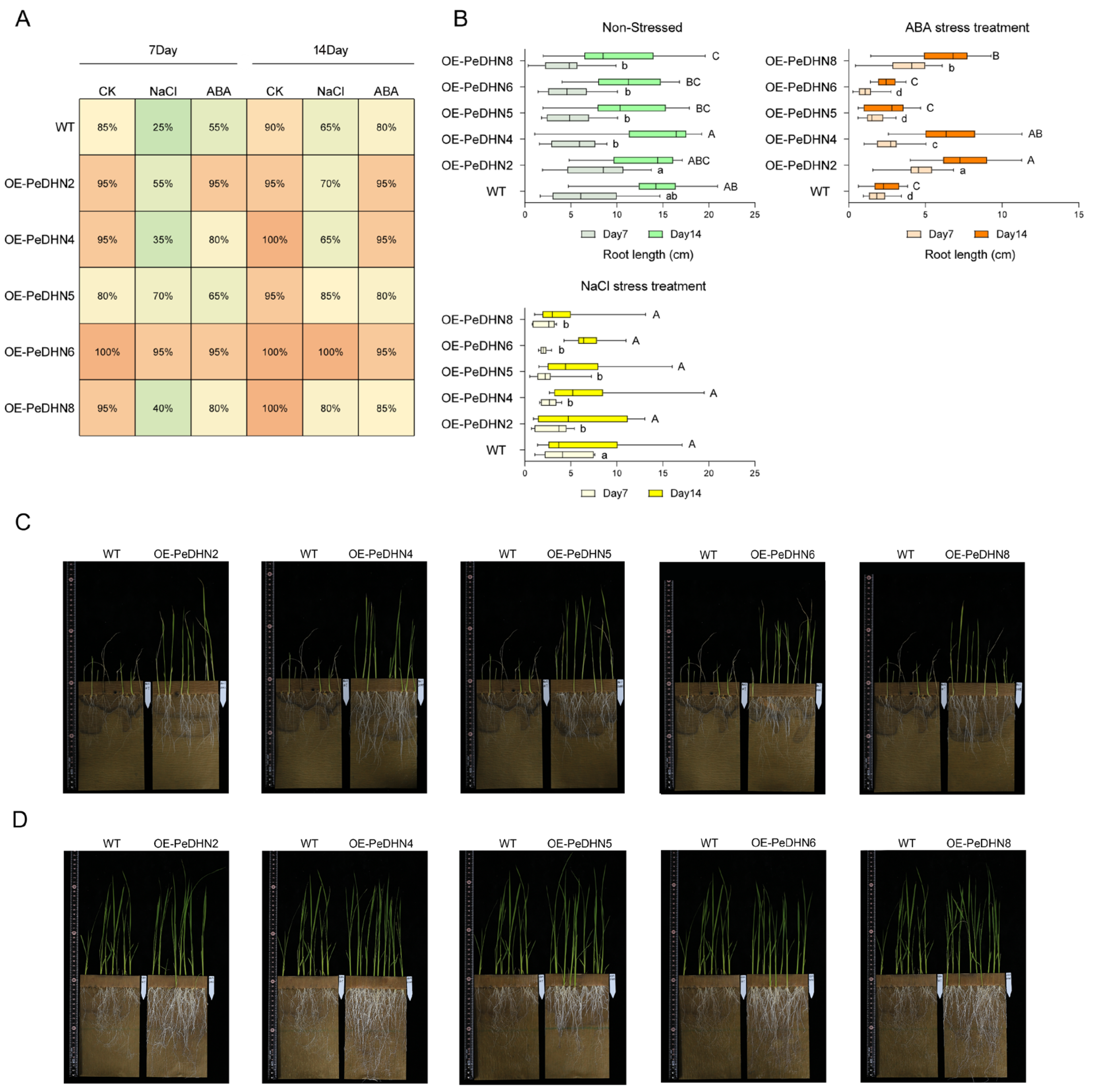
2.6. Evaluation of Stress Response Characteristics in DHN Transgenic Rice
2.7. PeDHN4 as a Target of PeABF1 in the ABA Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of the DHN Gene Family in Moso Bamboo
4.3. Protein Structures, Biochemical Properties, and Phylogenetic Analysis of PeDHNs
4.4. Transcriptome Expression Profiles of PeDHNs Under Various Treatments
4.5. Gene Cloning and Vector Construction
4.6. Subcellular Localization of PeDHNs
4.7. Real-Time Quantitative Reverse Transcription PCR of Five PeDHN Genes Following Stress Treatment
4.8. Genetic Transformation of Rice
4.9. Yeast Library Screening and Yeast One-Hybrid Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DHN | Dehydrins |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| WT | Wild type |
| ABRE | Abscisic acid response element |
| NJ | Neighbor-joining |
| PEG | Polyethylene glycol |
| MeJA | Methyl jasmonate |
| SMART | Simple modular architecture research tool |
| PFAM | Protein Families Database |
| NPS@ | Network Protein Sequence @analysis |
| iTOL | Interactive Tree Of Life |
| RT-qPCR | Real-time quantitative reverse transcription PCR |
| MEME | Multiple em for motif elicitation |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| HMMER | Hidden Markov Model Enhanced for Remote homology detection |
References
- Ahmad, Z.; Ding, Y.; Shahzad, A. (Eds.) Biotechnological Advances in Bamboo; Springer: Singapore, 2021. [Google Scholar]
- Tu, M.; Wang, W.; Yao, N.; Cai, C.; Liu, Y.; Lin, C.; Zuo, Z.; Zhu, Q. The transcriptional dynamics during de novo shoot organogenesis of Ma bamboo (Dendrocalamus latiflorus Munro): Implication of the contributions of the abiotic stress response in this process. Plant J. 2021, 107, 1513–1532. [Google Scholar] [CrossRef]
- Huang, R.; Gao, H.; Liu, J.; Li, X. WRKY transcription factors in moso bamboo that are responsive to abiotic stresses. J. Plant Biochem. Biotechnol. 2022, 31, 107–114. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, H.; Zhang, K.; Li, F.; Wu, M.; Xiang, Y. A moso bamboo transcription factor, Phehdz1, positively regulates the drought stress response of transgenic rice. Plant Cell Rep. 2021, 40, 187–204. [Google Scholar] [CrossRef]
- Li, T.; Li, H.; Zhu, C.; Yang, K.; Lin, Z.; Wang, J.; Gao, Z. Unveiling the biological function of Phyllostachys edulis FBA6 (PeFBA6) through the identification of the fructose-1, 6-bisphosphate aldolase gene. Plants 2024, 13, 968. [Google Scholar] [CrossRef]
- Wu, M.; Liu, H.; Gao, Y.; Shi, Y.; Pan, F.; Xiang, Y. The moso bamboo drought-induced 19 protein PheDi19-8 functions oppositely to its interacting partner, PheCDPK22, to modulate drought stress tolerance. Plant Sci. 2020, 299, 110605. [Google Scholar] [CrossRef]
- Zhu, C.; Lin, Z.; Yang, K.; Lou, Y.; Liu, Y.; Li, T.; Li, H.; Di, X.; Wang, J.; Sun, H.; et al. A bamboo‘PeSAPK4-PeMYB99-PeTIP4-3‘ regulatory model involved in water transport. New Phytol. 2024, 243, 195–212. [Google Scholar] [CrossRef]
