QTL-Seq and Fine-Mapping Analyses Identify QTL and Candidate Genes Controlling Snake-like Pod Surface Trait in Vegetable Cowpea Yardlong Bean
Abstract
1. Introduction
2. Results
2.1. Differences in Pod Appearance and Morphology Between Thua Ngu and Raya
2.2. Differences in Pod Chemical Contents Between Thua Ngu and Raya
2.3. Mendelian Inheritance of Snake-like Pod in F2 Population
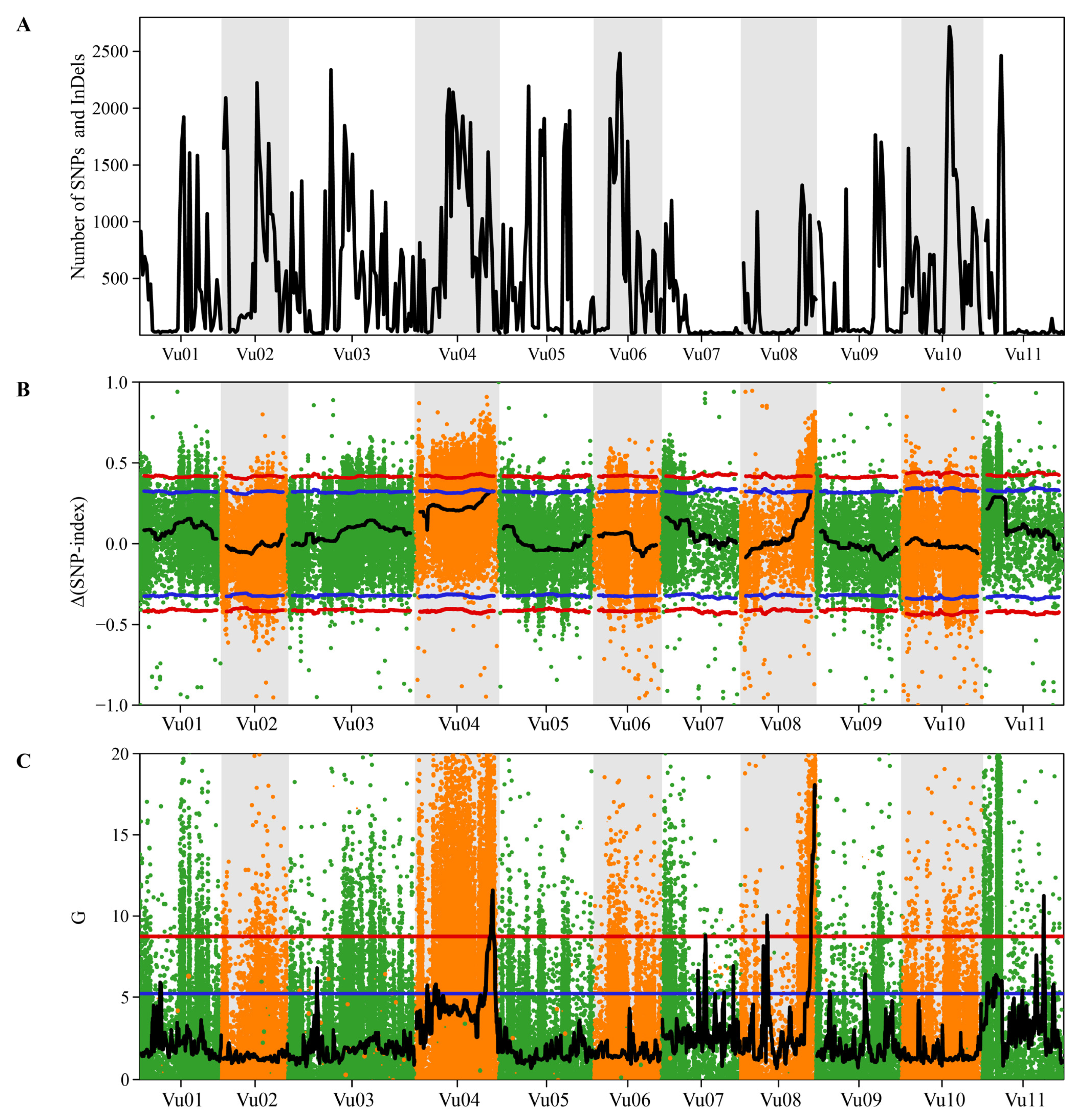
2.4. QTL-Seq Identified a Major Locus Controlling Snake-like Pod Surface Trait
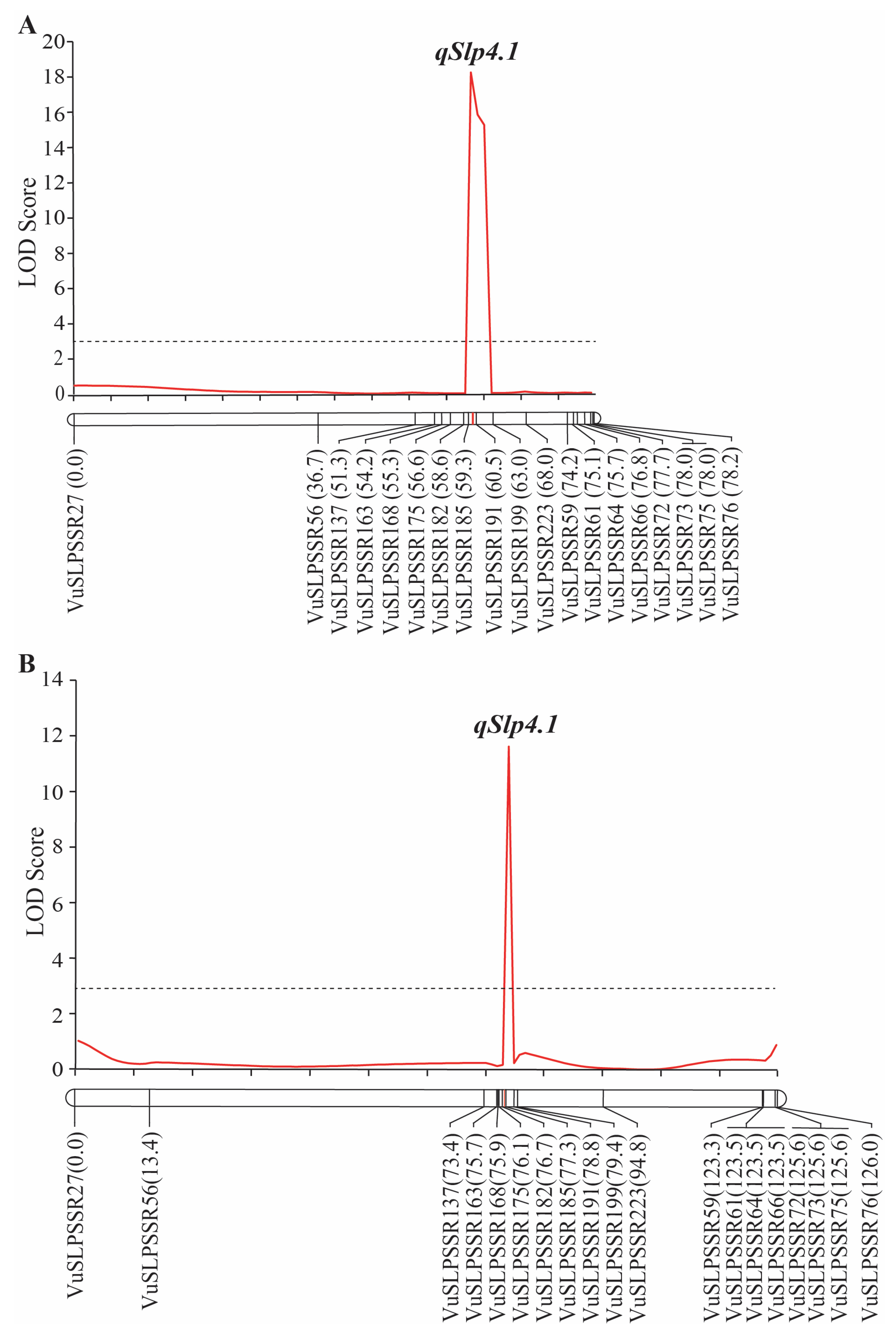
2.5. Fine-Mapping of qSlp4.1
2.6. Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Characterization of Pod Morphology
4.2. Determination of Pod Fiber Content
4.3. Population Mapping, Phenotyping of Pod Surface, and DNA Extraction
4.4. QTL Analysis for Pod Surface Trait by Bulked Segregant Analysis Coupled to Whole-Genome Sequencing (QTL-Seq)
4.5. Development of New DNA Markers and Fine-Mapping of QTL Controlling Pod Surface Trait
4.6. Identification and Sequencing of Candidate Genes
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QTL | Quantitative trait locus |
| ICIM | Inclusive composite interval mapping |
| FE-SEM | Field-emission scanning electron microscope |
| SSR | Simple sequence repeat |
| BLASTN | Basic local alignment search tool for nucleotide sequences |
References
- Singh, B.B. Cowpea [Vigna unguiculata (L.) Walp.]. In Genetic Resources Chromosome Engineering and Crop Improvement, 2nd ed.; Singh, R.J., Jauhar, P.P., Eds.; Taylor & Francis, LLC: Abingdon, UK, 2005; pp. 117–161. [Google Scholar]
- Verdcourt, B. Studies in the Leguminosae–Papilionoideae for the ‘Flora of Tropical East Africa’: IV. Kew Bull. 1970, 24, 507–569. [Google Scholar] [CrossRef]
