Genome-Wide Identification of the PHR Gene Family in Six Cucurbitaceae Species and Its Expression Analysis in Cucurbita moschata
Abstract
1. Introduction
2. Results
2.1. Identification of PHR Genes in Cucurbitaceae
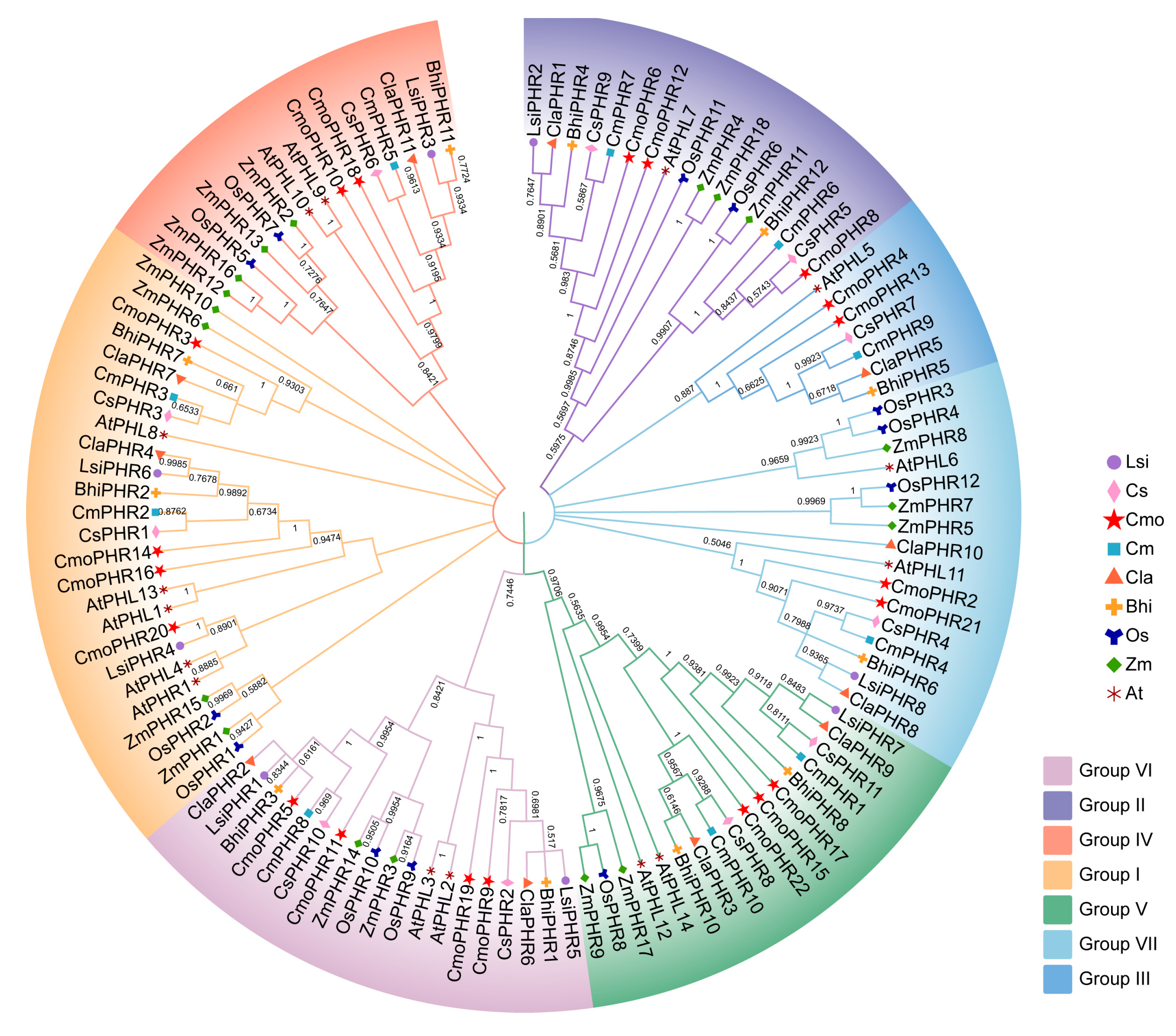
2.2. Evolutionary Analysis of the PHR Gene Family in Cucurbitaceae
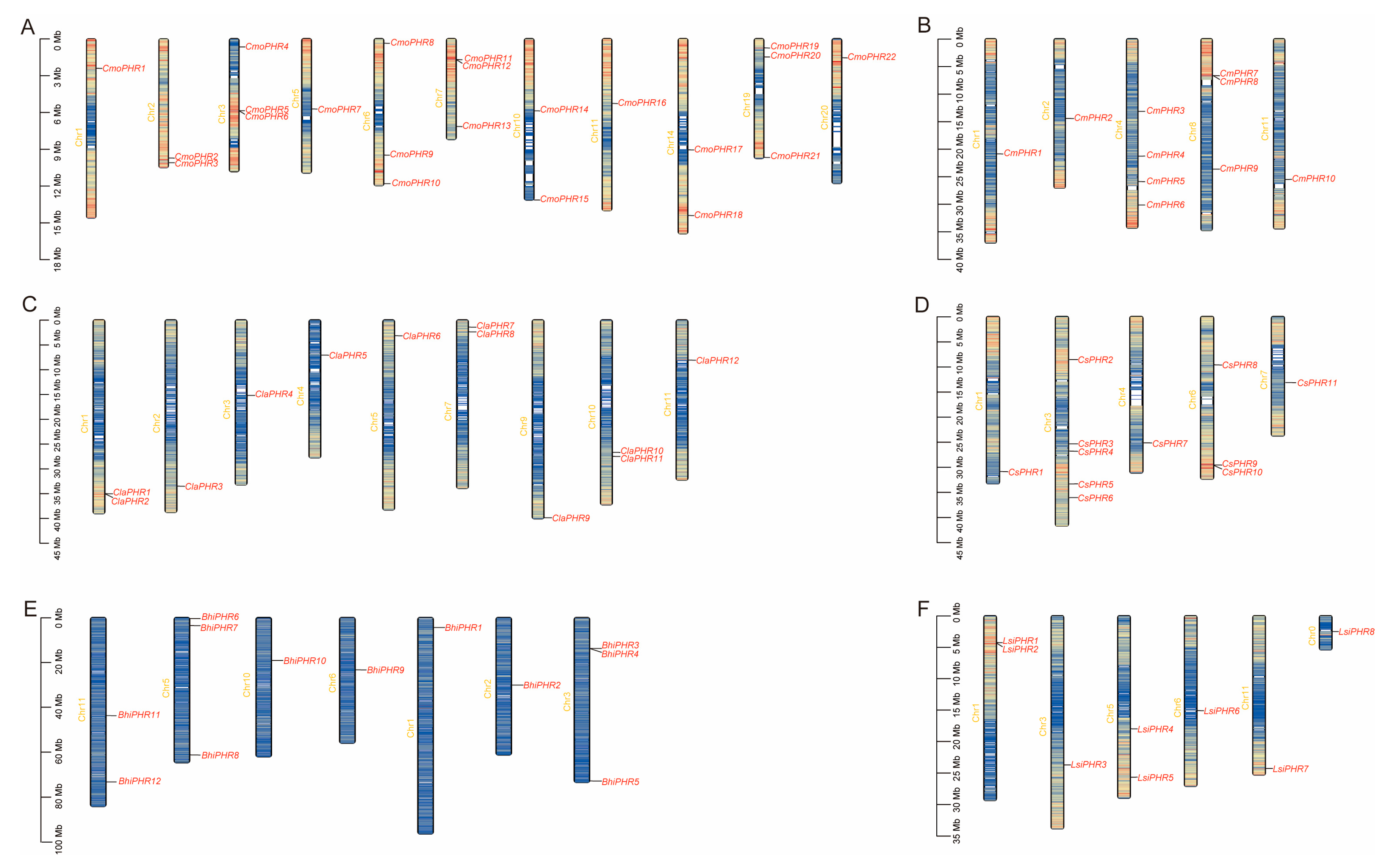
2.3. Chromosomal Distribution Heterogeneity of Cucurbitaceae PHR Genes
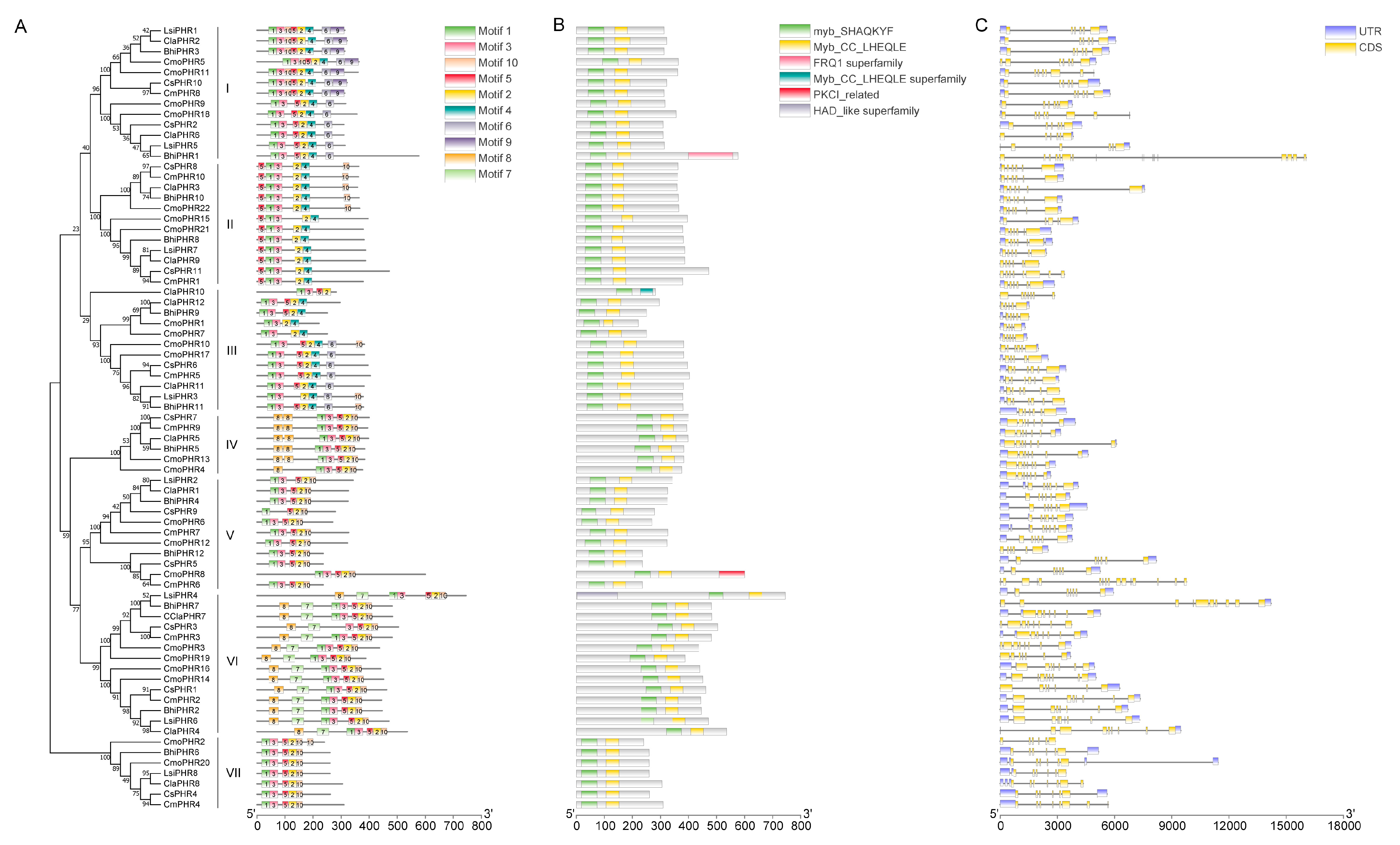
2.4. Gene Structures, Protein Motifs, and Conserved Domains of Cucurbitaceae PHR Genes
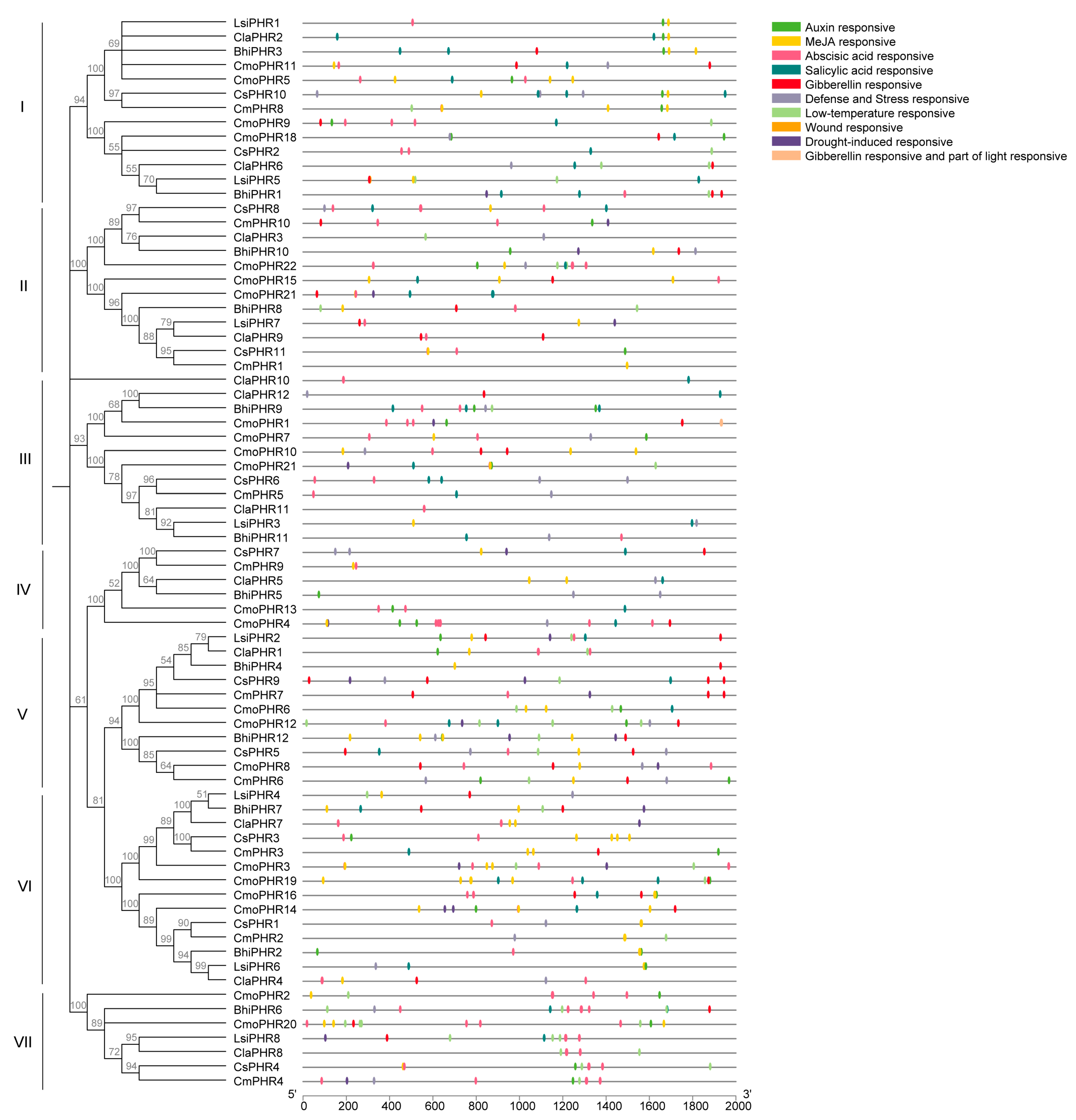
2.5. Cis-Regulatory Element Profiling in Cucurbitaceae PHR Promoters
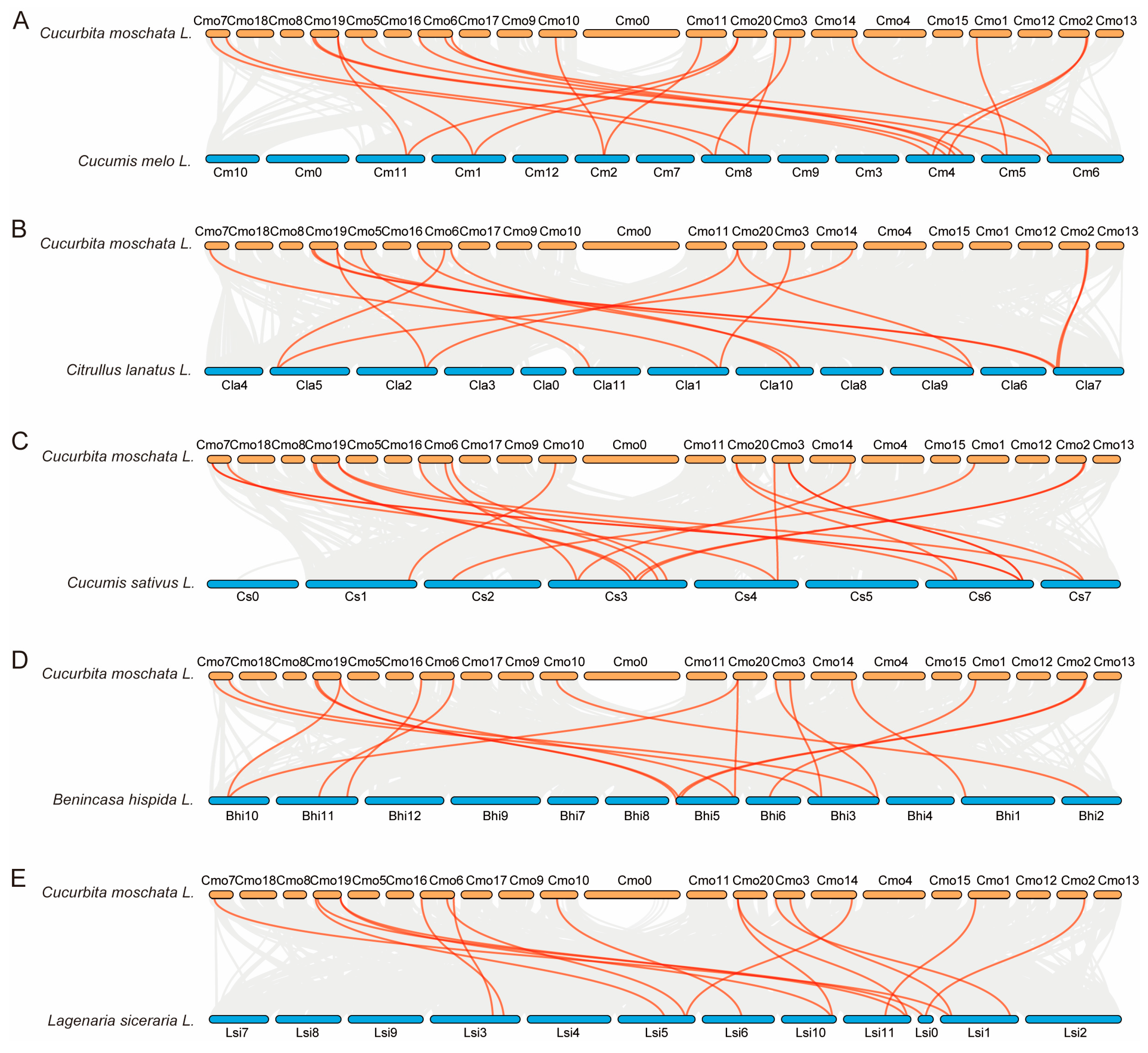
2.6. Duplication Events in PHR Genes Among Six Cucurbitaceae
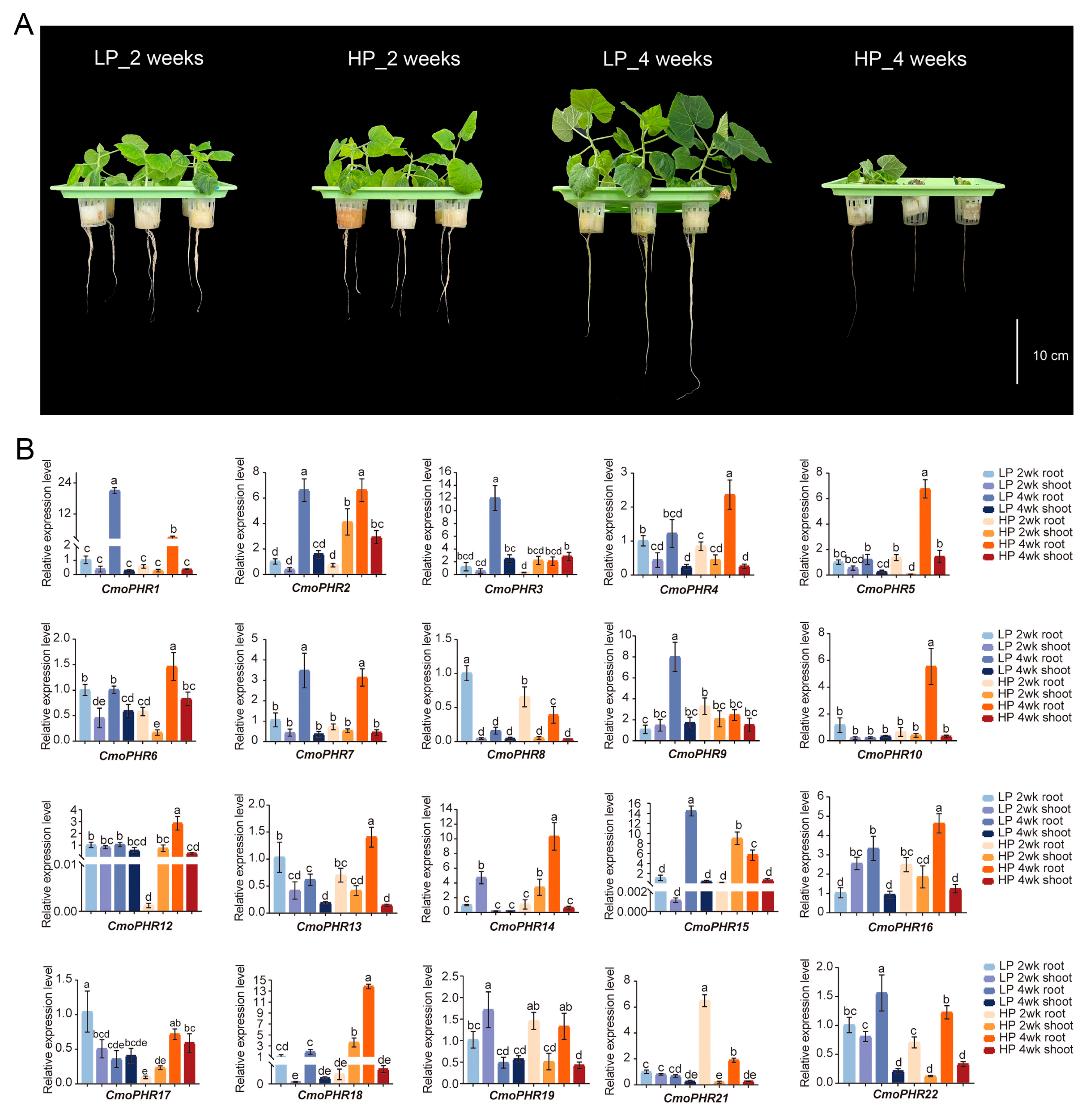
2.7. Expression Profiling of CmoPHR Genes in Pumpkin Under Different Pi Conditions
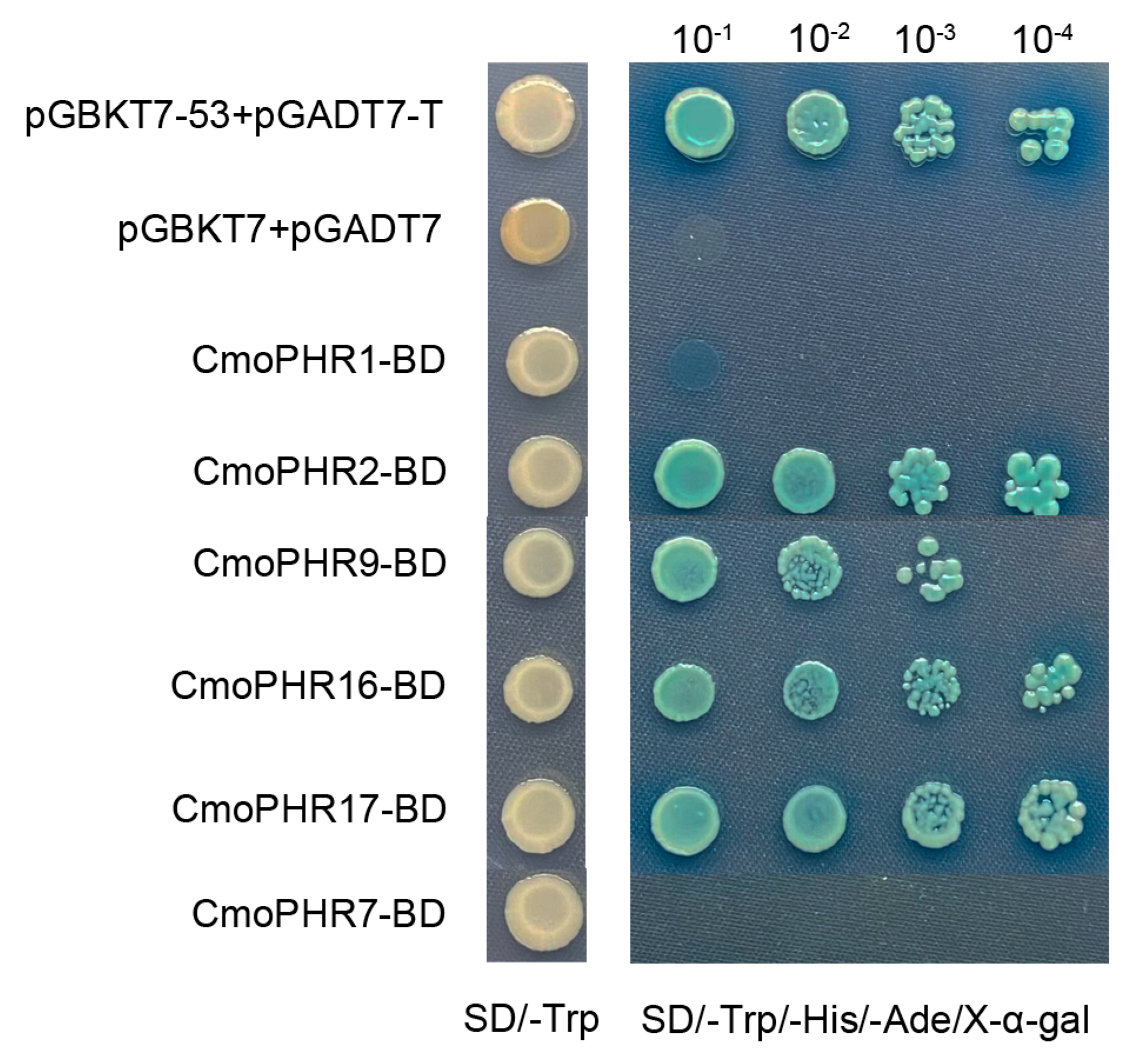
2.8. Transactivation Assay for CmoPHR Proteins
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of PHR Gene Family Members in Cucurbitaceae
4.2. Physicochemical Properties and Chromosomal Localization Analysis
4.3. Phylogenetic Analysis of the PHR Gene Family in Cucurbitaceae
4.4. Gene Structure, Conserved Motifs, and Cis-Regulatory Element Prediction
4.5. Plant Materials, Growth Conditions, and Treatments
4.6. RNA Isolation and qRT-PCR Analysis
4.7. Gene Cloning, Recombinant Plasmid Assembly, and Transcriptional Activation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate Nutrition: Improving Low-Phosphate Tolerance in Crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, I.; Biswas, A.K.; Parama, V.R.R. Phosphorus management in agriculture: A review. Agric. Rev. 2014, 35, 261. [Google Scholar] [CrossRef]
- Péret, B.; Clément, M.; Nussaume, L.; Desnos, T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011, 16, 442–450. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Chen, Q.; Lin, Y.; Tian, J.; Liang, C. Cell Wall Proteins Play Critical Roles in Plant Adaptation to Phosphorus Deficiency. Int. J. Mol. Sci. 2019, 20, 5259. [Google Scholar] [CrossRef]
- Richardson, A.E.; Simpson, R.J. Soil Microorganisms Mediating Phosphorus Availability Update on Microbial Phosphorus. Plant Physiol. 2011, 156, 989–996. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus Uptake by Plants: From Soil to Cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef]
