Characterization of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Its Response to Abiotic Stress in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
2.1. Whole-Genome Characterization of FARs in Rice
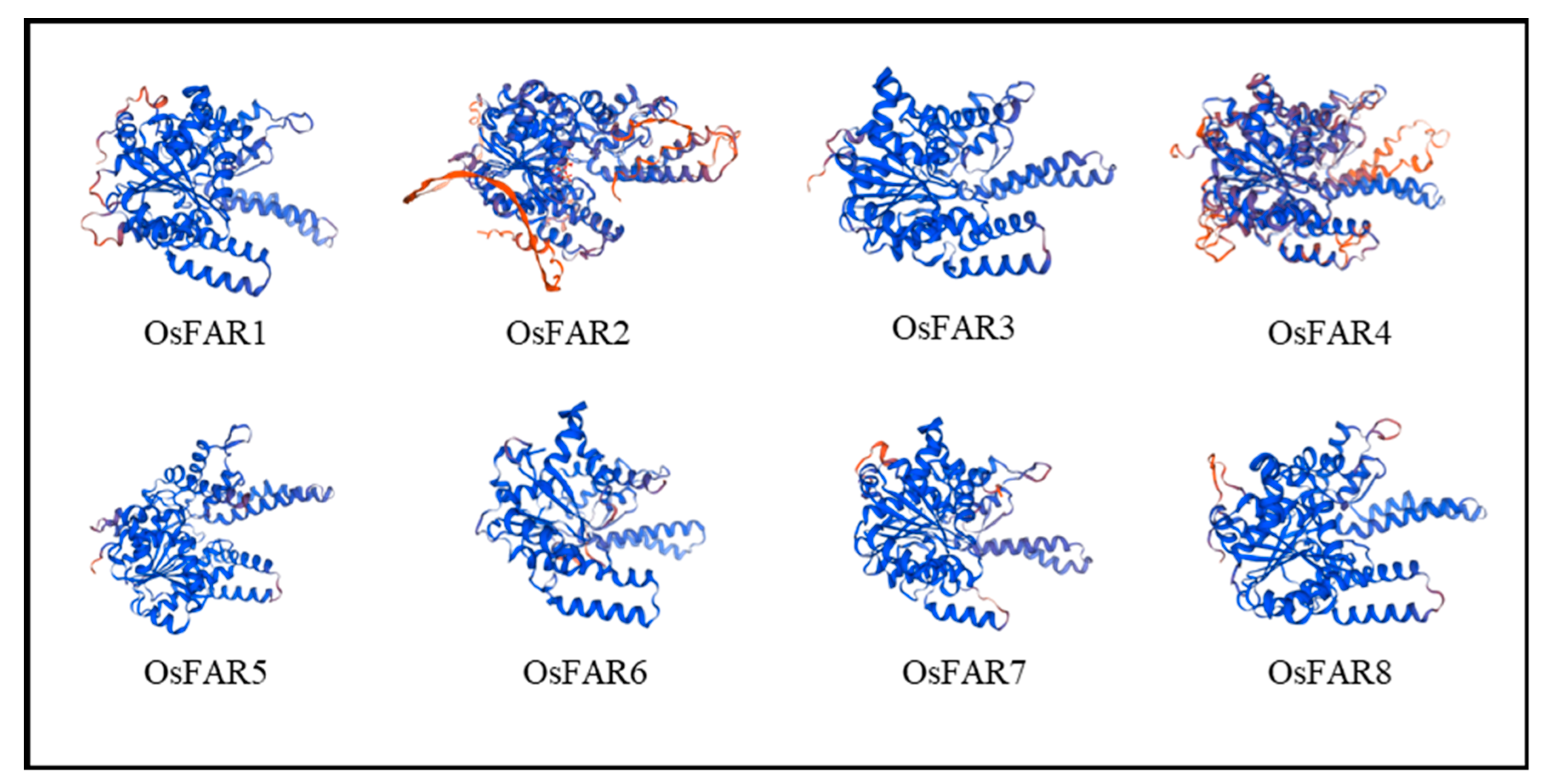
2.2. FAR Protein Structure Analysis
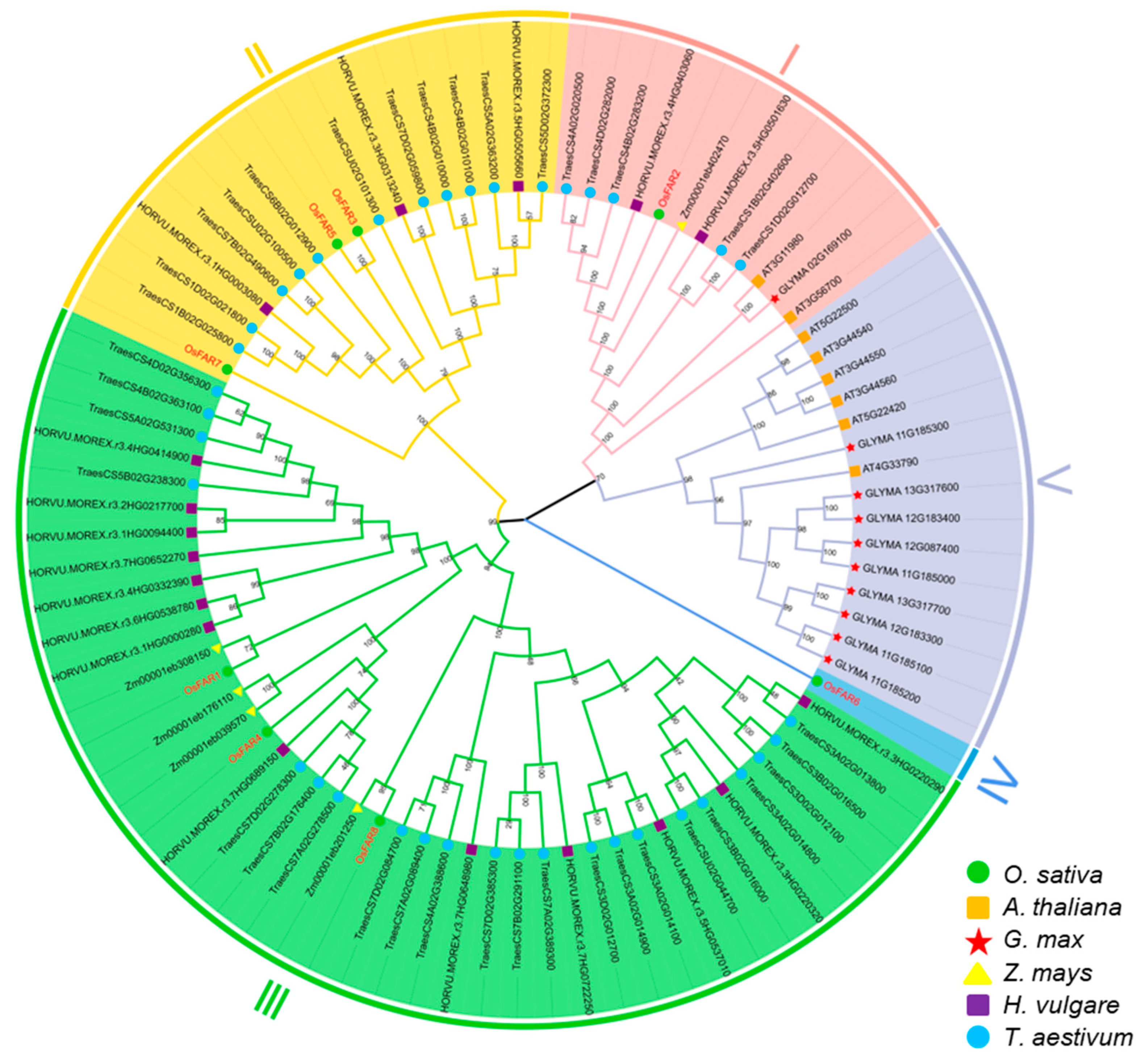
2.3. Multiple Sequence Alignment, Phylogenetic Analysis, and Classification of OsFAR Proteins
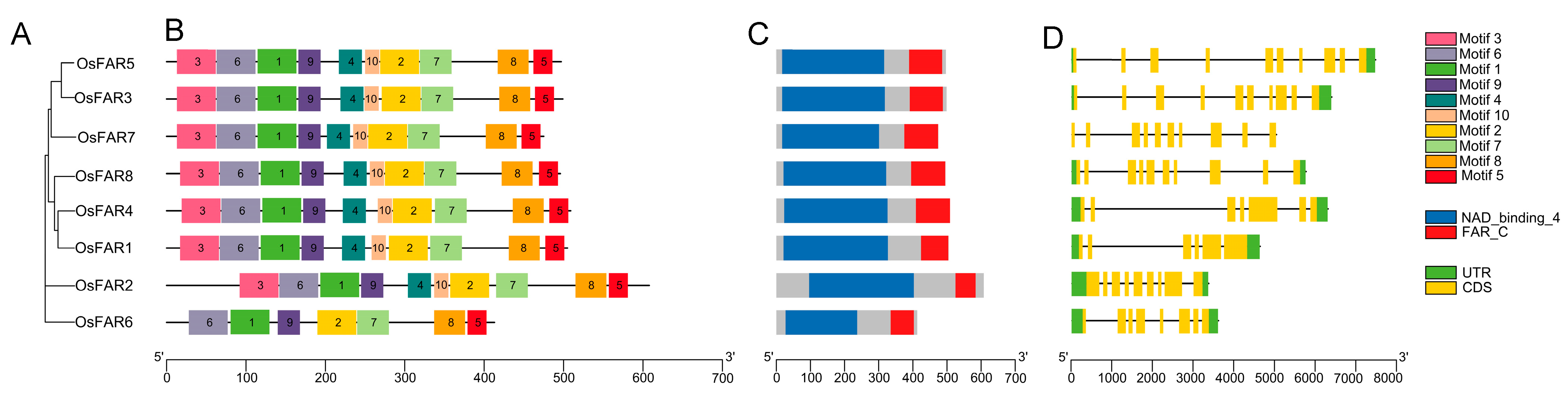
2.4. Conserved Motifs and Gene Structures of OsFAR Gene Family Members
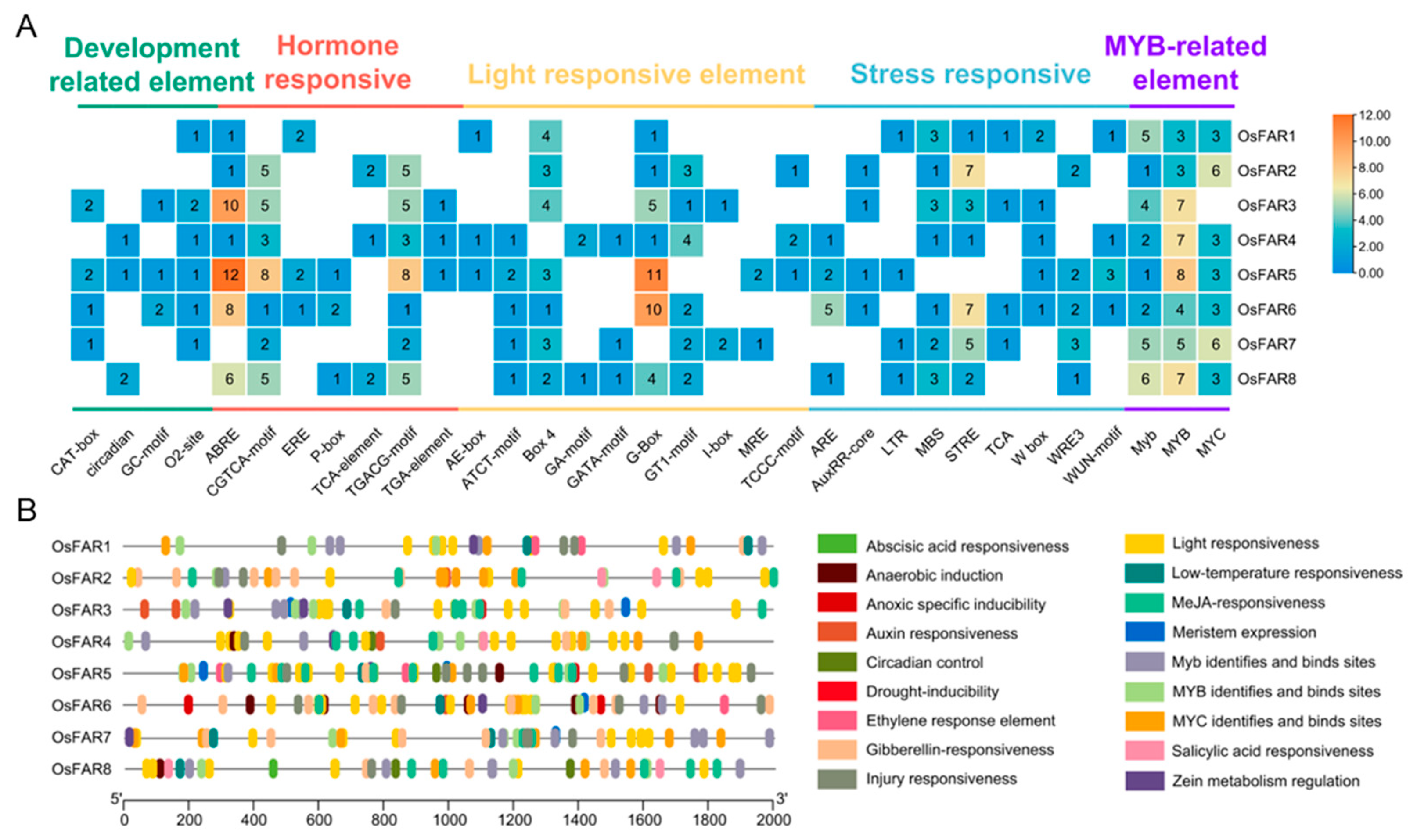
2.5. Analysis of Cis-Regulatory Element in the Promoter Region of OsFARs
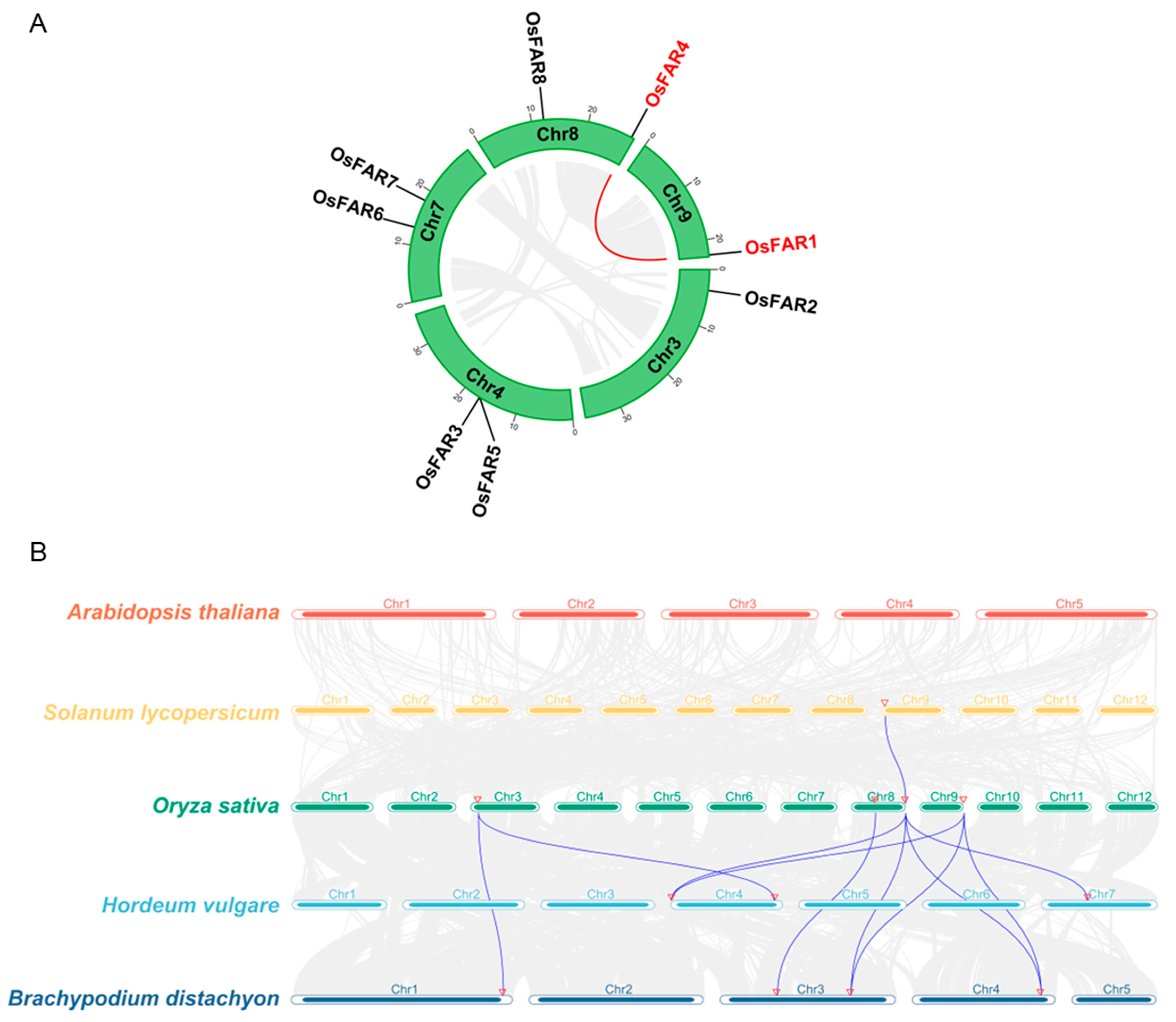
2.6. Analyses of Chromosomal Distribution and Gene Duplication of the OsFAR Genes
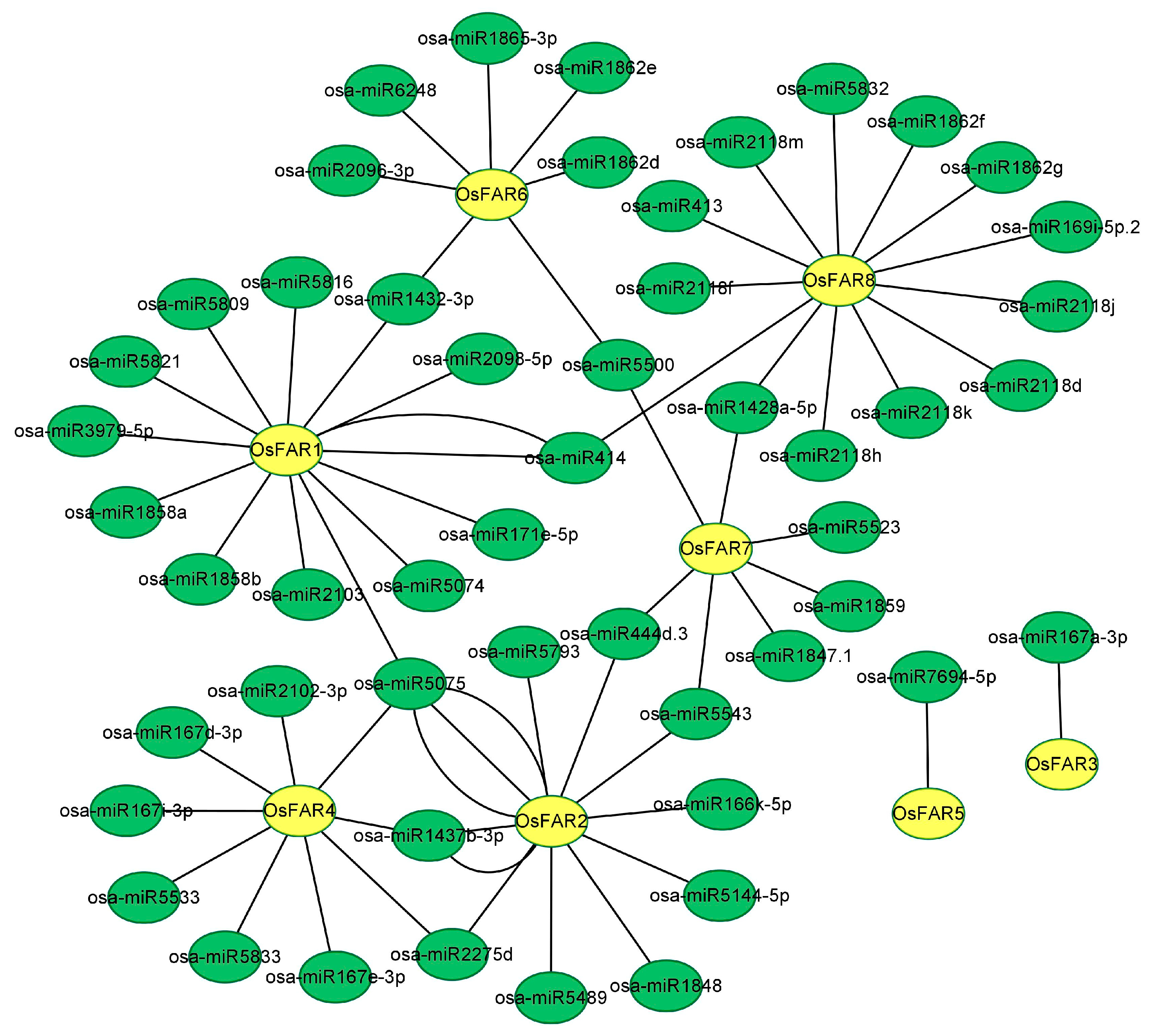
2.7. GO Enrichment Analysis and miRNA Targeting Prediction
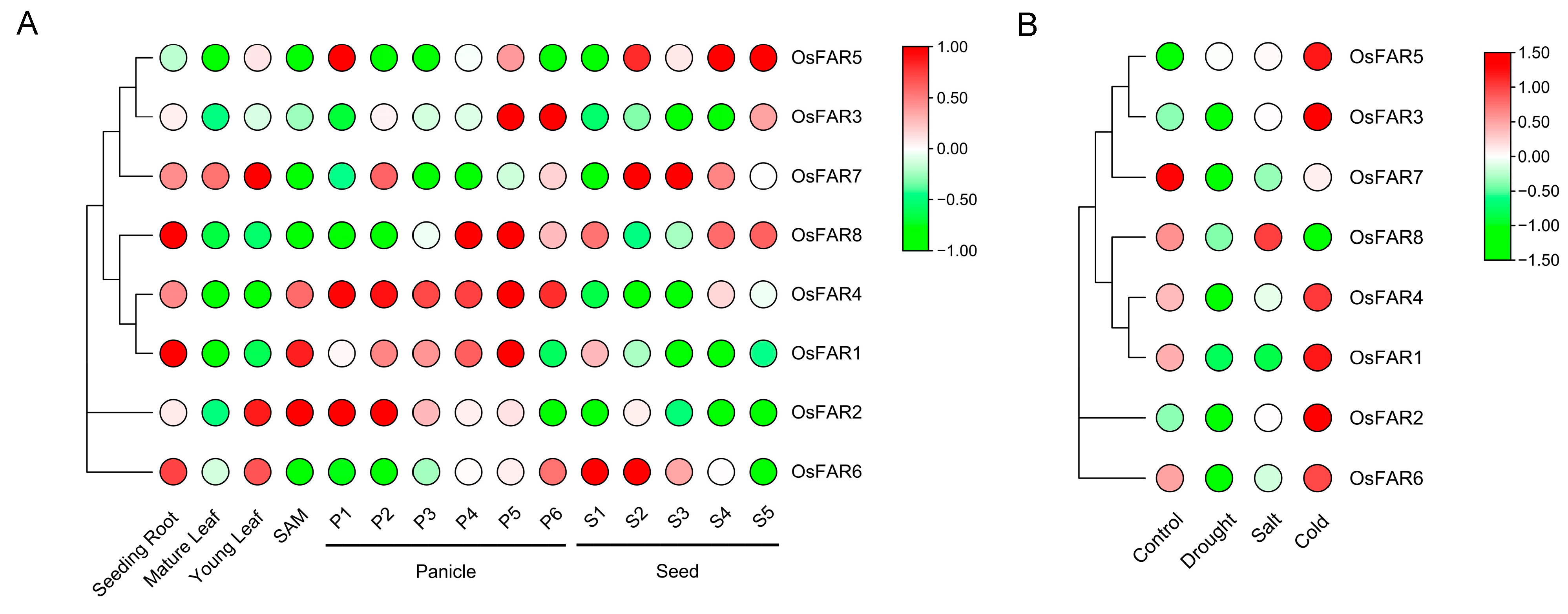
2.8. Expression Patterns of FARs in Different Tissues
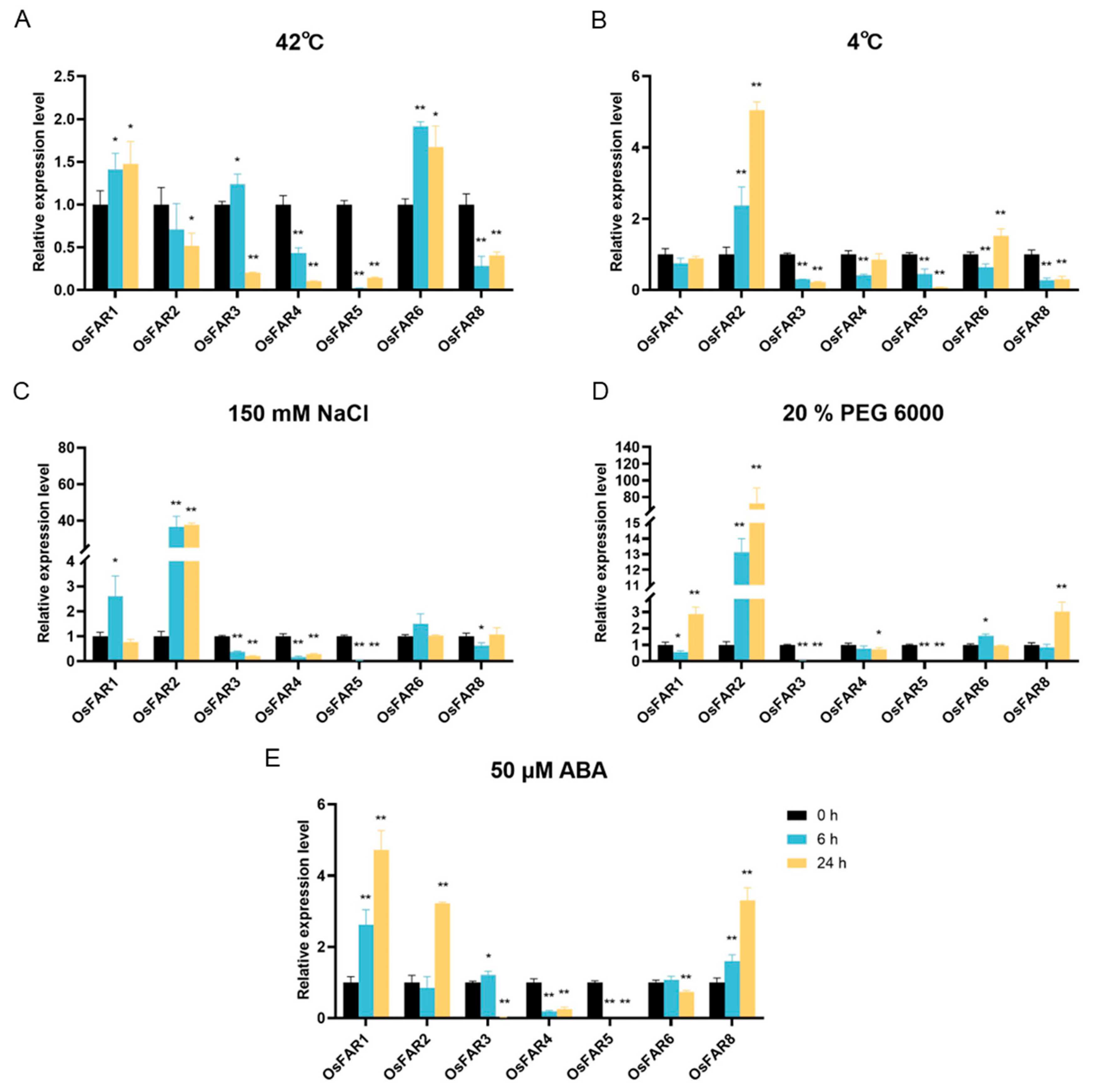
2.9. Expression Profiles of OsFARs under Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of FARs in Rice
4.2. Prediction of Secondary and Tertiary Structures
4.3. Multiple Alignments and Phylogenetic Analysis
4.4. Analysis of Conserved Motifs and Gene Structures
4.5. Cis-Regulatory Element Analysis of OsFARs
4.6. Chromosome Distribution, Gene Duplication, and Selective Pressure Analysis
4.7. GO Functional Annotation and miRNA Targeting Prediction
4.8. Gene Expression Profile Analysis
4.9. Stress Treatment, RNA Extraction, and RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Li, R.-J.; Li, L.-M.; Liu, X.-L.; Kim, J.-C.; Jenks, M.A.; Lü, S. Diurnal Regulation of Plant Epidermal Wax Synthesis through Antagonistic Roles of the Transcription Factors SPL9 and DEWAX. Plant Cell 2019, 31, 2711–2733. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Zhang, X.; Lu, X.; Chen, G.; Chen, Z.-H. Molecular and Evolutionary Mechanisms of Cuticular Wax for Plant Drought Tolerance. Front. Plant Sci. 2017, 8, 621. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax Biosynthesis in Response to Danger: Its Regulation upon Abiotic and Biotic Stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-Ketoacyl-CoA Synthase Is Involved in Rice Leaf Cuticular Wax Synthesis and Requires a CER2-LIKE Protein as a Cofactor. Plant Physiol. 2017, 173, 944–955. [Google Scholar] [CrossRef]
- Müller, Y.; Patwari, P.; Stöcker, T.; Zeisler-Diehl, V.; Steiner, U.; Campoli, C.; Grewe, L.; Kuczkowska, M.; Dierig, M.M.; Jose, S.; et al. Isolation and Characterization of the Gene HvFAR1 Encoding Acyl-CoA Reductase from the Cer-Za.227 Mutant of Barley (Hordeum vulgare) and Analysis of the Cuticular Barrier Functions. New Phytol. 2023, 239, 1903–1918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Ayaz, A.; Zhao, H.; Lü, S. The Plant Fatty Acyl Reductases. Int. J. Mol. Sci. 2022, 23, 16156. [Google Scholar] [CrossRef]
- Wang, M.; Wu, H.; Xu, J.; Li, C.; Wang, Y.; Wang, Z. Five Fatty Acyl-Coenzyme A Reductases Are Involved in the Biosynthesis of Primary Alcohols in Aegilops Tauschii Leaves. Front. Plant Sci. 2017, 8, 1012. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.G.; Pollard, M.R.; Anderson, L.; Hayes, T.R.; Lassner, M.W. Purification of a Jojoba Embryo Fatty Acyl-Coenzyme A Reductase and Expression of Its cDNA in High Erucic Acid Rapeseed. Plant Physiol. 2000, 122, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Sturtevant, D.; Lu, S.; Zhou, Z.-W.; Shen, Y.; Wang, S.; Song, J.-M.; Zhong, J.; Burks, D.J.; Yang, Z.-Q.; Yang, Q.-Y.; et al. The Genome of Jojoba (Simmondsia chinensis): A Taxonomically Isolated Species That Directs Wax Ester Accumulation in Its Seeds. Sci. Adv. 2020, 6, eaay3240. [Google Scholar] [CrossRef]
- Rowland, O.; Domergue, F. Plant Fatty Acyl Reductases: Enzymes Generating Fatty Alcohols for Protective Layers with Potential for Industrial Applications. Plant Sci. 2012, 193–194, 28–38. [Google Scholar] [CrossRef]