- Kongjaimun, A.; Somta, P.; Tomooka, N.; Kaga, A.; Vaughan, D.A.; Srinives, P. QTL mapping of pod tenderness and total soluble solid in yardlong bean [Vigna unguiculata (L.) Walp. subsp. unguiculata cv.-gr. sesquipedalis]. Euphytica 2013, 189, 217–223. [Google Scholar] [CrossRef]
- Steele, W.M.; Mehra, K.L. Structure, evolution, and adaptation to farming systems and environments in Vigna. In Advances in Legume Sciences, 2nd ed.; Summerfield, R.J., Bunting, A.H., Eds.; Royal Botanic Gardens: Kew, UK, 1980; pp. 393–404. [Google Scholar]
- Wu, X.; Hu, Z.; Zhang, Y.; Li, M.; Liao, N.; Dong, J.; Wang, B.; Wu, J.; Wu, X.; Wang, Y.; et al. Differential selection of yield and quality traits has shaped genomic signatures of cowpea domestication and improvement. Nat. Genet. 2024, 56, 992–1005. [Google Scholar] [CrossRef] [PubMed]
- Lazaridi, E.; Kapazoglou, A.; Gerakari, M.; Kleftogianni, K.; Passa, K.; Sarri, E.; Papasotiropoulos, V.; Tani, E.; Bebeli, P.J. Crop landraces and indigenous varieties: A valuable source of genes for plant breeding. Plants 2024, 13, 758. [Google Scholar] [CrossRef] [PubMed]
- Suanum, W.; Somta, P.; Kongjaimun, A.; Yimram, T.; Kaga, A.; Tomooka, N.; Takahashi, Y.; Srinives, P. Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 2016, 36, 80. [Google Scholar] [CrossRef]
- Watcharatpong, P.; Kaga, A.; Chen, X.; Somta, P. Narrowing down a major QTL region conferring pod fiber contents in yardlong bean (Vigna unguiculata), a vegetable cowpea. Genes 2020, 11, 363. [Google Scholar] [CrossRef]
- Kader, A.A. Maturity, ripening, and quality relationships of fruit-vegetable. Acta Hort. 1996, 434, 249–256. [Google Scholar] [CrossRef]
- Ozparpucu, M.; Ruggeberg, M.; Gierlinger, N.; Cesarino, I.; Vanholme, R.; Boerjan, W.; Burgert, I. Unravelling the impact of lignin on cell wall mechanics: A comprehensive study on young poplar trees downregulated for CINNAMYL ALCOHOL DEHYDROGENASE (CAD). Plant J. 2017, 91, 480–490. [Google Scholar] [CrossRef]