- Poirier, Y.; Jaskolowski, A.; Clúa, J. Phosphate acquisition and metabolism in plants. Curr. Biol. 2022, 32, R623–R629. [Google Scholar] [CrossRef]
- Hirsch, J.; Marin, E.; Floriani, M.; Chiarenza, S.; Richaud, P.; Nussaume, L.; Thibaud, M.C. Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 2006, 88, 1767–1771. [Google Scholar] [CrossRef]
- Aluwihare, Y.C.; Ishan, M.; Chamikara, M.D.M.; Weebadde, C.K.; Sirisena, D.N.; Samarasinghe, W.L.G.; Sooriyapathirana, S.D.S.S. Characterization and Selection of Phosphorus Deficiency Tolerant Rice Genotypes in Sri Lanka. Rice Sci. 2016, 23, 184–195. [Google Scholar] [CrossRef]
- Oropeza-Aburto, A.; Cruz-Ramírez, A.; Acevedo-Hernández, G.J.; Pérez-Torres, C.-A.; Caballero-Pérez, J.; Herrera-Estrella, L. Functional analysis of the Arabidopsis PLDZ2 promoter reveals an evolutionarily conserved low-Pi-responsive transcriptional enhancer element. J. Exp. Bot. 2012, 63, 2189–2202. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P. Symbiotic Nitrogen Fixation and Phosphorus Acquisition. Plant Nutrition in a World of Declining Renewable Resources. Plant Physiol. 2001, 127, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.B.; Moore, G.A.; Weatherley, A.J.; Arora, M. Key sustainability challenges for the global phosphorus resource, their implications for global food security, and options for mitigation. J. Clean. Prod. 2017, 140, 945–963. [Google Scholar] [CrossRef]
- Smith, V.H.; Schindler, D.W. Eutrophication science: Where do we go from here? Trends Ecol. Evol. 2009, 24, 201–207. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, D. Signaling Components Involved in Plant Responses to Phosphate Starvation. J. Integr. Plant Biol. 2008, 50, 849–859. [Google Scholar] [CrossRef]
- Puga, M.I.; Rojas-Triana, M.; de Lorenzo, L.; Leyva, A.; Rubio, V.; Paz-Ares, J. Novel signals in the regulation of Pi starvation responses in plants: Facts and promises. Curr. Opin. Plant Biol. 2017, 39, 40–49. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Guo, J.; Zhu, X.; Shi, J.; He, Q.; Liu, Y.; Wu, Y.; Zhang, L.; Lv, Q.; et al. Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 2018, 219, 135–148. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F.; et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540.e18. [Google Scholar] [CrossRef]