- Domergue, F.; Vishwanath, S.J.; Joubès, J.; Ono, J.; Lee, J.A.; Bourdon, M.; Alhattab, R.; Lowe, C.; Pascal, S.; Lessire, R.; et al. Three Arabidopsis Fatty Acyl-Coenzyme A Reductases, FAR1, FAR4, and FAR5, Generate Primary Fatty Alcohols Associated with Suberin Deposition. Plant Physiol. 2010, 153, 1539–1554. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 Encodes an Alcohol-Forming Fatty Acyl-Coenzyme A Reductase Involved in Cuticular Wax Production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.T.P.; Carlsson, A.S.; Hamberg, M.; Bülow, L.; Stymne, S.; Olsson, P. Functional Expression of Five Arabidopsis Fatty Acyl-CoA Reductase Genes in Escherichia Coli. J. Plant Physiol. 2009, 166, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Y.; Wu, H.; Xu, J.; Li, T.; Hegebarth, D.; Jetter, R.; Chen, L.; Wang, Z. Three TaFAR Genes Function in the Biosynthesis of Primary Alcohols and the Response to Abiotic Stresses in Triticum aestivum. Sci. Rep. 2016, 6, 25008. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, L.; Wang, B.; Wang, H.; Khan, I.; Zhang, S.; Wen, J.; Ma, C.; Dai, C.; Tu, J.; et al. BnA1.CER4 and BnC1.CER4 Are Redundantly Involved in Branched Primary Alcohols in the Cuticle Wax of Brassica napus. Theor. Appl. Genet. 2021, 134, 3051–3067. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Cuticular Wax Biosynthesis Is Up-Regulated by the MYB94 Transcription Factor in Arabidopsis. Plant Cell Physiol. 2015, 56, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Kong, L.; Yang, X.; Jiao, B.; Hu, J.; Zhang, Z.; Xu, C.; Luo, K. PtoMYB142, a Poplar R2R3-MYB Transcription Factor, Contributes to Drought Tolerance by Regulating Wax Biosynthesis. Tree Physiol. 2022, 42, 2133–2147. [Google Scholar] [CrossRef]