- Ozparpucu, M.; Gierlinger, N.; Cesarino, I.; Burgert, I.; Boerjan, W.; Ruggeberg, M. Significant influence of lignin on axial elastic modulus of poplar wood at low microfibril angles under wet conditions. J. Exp. Bot. 2019, 70, 4039–4047. [Google Scholar] [CrossRef]
- Selig, M.; Walz, K.; Lauer, J.; Rolauffs, C.; Hart, M.L. Therapeutic modulation of cell morphology and phenotype of diseased human cells towards a healthier cell state using lignin. Polymers 2023, 15, 3041. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. Plant fructokinases: Evolutionary, developmental, and metabolic aspects in sink tissues. Front. Plant Sci. 2018, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Stein, O.; Secchi, F.; German, M.A.; Damari-Weissler, H.; Aloni, R.; Holbrook, N.M.; Zwieniecky, M.A.; Granot, D. The tomato cytosolic fructokinase FRK1 is important for phloem fiber development. Biol. Plant. 2018, 62, 353–361. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 71447–71469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zou, W.; Lin, P.; Wang, Z.; Chen, Y.; Yang, X.; Zhao, W.; Zhang, Y.; Wang, D.; Que, Y.; et al. Evolution and function of MADS-box transcription factors in plants. Int. J. Mol. Sci. 2024, 25, 13278. [Google Scholar] [CrossRef]
- Cosio, C.; Ranocha, P.; Francoz, E.; Burlat, V.; Zheng, Y.; Perry, S.E.; Ripoll, J.J.; Yanofsky, M.; Dunand, C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2017, 213, 250–263. [Google Scholar] [CrossRef]
- Vasantrao, J.M.; Baruah, I.K.; Panda, D.; Bhattacharjee, M.; Acharjee, S.; Sarmah, B.K. Transcript profiling of chickpea pod wall revealed the expression of floral homeotic gene AGAMOUS-like X2 (CaAGLX2). Mol. Biol. Rep. 2019, 46, 5713–5722. [Google Scholar] [CrossRef]