- Guo, M.; Ruan, W.; Li, C.; Huang, F.; Zeng, M.; Liu, Y.; Yu, Y.; Ding, X.; Wu, Y.; Wu, Z.; et al. Integrative Comparison of the Role of the PHOSPHATE RESPONSE1 Subfamily in Phosphate Signaling and Homeostasis in Rice. Plant Physiol. 2015, 168, 1762–1776. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.; Jiang, H.; Jiang, C.; Du, Y.; Gong, C.; Wang, W.; Zhu, S.; Han, G.; Cheng, B. Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef] [PubMed]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed]
- Bajhaiya, A.K.; Dean, A.P.; Zeef, L.A.H.; Webster, R.E.; Pittman, J.K. PSR1 Is a Global Transcriptional Regulator of Phosphorus Deficiency Responses and Carbon Storage Metabolism in Chlamydomonas reinhardtii. Plant Physiol. 2016, 170, 1216–1234. [Google Scholar] [CrossRef]
- Shimogawara, K.; Wykoff, D.D.; Usuda, H.; Grossman, A.R. Chlamydomonas reinhardtii Mutants Abnormal in Their Responses to Phosphorus Deprivation1. Plant Physiol. 1999, 120, 685–694. [Google Scholar] [CrossRef]
- Nilsson, L.; MÜLler, R.; Nielsen, T.H. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 2007, 30, 1499–1512. [Google Scholar] [CrossRef]
- Zhou, J.; Jiao, F.; Wu, Z.; Li, Y.; Wang, X.; He, X.; Zhong, W.; Wu, P. OsPHR2 Is Involved in Phosphate-Starvation Signaling and Excessive Phosphate Accumulation in Shoots of Plants. Plant Physiol. 2008, 146, 1673–1686. [Google Scholar] [CrossRef]
- Wu, P.; Wang, X.M. Role of OsPHR2 on phosphorus homoestasis and root hairs development in rice (Oryza sativa L.). Plant Signal. Behav. 2008, 3, 674–675. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Miao, J.; Guo, J.; Shi, Z.; He, M.; Chen, Y. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann. Bot. 2013, 111, 1139–1153. [Google Scholar] [CrossRef]
- Wu, W.; Zhao, H.; Deng, Q.; Yang, H.; Guan, X.; Qi, R.; Shi, P.; Yang, J.; Zhang, M.; Hu, Z. The Novel Cucurbitaceae miRNA ClmiR86 Is Involved in Grafting-Enhanced Phosphate Utilization and Phosphate Starvation Tolerance in Watermelon. Plants 2021, 10, 2133. [Google Scholar] [CrossRef]
- Uygur, V.; Yetisir, H. Effects of Rootstocks on Some Growth Parameters, Phosphorous and Nitrogen Uptake Watermelon under Salt Stress. J. Plant Nutr. 2009, 32, 629–643. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, F.; Han, G.; Cheng, B. Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Rep. 2018, 37, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Han, X.; Du, H.; Mu, Z. Genome-Wide Identification and Characterization of MYB-CC Gene Factor Family in Sorghum and Their Response to Phosphate Starvation. Russ. J. Plant Physiol. 2022, 69, 120. [Google Scholar] [CrossRef]