- Kirungu, J.N.; Magwanga, R.O.; Pu, L.; Cai, X.; Xu, Y.; Hou, Y.; Zhou, Y.; Cai, Y.; Hao, F.; Zhou, Z.; et al. Knockdown of Gh_A05G1554 (GhDHN_03) and Gh_D05G1729 (GhDHN_04) Dehydrin genes, Reveals their potential role in enhancing osmotic and salt tolerance in cotton. Genomics 2020, 112, 1902–1915. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Wang, X.; Zhang, M.; Xi, Y.; Wang, A.; Zhu, J. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 1996, 97, 795–803. [Google Scholar] [CrossRef]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-Segments of the Wheat Dehydrin DHN-5 are Essential for the Protection of Lactate Dehydrogenase and β-Glucosidase Activities In Vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Zhang, T.; Yang, X.; Li, D. Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Sci. Rep. 2017, 7, 7361. [Google Scholar] [CrossRef]
- Richard Strimbeck, G. Hiding in plain sight: The F segment and other conserved features of seed plant SK n dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef]
- Verma, G.; Dhar, Y.V.; Srivastava, D.; Kidwai, M.; Chauhan, P.S.; Bag, S.K.; Asif, M.H.; Chakrabarty, D. Genome-wide analysis of rice dehydrin gene family: Its evolutionary conservedness and expression pattern in response to PEG induced dehydration stress. PLoS ONE 2017, 12, e0176399. [Google Scholar] [CrossRef]
- Mei, Y.; Liu, Z.; Zheng, J.; Wang, W.; Zu, Y.; Wu, Y.; Zhang, L.; Feng, R.; Shen, F. Genome-Wide Identification and Expression Pattern Analysis of the TCP Gene Family in Radish (Raphanus sativus L.). Horticulturae 2022, 8, 656. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, M.; Cui, Y.; Kong, L.; Wang, H. Genome-wide Survey of the Dehydrin Genes in Bread Wheat (Triticum aestivum L.) and its Relatives: Identification, Evolution and Expression Profiling Under Various Abiotic Stresses. BMC Genomic. 2022, 23, 73. [Google Scholar] [CrossRef]
- Jing, H.; Li, C.; Ma, F.; Ma, J.-H.; Khan, A.; Wang, X.; Zhao, L.-Y.; Gong, Z.-H.; Chen, R.-G. Genome-Wide Identification, Expression Diversication of Dehydrin Gene Family and Characterization of CaDHN3 in Pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef]
- Hara, M.; Fujinaga, M.; Kuboi, T. Radical scavenging activity and oxidative modification of citrus dehydrin. Plant Physiol. Biochem. 2004, 42, 657–662. [Google Scholar] [CrossRef]
- Zhou, M.; Peng, N.; Yang, C.; Wang, C. The Poplar (Populus trichocarpa) Dehydrin Gene PtrDHN-3 Enhances Tolerance to Salt Stress in Arabidopsis. Plants 2022, 11, 2700. [Google Scholar] [CrossRef]
- Liu, J.; Dai, M.; Li, J.; Zhang, Y.; Ren, Y.; Xu, J.; Gao, W.; Guo, S. Expression, purification, and preliminary protection study of dehydrin PicW1 from the biomass of Picea wilsonii. Front. Bioeng. Biotechnol. 2022, 10, 870672. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Huang, Z.; Zhu, P.; Zhong, X.; Qiu, J.; Xu, W.; Song, L. Transcriptome analysis of Moso bamboo (Phyllostachys edulis) reveals candidate genes involved in response to dehydration and cold stresses. Front. Plant Sci. 2022, 13, 960302. [Google Scholar] [CrossRef]