- Kong, X.; Wang, F.; Geng, S.; Guan, J.; Tao, S.; Jia, M.; Sun, G.; Wang, Z.; Wang, K.; Ye, X.; et al. The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 2022, 20, 75–88. [Google Scholar] [CrossRef]
- Li, H.; Liang, W.; Jia, R.; Yin, C.; Zong, J.; Kong, H.; Zhang, D. The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 2010, 20, 299–313. [Google Scholar] [CrossRef]
- Hsu, H.F.; Chen, W.H.; Shen, Y.H.; Hsu, W.H.; Mao, W.T.; Yang, C.H. Multifunctional evolution of B and AGL6 MADS box genes in orchids. Nat. Commun. 2021, 12, 902. [Google Scholar] [CrossRef]
- Schomburg, D.; Schomburg, I. Gibberellin-44 dioxygenase. In Class 1 Oxidoreductases XI. Springer Handbook of Enzymes; Springer: Berlin/Heidelberg, Germany, 2006; Volume 26. [Google Scholar] [CrossRef]
- Zanewich, K.P.; Rood, S.B. Gibberellins and heterosis in crops and trees: An integrative review and preliminary study with brassica. Plants 2020, 9, 139. [Google Scholar] [CrossRef]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J.M. Plant development and crop yield: The role of gibberellins. Plants 2022, 11, 2650. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Giraldo, L.; García-Martínez, J.L.; Moritz, T.; López-Díaz, I. Flowering in tobacco needs gibberellins but is not promoted by the levels of active GA1 and GA4 in the apical shoot. Plant Cell Physiol. 2007, 48, 615–625. [Google Scholar] [CrossRef]
- Paopun, Y.; Thanomchat, P. Freeze drying versus chemical fixation technique for scanning electron microscope of succulent and aquatic plant leaves. Microsc. Microanal. Res. 2017, 1, 28–32. [Google Scholar] [CrossRef]