- Wang, X.; Qu, R.; Wang, S.; Peng, J.; Guo, J.; Cui, G.; Chen, T.; Chen, M.; Shen, Y. Genome-wide identification of the SmPHR gene family in Salvia miltiorrhiza and SmPHR7-mediated response to phosphate starvation in Arabidopsis thaliana. Plant Cell Rep. 2025, 44, 73. [Google Scholar] [CrossRef] [PubMed]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef]
- Doornbos, R.F.; Geraats, B.P.J.; Kuramae, E.E.; Van Loon, L.C.; Bakker, P. Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2011, 24, 395–407. [Google Scholar] [CrossRef]
- Jiang, C.; Gao, X.; Liao, L.; Harberd, N.P.; Fu, X. Phosphate Starvation Root Architecture and Anthocyanin Accumulation Responses Are Modulated by the Gibberellin-DELLA Signaling Pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary Genetics of Genome Merger and Doubling in Plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype Stability and Unbiased Fractionation in the Paleo-Allotetraploid Cucurbita Genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Z.; Zhu, Y.; Kong, S.; Liu, D. Phosphate response 1 family members act distinctly to regulate transcriptional responses to phosphate starvation. Plant Physiol. 2022, 191, 1324–1343. [Google Scholar] [CrossRef]
- Lv, Q.; Zhong, Y.; Wang, Y.; Wang, Z.; Zhang, L.; Shi, J.; Wu, Z.; Liu, Y.; Mao, C.; Yi, K.; et al. SPX4 Negatively Regulates Phosphate Signaling and Homeostasis through Its Interaction with PHR2 in Rice. Plant Cell 2014, 26, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Ried, M.K.; Wild, R.; Zhu, J.; Pipercevic, J.; Sturm, K.; Broger, L.; Harmel, R.K.; Abriata, L.A.; Hothorn, L.A.; Fiedler, D.; et al. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 2021, 12, 384. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-B.; Sun, X.-Y.; Zheng, M.-Y.; Liu, Y.-N.; Zhang, J.-X.; Zhou, Q.-F.; Pei, D.-L.; Liu, D.-M.; Chen, Y.-W.; Gao, H.; et al. Transcription factor CaPHR3 enhances phosphate starvation tolerance by up-regulating the expression of the CaPHT1;4 phosphate transporter gene in pepper. Int. J. Biol. Macromol. 2025, 292, 139315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, F.; Cui, W.; Chen, K.; Zhao, R.; Zhang, Z. The FvPHR1 transcription factor control phosphate homeostasis by transcriptionally regulating miR399a in woodland strawberry. Plant Sci. 2019, 280, 258–268. [Google Scholar] [CrossRef]