- Vilardell, J.; Goday, A.; Freire, M.A.; Torrent, M.; Martínez, M.C.; Torne, J.M.; Pagès, M. Gene sequence, developmental expression, and protein phosphorylation of RAB-17 in maize. Plant Mol. Biol. 1990, 14, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Godoy, J.A.; Lunar, R.; Torres-Schumann, S.; Rodrigo, R.M.; Godoy, J.A.; Pintor-Toro, J.A. Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol. Biol. 1994, 26, 1921–1934. [Google Scholar] [CrossRef]
- Jensen, A.B.; Goday, A.; Figueras, M.; Jessop, A.C.; Pagès, M. Phosphorylation mediates the nuclear targeting of the maize Rab17 protein. Plant J. 1998, 13, 691–697. [Google Scholar] [CrossRef]
- Lv, A.; Wen, W.; Fan, N.; Su, L.; Zhou, P.; An, Y. Dehydrin MsDHN1 improves aluminum tolerance of alfalfa (Medicago sativa L.) by affecting oxalate exudation from root tips. Plant J. 2021, 108, 441–458. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Ali, Y.; Aslam, Z.; Ashraf, M.Y.; Tahir, G.R. Effect of salinity on chlorophyll concentration, leaf area, yield and yield components of rice genotypes grown under saline environment. Int. J. Environ. Sci. Technol. 2004, 1, 221–225. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Maszkowska, J.; Szymańska, K.P.; Kasztelan, A.; Krzywińska, E.; Sztatelman, O.; Dobrowolska, G. The multifaceted regulation of SnRK2 kinases. Cells 2021, 10, 2180. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Li, F.; Mei, F.; Zhang, Y.; Li, S.; Kang, Z.; Mao, H. Genome-wide analysis of the AREB/ABF gene lineage in land plants and functional analysis of TaABF3 in Arabidopsis. BMC Plant Biol. 2020, 20, 558. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Ma, J.; Wang, X.; Haq, S.U.; Meng, Y.; Zhang, Y.; Chen, R. CaDHN4, a salt and cold stress-responsive dehydrin gene from pepper decreases abscisic acid sensitivity in Arabidopsis. Int. J. Mol. Sci. 2019, 21, 26. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, D.; Lu, X.; Zhang, L.; Yu, Z.; Lv, H.; Zhang, H. Characterisation of an SKn-type Dehydrin Promoter from Wheat and Its Responsiveness to Various Abiotic and Biotic Stresses. Plant Mol. Biol. Rep. 2014, 32, 664–678. [Google Scholar] [CrossRef]
- Dutta, M.; Raturi, V.; Gahlaut, V.; Kumar, A.; Sharma, P.; Verma, V.; Gupta, V.K.; Sood, S.; Zinta, G. The interplay of DNA methyltransferases and demethylases with tuberization genes in potato (Solanum tuberosum L.) genotypes under high temperature. Front. Plant Sci. 2022, 13, 933740. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Zhang, L. Identification of upstream transcription factors and an interacting PP2C protein of dehydrin WZY2 gene in wheat. Plant Signal. Behav. 2019, 14, 1678370. [Google Scholar] [CrossRef]