- Guan, L.; Xia, D.; Hu, N.; Zhang, H.; Wu, H.; Jiang, Q.; Li, X.; Sun, Y.; Wang, Y.; Wang, Z. OsFAR1 Is Involved in Primary Fatty Alcohol Biosynthesis and Promotes Drought Tolerance in Rice. Planta 2023, 258, 24. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid Metabolism: Critical Roles in Male Fertility and Other Aspects of Reproductive Development in Plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Chen, W.; Yu, X.-H.; Zhang, K.; Shi, J.; De Oliveira, S.; Schreiber, L.; Shanklin, J.; Zhang, D. Male Sterile2 Encodes a Plastid-Localized Fatty Acyl Carrier Protein Reductase Required for Pollen Exine Development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef]
- Shi, J.; Tan, H.; Yu, X.-H.; Liu, Y.; Liang, W.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.; Kai, G.; et al. Defective Pollen Wall Is Required for Anther and Microspore Development in Rice and Encodes a Fatty Acyl Carrier Protein Reductase[C][W][OA]. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xiao, S.; Liu, J.; Somaratne, Y.; Zhang, H.; Wang, M.; Zhang, H.; Zhao, L.; Chen, H. MALE STERILE6021 (MS6021) Is Required for the Development of Anther Cuticle and Pollen Exine in Maize. Sci. Rep. 2017, 7, 16736. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; You, Q.; Luo, W.; Wang, C.; Zhao, S.; Chai, G.; Li, T.; Shi, X.; Li, C.; et al. Three Fatty Acyl-Coenzyme A Reductases, BdFAR1, BdFAR2 and BdFAR3, Are Involved in Cuticular Wax Primary Alcohol Biosynthesis in Brachypodium distachyon. Plant Cell Physiol. 2018, 59, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Li, C.; Xu, F.; Li, Y.; Shi, X.; Wang, Y.; Wang, Z. Three Endoplasmic Reticulum-Associated Fatty Acyl-Coenzyme a Reductases Were Involved in the Production of Primary Alcohols in Hexaploid Wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cheng, X.; Jia, M.; Zhang, X.; Xue, F.; Li, Y.; Sun, J.; Liu, F. Silencing GhFAR3.1 Reduces Wax Accumulation in Cotton Leaves and Leads to Increased Susceptibility to Drought Stress. Plant Direct 2021, 5, e00313. [Google Scholar] [CrossRef] [PubMed]