- Chai, M.; Zhou, C.; Molina, I.; Fu, C.; Nakashima, J.; Li, G.; Zhang, W.; Park, J.; Tang, Y.; Jiang, Q.; et al. A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. USA 2016, 113, 6997–7002. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.A.; Ye, G.N.; Weeden, N.F.; Reisch, B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994, 12, 6–13. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R package for bulk segregant analysis with next-generation sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef] [PubMed]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Yundaeng, C.; Somta, P.; Chen, J.; Yuan, X.; Chankaew, S.; Chen, X. Fine mapping of QTL conferring Cercospora leaf spot disease resistance in mungbean revealed TAF5 as candidate gene for the resistance. Theor. Appl. Genet. 2021, 134, 701–714. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Van Os, H.; Stam, P.; Visser, R.G.; Van Eck, H.J. RECORD: A novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 2005, 112, 30–40. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics 2007, 175, 361–374. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, version 4.2.0; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]


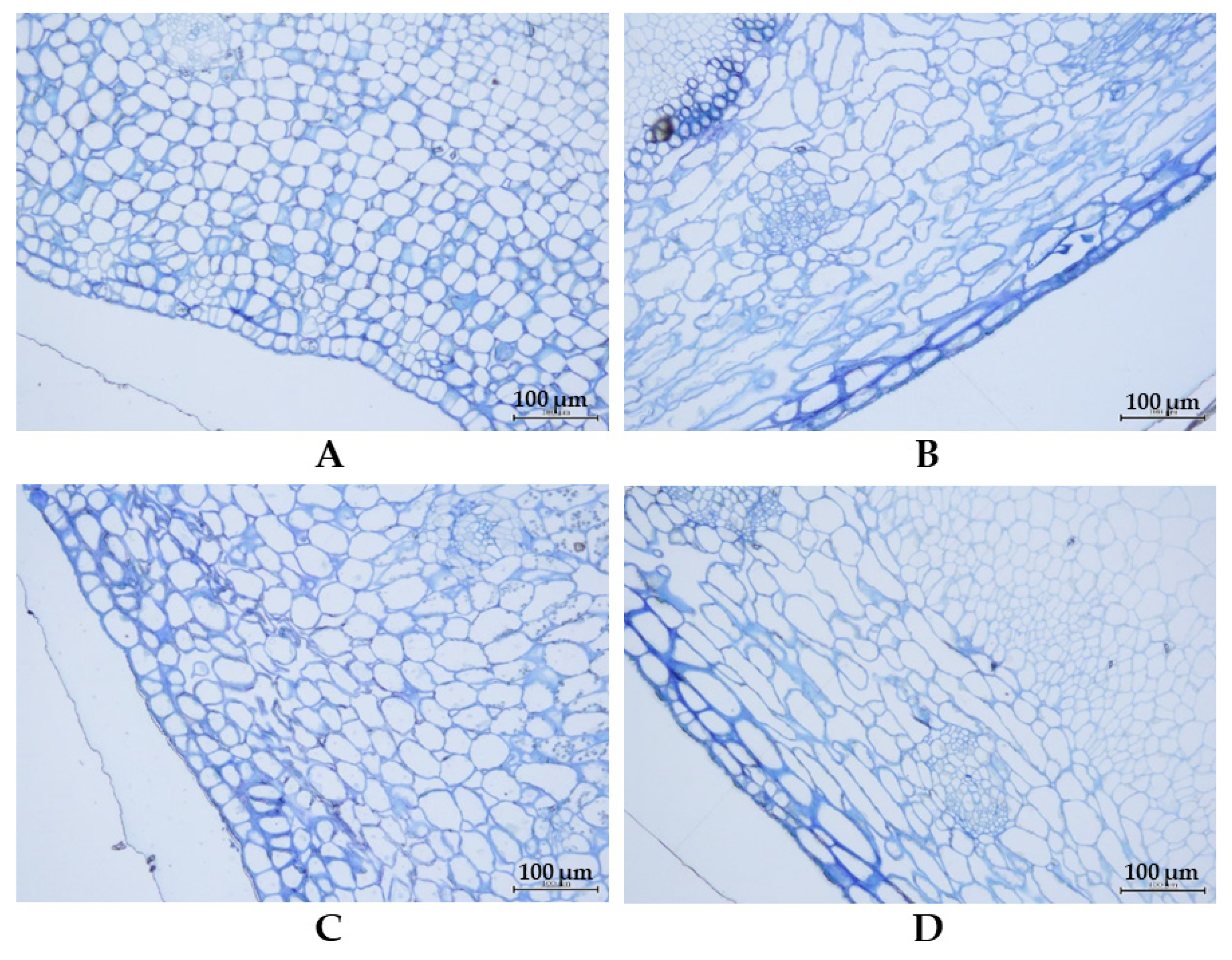
| Type of Fiber | Stage of Pod | Thua Ngu | Raya | t-Test | p-Value | ||
|---|---|---|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | ||||
| Cellulose | Immature | 9.28–10.88 | 9.84 ± 0.70 | 10.17–11.36 | 10.70 ± 0.53 | −2.39 | 0.0378 |
| Mature | 12.73–21.32 | 16.61 ± 3.48 | 18.22–22.53 | 20.68 ± 1.86 | −2.52 | 0.0304 | |
| Hemicellulose | Immature | 4.58–5.77 | 4.94 ± 0.44 | 3.41–4.67 | 4.33 ± 0.47 | 2.35 | 0.0406 |
| Mature | 7.41–12.73 | 10.05 ± 1.90 | 8.90–12.29 | 11.12 ± 1.49 | −1.08 | 0.3035 | |
| Lignin | Immature | 0.07–0.64 | 0.34 ± 0.24 | 0.35–0.95 | 0.70 ± 0.25 | −2.50 | 0.0316 |
| Mature | 0.76–1.43 | 1.10 ± 0.26 | 2.94–4.44 | 3.81 ± 0.53 | −11.25 | 0.0005 | |
| Population | No. of Plants | No. Observed Plants 1 | No. Expected Plants 1 | χ2 Value | p-Value |
|---|---|---|---|---|---|
| F2 | 227 | 56:129:42 | 1:2:1 | 5.96 | 0.05 |