- Barragán-Rosillo, A.C.; Peralta-Alvarez, C.A.; Ojeda-Rivera, J.O.; Arzate-Mejía, R.G.; Recillas-Targa, F.; Herrera-Estrella, L. Genome accessibility dynamics in response to phosphate limitation is controlled by the PHR1 family of transcription factors in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2107558118. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Z.; Liu, D. Comparative functional analyses of PHR1, PHL1, and PHL4 transcription factors in regulating Arabidopsis responses to phosphate starvation. Front. Plant Sci. 2024, 15, 1379562. [Google Scholar] [CrossRef]
- Huang, W.; Xian, Z.; Kang, X.; Tang, N.; Li, Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 2015, 15, 209. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Zhang, M.; Chen, M.; Ni, Y.; Xu, X.; Xu, S.; Li, Y.; Zhang, X. Genome-Wide Identification of BES1 Gene Family in Six Cucurbitaceae Species and Its Expression Analysis in Cucurbita moschata. Int. J. Mol. Sci. 2023, 24, 2287. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Chromosome Location | Exon Number | CDS Length (bp) | Protein Length (aa) | MW (kDa) | pI |
|---|---|---|---|---|---|---|---|
| CsPHR1 | CsGy1G031810.1 | chr1:30837491-30843757 (−) | 7 | 1389 | 462 | 51.36 | 5.14 |
| CsPHR2 | CsGy3G010900.1 | chr3:8526938-8531213 (−) | 6 | 933 | 310 | 33.65 | 6.02 |
| CsPHR3 | CsGy3G024970.1 | chr3:25330062-25333810 (−) | 8 | 1515 | 504 | 55.89 | 5.34 |
| CsPHR4 | CsGy3G026010.1 | chr3:26761363-26766955 (−) | 7 | 786 | 261 | 29.53 | 5.92 |
| CsPHR5 | CsGy3G034630.1 | chr3:33313222-33318459 (+) | 7 | 711 | 236 | 25.67 | 9.18 |
| CsPHR6 | CsGy3G038310.1 | chr3:36043617-36047056 (+) | 7 | 1191 | 396 | 43.01 | 8.85 |
| CsPHR7 | CsGy4G019000.1 | chr4:25077536-25081462 (+) | 7 | 1203 | 400 | 44.66 | 7.95 |
| CsPHR8 | CsGy6G011140.1 | chr6:9566650-9569975 (−) | 6 | 1092 | 363 | 40.00 | 8.86 |
| CsPHR9 | CsGy6G032210.1 | chr6:29533121-29536937 (−) | 7 | 840 | 279 | 30.90 | 8.93 |
| CsPHR10 | CsGy6G032220.1 | chr6:29538118-29543334 (−) | 6 | 969 | 322 | 35.20 | 6.56 |
| CsPHR11 | CsGy7G010430.1 | chr7:13099901-13103260 (+) | 8 | 1419 | 472 | 52.89 | 8.96 |
| CmPHR1 | MELO3C012590.2.1 | chr01:20867349-20870178 (−) | 7 | 1140 | 379 | 42.04 | 7.78 |
| CmPHR2 | MELO3C010134.2.1 | chr02:14418218-14425559 (−) | 8 | 1335 | 444 | 49.31 | 5.13 |
| CmPHR3 | MELO3C012848.2.1 | chr04:13120451-13125000 (+) | 8 | 1449 | 482 | 53.61 | 5.34 |
| CmPHR4 | MELO3C022765.2.1 | chr04:21256362-21262033 (−) | 8 | 933 | 310 | 34.79 | 8.28 |
| CmPHR5 | MELO3C026896.2.1 | chr04:25859900-25862961 (−) | 7 | 1215 | 404 | 43.83 | 8.85 |
| CmPHR6 | MELO3C009690.2.1 | chr04:30135365-30141300 (−) | 7 | 711 | 236 | 25.74 | 9.30 |
| CmPHR7 | MELO3C007988.2.1 | chr08:6623374-6627144 (−) | 8 | 813 | 270 | 29.67 | 8.71 |