- Ran, C.; Zhang, Y.; Chang, F.; Yang, X.; Liu, Y.; Wang, Q.; Zhu, W. Genome-Wide Analyses of SlFWL Family Genes and Their Expression Profiles under Cold, Heat, Salt and Drought Stress in Tomato. Int. J. Mol. Sci. 2023, 24, 11783. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.-K. UTR Dependent Control of Gene Expression in Plants. Trends Plant Sci. 2018, 23, 248–259. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Ngatia, J.N.; Wang, Y.; Khoso, M.A.; Farooq, U.; Chen, S. AP2/ERF, an Important Cold Stress-Related Transcription Factor Family in Plants: A Review. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2021, 27, 1953–1968. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y.; et al. Genome-Wide Identification and Expression Analysis of the bHLH Gene Family in Passion Fruit (Passiflora edulis) and Its Response to Abiotic Stress. Int. J. Biol. Macromol. 2023, 225, 389–403. [Google Scholar] [CrossRef]
- Yu, J.; Xie, Q.; Li, C.; Dong, Y.; Zhu, S.; Chen, J. Comprehensive Characterization and Gene Expression Patterns of LBD Gene Family in Gossypium. Planta 2020, 251, 81. [Google Scholar] [CrossRef]
- Yuan, T.; Liang, J.; Dai, J.; Zhou, X.-R.; Liao, W.; Guo, M.; Aslam, M.; Li, S.; Cao, G.; Cao, S. Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses. Int. J. Mol. Sci. 2022, 23, 8044. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.; Ding, A.; Li, P.; Wang, J.; Cheng, T.; Bao, F.; Zhang, Q. Genome-Wide Identification and Low-Temperature Expression Analysis of bHLH Genes in Prunus Mume. Front. Genet. 2021, 12, 762135. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ma, C.; Zheng, C.; Yao, Y.; Du, Y. Advances in the Regulation of Plant Salt-Stress Tolerance by miRNA. Mol. Biol. Rep. 2022, 49, 5041–5055. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.T.P.; Domergue, F.; Fournier, A.E.; Vishwanath, S.J.; Rowland, O.; Moreau, P.; Wood, C.C.; Carlsson, A.S.; Hamberg, M.; Hofvander, P. Biochemical Characterization of a Chloroplast Localized Fatty Acid Reductase from Arabidopsis thaliana. Biochim. Biophys. Acta 2012, 1821, 1244–1255. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Xia, Y.; Zhang, G.; Wang, J.; Ma, S.; Song, Y.; Yang, Z.; Dennis, E.S.; Niu, N. The Transcription Factors TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a in Wheat, Which Positively Regulates the Development of Microspore in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 7996. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, J.; He, Z.; Hu, N.; Luo, W.; Liu, X.; Shi, X.; Liu, T.; Jiang, Q.; An, P.; et al. BdFAR4, a Root-Specific Fatty Acyl-Coenzyme A Reductase, Is Involved in Fatty Alcohol Synthesis of Root Suberin Polyester in Brachypodium distachyon. Plant J. Cell Mol. Biol. 2021, 106, 1468–1483. [Google Scholar] [CrossRef]
- DeAndrés-Gil, C.; Moreno-Pérez, A.J.; Villoslada-Valbuena, M.; Halsey, K.; Martínez-Force, E.; Garcés, R.; Kurup, S.; Beaudoin, F.; Salas, J.J.; Venegas-Calerón, M. Characterisation of Fatty Acyl Reductases of Sunflower (Helianthus annuus L.) Seed. Plant Sci. 2024, 341, 111992. [Google Scholar] [CrossRef] [PubMed]
- Chacón, M.G.; Fournier, A.E.; Tran, F.; Dittrich-Domergue, F.; Pulsifer, I.P.; Domergue, F.; Rowland, O. Identification of Amino Acids Conferring Chain Length Substrate Specificities on Fatty Alcohol-Forming Reductases FAR5 and FAR8 from Arabidopsis thaliana. J. Biol. Chem. 2013, 288, 30345–30355. [Google Scholar] [CrossRef]
- Chang, D.; Duda, T.F., Jr. Extensive and Continuous Duplication Facilitates Rapid Evolution and Diversification of Gene Families. Mol. Biol. Evol. 2012, 29, 2019–2029. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica rapa Shed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Sun, Y.; Wang, Y.; Li, T.; Chai, G.; Jiang, W.; Shan, L.; Li, C.; Xiao, E.; et al. FAR5, a Fatty Acyl-Coenzyme A Reductase, Is Involved in Primary Alcohol Biosynthesis of the Leaf Blade Cuticular Wax in Wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Balbontín, C.; Gutiérrez, C.; Schreiber, L.; Zeisler-Diehl, V.V.; Marín, J.C.; Urrutia, V.; Hirzel, J.; Figueroa, C.R. Alkane Biosynthesis Is Promoted in Methyl Jasmonate-Treated Sweet Cherry (Prunus avium) Fruit Cuticles. J. Sci. Food Agric. 