| F2:3 | 260 | 59:134:67 | 1:2:1 | 0.74 | 0.69 |
| Sample | Total Reads | Raw Data (Gb) | Clean Reads | Mapping Rate (%) | Genome Coverage (%) | Average Depth |
|---|---|---|---|---|---|---|
| Ngu | 157,417,310 | 23.6 | 146,209,198 | 84.5 | 97.39 | 30.49 |
| Raya | 141,492,570 | 21.2 | 130,397,263 | 84.3 | 92.08 | 35.64 |
| SLPD | 206,626,290 | 21.0 | 191,749,197 | 89.4 | 97.18 | 40.47 |
| NPD | 169,252,490 | 25.4 | 156,981,684 | 89.4 | 97.20 | 32.92 |
| Method | Chromosome | Position (bp) | Length (bp) | No. of SNPs and InDels | Peak of Δ(SNP-Index) | Maximum of G-Stat |
|---|---|---|---|---|---|---|
| Δ(SNP-index) | 4 | 36,200,001–42,731,077 | 6,531,076 | 3577 | 0.3253 | - |
| G′-statistic | 4 | 38,000,001–40,500,000 | 2,499,999 | 1083 | - | 11.5729 |
| 7 | 22,100,001–23,100,000 | 999,999 | 22 | - | 8.7993 | |
| 8 | 12,800,001–14,200,000 | 1,399,999 | 17 | - | 10.0492 | |
| 35,300,001–38,363,498 | 3,063,497 | 757 | - | 18.0799 | ||
| 11 | 30,700,001–32,000,000 | 1,299,999 | 19 | - | 11.244 |
| Population | Location (cM) | Marker Interval | LOD Score | Percentage of Variance Explained by QTL | Additive Effect | Dominant Effect |
|---|---|---|---|---|---|---|
| F2 | 9.0 | VuSLPSSR185–VuSLPSSR191 | 18.19 | 31.40 | 0.45 | −0.28 |
| F2:3 | 77.00 | VuSLPSSR182–VuSLPSSR185 | 11.63 | 19.24 | 0.42 | −0.06 |
| Gene | Location on Cowpea Chromosome 4 | Encoded Protein | Homologous Gene in Arabidopsis thaliana |
|---|---|---|---|
| Vigun04g163400 | 38729477..38732599 | Sugar kinase | AT5G51830 (FRK1, FRUCTOKINASE 1, FRK7, and FRUCTOKINASE 7) |
| Vigun04g163600 | 38737147..38745823 | MADS-box protein | AT2G45650 (AGL6 and AGAMOUS-LIKE 6) |
| Vigun04g163700 | 38794766..38796717 | Gibberellin A44 oxidase | AT4G25420 (GA20OX1) |
| Vigun04g163800 | 38805392..38806670 | Carboxylesterase 9-related protein | AT5G62180 (CXE20 and CARBOXYLESTERASE 20) |