| CmPHR8 | MELO3C007989.2.1 | chr08:6628818-6634578 (−) | 6 | 942 | 313 | 34.39 | 6.22 |
| CmPHR9 | MELO3C008929.2.1 | chr08:23621008-23624170 (+) | 7 | 1185 | 394 | 44.07 | 8.41 |
| CmPHR10 | MELO3C019621.2.1 | chr11:25490228-25493530 (+) | 6 | 1089 | 362 | 40.03 | 8.87 |
| LsiPHR1 | Lsi01G005190.1 | chr01:4238229-4243824 (+) | 6 | 942 | 313 | 34.44 | 6.56 |
| LsiPHR2 | Lsi01G005200.1 | chr01:4247675-4251767 (+) | 8 | 1032 | 343 | 38.03 | 6.93 |
| LsiPHR3 | Lsi03G012900.1 | chr03:23709912-23713273 (−) | 7 | 1137 | 379 | 41.49 | 7.73 |
| LsiPHR4 | Lsi05G010460.1 | chr05:17974512-17988719 (−) | 11 | 2238 | 745 | 83.31 | 6.49 |
| LsiPHR5 | Lsi05G018380.1 | chr05:25663614-25670407 (+) | 6 | 945 | 314 | 34.06 | 6.26 |
| LsiPHR6 | Lsi06G007750.1 | chr06:15079402-15086717 (−) | 9 | 1416 | 471 | 52.28 | 5.23 |
| LsiPHR7 | Lsi11G015860.1 | chr11:24259033-24261452 (−) | 6 | 1164 | 387 | 42.74 | 7.78 |
| LsiPHR8 | LsiUNG000820.1 | chr00:2469195-2472643 (+) | 7 | 783 | 260 | 29.38 | 8.61 |
| ClaPHR1 | ClCG01G021020.1 | chr01:35006738-35010386 (−) | 7 | 981 | 326 | 36.07 | 8.60 |
| ClaPHR2 | ClCG01G021030.1 | chr01:35013406-35019457 (−) | 6 | 969 | 322 | 35.39 | 6.38 |
| ClaPHR3 | ClCG02G018740.1 | chr02:33478656-33486223 (+) | 6 | 1080 | 359 | 39.60 | 9.03 |
| ClaPHR4 | ClCG03G009700.1 | chr03:15190181-15199642 (+) | 9 | 1611 | 536 | 59.49 | 5.62 |
| ClaPHR5 | ClCG04G002100.1 | chr04:7040908-7046999 (−) | 7 | 1197 | 398 | 44.75 | 6.52 |
| ClaPHR6 | ClCG05G003320.1 | chr05:3139327-3143142 (−) | 6 | 933 | 310 | 33.83 | 6.20 |
| ClaPHR7 | ClCG07G001320.1 | chr07:1402922-1406979 (−) | 7 | 1452 | 483 | 53.53 | 5.32 |
| ClaPHR8 | ClCG07G002310.1 | chr07:2352358-2356712 (+) | 9 | 918 | 305 | 34.20 | 6.05 |
| ClaPHR9 | ClCG09G022860.1 | chr09:39854276-39856296 (+) | 6 | 1164 | 387 | 42.77 | 7.30 |
| ClaPHR10 | ClCG10G012620.1 | chr10:26664138-26666981 (−) | 7 | 846 | 282 | 33.14 | 6.34 |
| ClaPHR11 | ClCG10G013290.1 | chr10:27482619-27485716 (−) | 7 | 1149 | 382 | 42.07 | 8.43 |
| ClaPHR12 | ClCG11G007240.1 | chr11:8067404-8068916 (+) | 5 | 891 | 296 | 33.60 | 6.06 |
| BhiPHR1 | Bhi01M000162 | chr1:4358307-4374348 (−) | 17 | 1734 | 577 | 5.94 | 64.66 |
| BhiPHR2 | Bhi02M001052 | chr2:30073057-30079744 (+) | 8 | 1341 | 446 | 5.32 | 49.72 |
| BhiPHR3 | Bhi03M000545 | chr3:13712964-13718673 (+) | 6 | 942 | 313 | 6.60 | 34.44 |
| BhiPHR4 | Bhi03M000547 | chr3:13774321-13778870 (+) | 7 | 978 | 325 | 7.77 | 35.90 |
| BhiPHR5 | Bhi03M002344 | chr3:72814246-72818856 (−) | 7 | 1155 | 384 | 6.52 | 43.24 |