- Fiallos-Salguero, M.S.; Li, J.; Li, Y.; Xu, J.; Fang, P.; Wang, Y.; Zhang, L.; Tao, A. Identification of AREB/ABF Gene Family Involved in the Response of ABA under Salt and Drought Stresses in Jute (Corchorus olitorius L.). Plants 2023, 12, 1161. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Zhang, Q.; Yan, X.; Zuo, C.; Song, X.; Yuan, M. Comprehensive identification of GSK3 gene family in celery and functional characterization of AgSK22 involvement in salt stress. Veg. Res. 2023, 3, 23. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer Premier: Program for Design of Degenerate Primers from a Protein Sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Bai, Y.; Cai, M.; Mu, C.; Zheng, H.; Cheng, Z.; Xie, Y.; Gao, J. Integrative analysis of exogenous auxin mediated plant height regulation in Moso bamboo (Phyllostachys edulis). Ind. Crops Prod. 2023, 200, 116852. [Google Scholar] [CrossRef]
- Iwema, C. LibGuides: HSLS MolBio Available Software; SnapGene. 2020. Available online: https://hsls.libguides.com/MBISsoftware/snapgene (accessed on 24 October 2023).



| Protein | Y Segment | S Segment | K Segment | Subclass |
|---|---|---|---|---|
| PeDHN1 | 1 segment VDEYGNP | 1 segment SSSSSSS | 2 segments EKKGLMDKIKEKLPG, EKKGIKEKIKEKLPG | YK2S |
| PeDHN2 | 1 segment VDEYGNP | 1 segment SSSSSSS | 2 segments EKKGLMDKIKEKLPG, EKKGIKEKIKEKLPG | YK2S |
| PeDHN3 | 1 segment VDEYGNP | 1 segment SSSSSSS | 2 segments EKKGVMEKIKEKLPG, EKKGIKEKIKEKLPG | YK2S |
| PeDHN4 | 0 segments | 1 segment SSSSSSSSS | 2 segments EKKGILGKIMEKLPG, EKKGIKEKIKEKLPG | K2S |
| PeDHN5 | 1 segment VDEYGNP | 1 segment SSSSSSS | 2 segments EKKGVMEKIKEKLPG, EKKGIKEKIKEKLPG | YK2S |
| PeDHN6 | 1 segment VDGSGNP | 1 segment SSSSSSS | 2 segments EKKGIMDKIKEKLPG, EKKGIKEKIKEKLPG | YK2S |
| PeDHN7 | 2 segments VDQYGNP | 1 segment SSSSSSS | 2 segments EKKGIMEKIKEKLPG, EKKSIKDKIKEKLPG | Y2K2S |
| PeDHN8 | 0 segment | 1 segment SSSSSSSSS | 2 segments EKKGILGKIMEKLPG, EKKGIKEKIKEKLPG | K2S |
| PeDHN9 | 0 segment | 1 segment SSSSSSS | 2 segments EKKGAMDKIKEKLPG, EKKGIKEKIKEKLPG | K2S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Chang, Y.; Zhang, W.; Chu, T.; Tian, H.; Deng, Y.; Jiang, Z.; Ma, Y.; Hu, T. Identification and Functional Validation of the PeDHN Gene Family in Moso Bamboo. Plants 2025, 14, 1520. https://doi.org/10.3390/plants14101520
Ye Y, Chang Y, Zhang W, Chu T, Tian H, Deng Y, Jiang Z, Ma Y, Hu T. Identification and Functional Validation of the PeDHN Gene Family in Moso Bamboo. Plants. 2025; 14(10):1520. https://doi.org/10.3390/plants14101520
Chicago/Turabian StyleYe, Yaqin, Yanting Chang, Wenbo Zhang, Tiankui Chu, Hanchen Tian, Yayun Deng, Zehui Jiang, Yanjun Ma, and Tao Hu. 2025. "Identification and Functional Validation of the PeDHN Gene Family in Moso Bamboo" Plants 14, no. 10: 1520. https://doi.org/10.3390/plants14101520
APA StyleYe, Y., Chang, Y., Zhang, W., Chu, T., Tian, H., Deng, Y., Jiang, Z., Ma, Y., & Hu, T. (2025). Identification and Functional Validation of the PeDHN Gene Family in Moso Bamboo. Plants, 14(10), 1520. https://doi.org/10.3390/plants14101520