| Vigun04g163900 | 38812815..38814262 | Carboxylesterase 9-related protein | AT5G62180 (CXE20 and CARBOXYLESTERASE 20) |
| Vigun04g164000 | 38820160..38821585 | Carboxylesterase 9-related protein | AT5G62180 (CXE20 and CARBOXYLESTERASE 20) |
| Vigun04g164100 | 38826706..38830585 | Pentatricopeptide repeat | AT1G80880 (TETRATRICOPEPTIDE REPEAT (TPR)-LIKE SUPERFAMILY PROTEIN) |
| Vigun04g164200 | 38830603..38832039 | Carboxylesterase 9-related protein | AT5G62180 (CXE20 and CARBOXYLESTERASE 20) |
| Vigun04g164300 | 38848168..38852702 | Hypothetical protein | AT5G51800 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thepphomwong, K.; Srichan, M.; Deeroum, A.; Laosatit, K.; Somta, P. QTL-Seq and Fine-Mapping Analyses Identify QTL and Candidate Genes Controlling Snake-like Pod Surface Trait in Vegetable Cowpea Yardlong Bean. Plants 2025, 14, 1447. https://doi.org/10.3390/plants14101447
Thepphomwong K, Srichan M, Deeroum A, Laosatit K, Somta P. QTL-Seq and Fine-Mapping Analyses Identify QTL and Candidate Genes Controlling Snake-like Pod Surface Trait in Vegetable Cowpea Yardlong Bean. Plants. 2025; 14(10):1447. https://doi.org/10.3390/plants14101447
Chicago/Turabian StyleThepphomwong, Khwanruedee, Makawan Srichan, Artitaya Deeroum, Kularb Laosatit, and Prakit Somta. 2025. "QTL-Seq and Fine-Mapping Analyses Identify QTL and Candidate Genes Controlling Snake-like Pod Surface Trait in Vegetable Cowpea Yardlong Bean" Plants 14, no. 10: 1447. https://doi.org/10.3390/plants14101447
APA StyleThepphomwong, K., Srichan, M., Deeroum, A., Laosatit, K., & Somta, P. (2025). QTL-Seq and Fine-Mapping Analyses Identify QTL and Candidate Genes Controlling Snake-like Pod Surface Trait in Vegetable Cowpea Yardlong Bean. Plants, 14(10), 1447. https://doi.org/10.3390/plants14101447