2023, 104, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Meng, G.; Zamin, I.; Wei, T.; Ma, D.; An, L.; Yue, X. Genome-Wide Identification and Functional Analysis of the TIFY Family Genes in Response to Abiotic Stresses and Hormone Treatments in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2023, 24, 10916. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Mao, L.; Wei, X.; Xia, M.; Xu, C. MYB41, MYB107, and MYC2 Promote ABA-Mediated Primary Fatty Alcohol Accumulation via Activation of AchnFAR in Wound Suberization in Kiwifruit. Hortic. Res. 2020, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Yan, W.; Chang, Z.; Xu, Y.; Luo, M.; Xu, C.; Chen, Z.; Wu, J.; Tang, X. OsMYB80 Regulates Anther Development and Pollen Fertility by Targeting Multiple Biological Pathways. Plant Cell Physiol. 2020, 61, 988–1004. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Santosh Kumar, V.V.; Verma, R.K.; Yadav, P.; Saroha, A.; Wankhede, D.P.; Chaudhary, B.; Chinnusamy, V. Genome-Wide Identification and Characterization of ABA Receptor PYL Gene Family in Rice. BMC Genom. 2020, 21, 676. [Google Scholar] [CrossRef]
- Parmar, S.; Gharat, S.A.; Tagirasa, R.; Chandra, T.; Behera, L.; Dash, S.K.; Shaw, B.P. Identification and Expression Analysis of miRNAs and Elucidation of Their Role in Salt Tolerance in Rice Varieties Susceptible and Tolerant to Salinity. PLoS ONE 2020, 15, e0230958. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, A.; Elango, T.; Li, X.; Guo, G. Utilization of microRNAs and Their Regulatory Functions for Improving Biotic Stress Tolerance in Tea Plant [Camellia sinensis (L.) O. Kuntze]. RNA Biol. 2020, 17, 1365–1382. [Google Scholar] [CrossRef]