| BhiPHR6 | Bhi05M000006 | chr5:339928-345085 (+) | 7 | 783 | 260 | 6.47 | 29.13 |
| BhiPHR7 | Bhi05M000125 | chr5:3492888-3498142 (+) | 8 | 1449 | 482 | 5.39 | 53.54 |
| BhiPHR8 | Bhi05M001707 | chr5:61216044-61218783 (−) | 7 | 1149 | 382 | 7.78 | 41.95 |
| BhiPHR9 | Bhi06M000772 | chr6:23276891-23278379 (−) | 6 | 753 | 251 | 8.20 | 28.35 |
| BhiPHR10 | Bhi10M000727 | chr10:19061719-19064970 (−) | 6 | 1095 | 364 | 8.87 | 39.99 |
| BhiPHR11 | Bhi11M001288 | chr11:43469320-43472785 (−) | 7 | 1143 | 380 | 8.43 | 41.69 |
| BhiPHR12 | Bhi11M002259 | chr11:73208843-73217031 (+) | 7 | 711 | 236 | 9.37 | 25.80 |
| CmoPHR1 | CmoCh01G005130 | chr01:2468014-2469314 (+) | 4 | 666 | 221 | 25.51 | 8.91 |
| CmoPHR2 | CmoCh02G016940 | chr02:9712038-9714929 (+) | 6 | 723 | 240 | 27.29 | 8.41 |
| CmoPHR3 | CmoCh02G017700 | chr02:10102355-10106076 (+) | 8 | 1314 | 437 | 48.55 | 5.13 |
| CmoPHR4 | CmoCh03G000340 | chr03:656888-659538 (−) | 7 | 1131 | 376 | 42.11 | 6.16 |
| CmoPHR5 | CmoCh03G006380 | chr03:5845977-5850993 (+) | 7 | 1095 | 364 | 40.86 | 6.43 |
| CmoPHR6 | CmoCh03G006390 | chr03:5852404-5856186 (+) | 7 | 984 | 327 | 36.12 | 7.17 |
| CmoPHR7 | CmoCh05G008680 | chr05:5707748-5709145 (+) | 5 | 756 | 251 | 28.19 | 6.64 |
| CmoPHR8 | CmoCh06G000560 | chr06:349595-359369 (−) | 16 | 1803 | 600 | 67.55 | 9.25 |
| CmoPHR9 | CmoCh06G012530 | chr06:9468323-9472100 (−) | 6 | 951 | 316 | 34.29 | 6.09 |
| CmoPHR10 | CmoCh06G017240 | chr06:11843093-11845080 (−) | 6 | 1152 | 383 | 41.82 | 8.69 |
| CmoPHR11 | CmoCh07G003670 | chr07:1675620-1680532 (+) | 8 | 1086 | 361 | 39.81 | 8.43 |
| CmoPHR12 | CmoCh07G003680 | chr07:1680538-1683042 (+) | 6 | 972 | 323 | 35.51 | 8.30 |
| CmoPHR13 | CmoCh07G012910 | chr07:7134301-7137195 (−) | 7 | 1155 | 384 | 43.29 | 6.11 |
| CmoPHR14 | CmoCh10G010790 | chr10:5854637-5859651 (+) | 9 | 1356 | 451 | 50.19 | 5.31 |
| CmoPHR15 | CmoCh10G012730 | chr10:13133524-13137613 (+) | 5 | 1191 | 396 | 44.27 | 8.27 |
| CmoPHR16 | CmoCh11G009860 | chr11:5250875-5255809 (+) | 8 | 1326 | 441 | 49.06 | 5.09 |
| CmoPHR17 | CmoCh14G011540 | chr14:9041919-9044422 (+) | 6 | 1152 | 383 | 41.95 | 8.16 |
| CmoPHR18 | CmoCh14G019460 | chr14:14393688-14400483 (+) | 8 | 1071 | 356 | 38.47 | 6.04 |
| CmoPHR19 | CmoCh19G001230 | chr19:723628-727308 (−) | 7 | 1167 | 388 | 43.63 | 5.43 |
| CmoPHR20 | CmoCh19G002200 | chr19:1459343-1470768 (−) | 9 | 783 | 260 | 29.29 | 6.17 |
| CmoPHR21 | CmoCh19G011500 | chr19:9659420-9662090 (+) | 6 | 1140 | 379 | 41.48 | 7.75 |
| CmoPHR22 | CmoCh20G003120 | chr20:1530207-1533405 (+) | 6 | 1101 | 366 | 40.43 | 8.87 |
| Pumpkin | Melon | Watermelon | Cucumber | Wax Gourd | Bottle Gourd |
|---|---|---|---|---|---|
| CmoPHR1 | - | - | - | BhiPHR9 | - |
| CmoPHR2 | CmPHR4 | ClaPHR8 | CsPHR4 | BhiPHR6 | LsiPHR8 |
| CmoPHR3 | CmPHR3 | ClaPHR7/ClaPHR19 | CsPHR3 | BhiPHR7 | - |
| CmoPHR4 | CmPHR9 | - | CsPHR7 | BhiPHR5 | - |
| CmoPHR5 | CmPHR7 | ClaPHR1 | CsPHR10 | BhiPHR3 | LsiPHR1 |
| CmoPHR6 | - | - | CsPHR9 | - | - |
| CmoPHR7 | - | ClaPHR12 | - | - | - |
| CmoPHR8 | - | - | CsPHR5 | BhiPHR12 | - |
| CmoPHR9 | - | ClaPHR6 | CsPHR2 | - | LsiPHR5 |
| CmoPHR10 | CmPHR5 | ClaPHR11 | CsPHR6 | BhiPHR11 | LsiPHR3 |
| CmoPHR11 | CmPHR7 | ClaPHR1 | CsPHR10 | BhiPHR3 | LsiPHR1 |
| CmoPHR12 | - | - | CsPHR9 | - | - |
| CmoPHR13 | CmPHR9 | - | CsPHR7 | BhiPHR5 | - |
| CmoPHR14 | CmPHR2 | - | CsPHR1 | BhiPHR2 | LsiPHR6 |
| CmoPHR16 | CmPHR2 | - | - | - | - |
| CmoPHR18 | - | ClaPHR6 | CsPHR2 | BhiPHR1 | LsiPHR5 |
| CmoPHR19 | CmPHR3 | - | CsPHR3 | BhiPHR7 | LsiPHR4 |
| CmoPHR20 | CmPHR4 | ClaPHR8 | CsPHR4 | BhiPHR6 | LsiPHR8 |
| CmoPHR21 | CmPHR1/CmPHR10 | ClaPHR3/ClaPHR9 | CsPHR8/CsPHR11 | BhiPHR8/BhiPHR10 | LsiPHR7 |
| CmoPHR22 | CmPHR1/CmPHR10 | ClaPHR3/ClaPHR9 | CsPHR8 | BhiPHR8/BhiPHR10 | LsiPHR7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Y.; Xie, K.; Shi, M.; Shan, H.; Wu, W.; Wang, W.; Cheng, B.; Li, X. Genome-Wide Identification of the PHR Gene Family in Six Cucurbitaceae Species and Its Expression Analysis in Cucurbita moschata. Plants 2025, 14, 1443. https://doi.org/10.3390/plants14101443
Ni Y, Xie K, Shi M, Shan H, Wu W, Wang W, Cheng B, Li X. Genome-Wide Identification of the PHR Gene Family in Six Cucurbitaceae Species and Its Expression Analysis in Cucurbita moschata. Plants. 2025; 14(10):1443. https://doi.org/10.3390/plants14101443
Chicago/Turabian StyleNi, Ying, Kailing Xie, Minghui Shi, Hanchen Shan, Wenxiang Wu, Weiwei Wang, Beijiu Cheng, and Xiaoyu Li. 2025. "Genome-Wide Identification of the PHR Gene Family in Six Cucurbitaceae Species and Its Expression Analysis in Cucurbita moschata" Plants 14, no. 10: 1443. https://doi.org/10.3390/plants14101443
APA StyleNi, Y., Xie, K., Shi, M., Shan, H., Wu, W., Wang, W., Cheng, B., & Li, X. (2025). Genome-Wide Identification of the PHR Gene Family in Six Cucurbitaceae Species and Its Expression Analysis in Cucurbita moschata. Plants, 14(10), 1443. https://doi.org/10.3390/plants14101443