- Wan, J.; Meng, S.; Wang, Q.; Zhao, J.; Qiu, X.; Wang, L.; Li, J.; Lin, Y.; Mu, L.; Dang, K.; et al. Suppression of microRNA168 Enhances Salt Tolerance in Rice (Oryza sativa L.). BMC Plant Biol. 2022, 22, 563. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.; Kumar, I.S. Drought Response in Rice: The miRNA Story. Int. J. Mol. Sci. 2019, 20, 3766. [Google Scholar] [CrossRef]
- Han, Y.; Zhao, Y.; Wang, H.; Zhang, Y.; Ding, Q.; Ma, L. Identification of ceRNA and Candidate Genes Related to Fertility Conversion of TCMS Line YS3038 in Wheat. Plant Physiol. Biochem. 2021, 158, 190–207. [Google Scholar] [CrossRef]
- Divya, K.; Palakolanu, S.R.; Kavi Kishor, P.; Rajesh, A.S.; Vadez, V.; Sharma, K.K.; Mathur, P.B. Functional Characterization of Late Embryogenesis Abundant Genes and Promoters in Pearl Millet (Pennisetum glaucum L.) for Abiotic Stress Tolerance. Physiol. Plant. 2021, 173, 1616–1628. [Google Scholar] [CrossRef] [PubMed]
- Cheah, B.H.; Jadhao, S.; Vasudevan, M.; Wickneswari, R.; Nadarajah, K. Identification of Functionally Important microRNAs from Rice Inflorescence at Heading Stage of a qDTY4.1-QTL Bearing Near Isogenic Line under Drought Conditions. PLoS ONE 2017, 12, e0186382. [Google Scholar] [CrossRef] [PubMed]
- Kushawaha, A.K.; Khan, A.; Sopory, S.K.; Sanan-Mishra, N. Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice. Life 2021, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-Wide Changes in microRNA Expression during Short and Prolonged Heat Stress and Recovery in Contrasting Rice Cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Su, Q.; Wu, Z.; Huang, Z.; Bao, J.; Li, J.; Tu, H.; Zeng, C.; Fu, J.; He, H. Genome-Wide Characterization of MATE Gene Family and Expression Profiles in Response to Abiotic Stresses in Rice (Oryza sativa). BMC Ecol. Evol. 2021, 21, 141. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Xiao, Y.; Wen, Y.; Li, K.; Ma, Z.; Yang, L.; Zhu, Y.; Yin, J. Genome-Wide Characterization and Function Analysis Uncovered Roles of Wheat LIMs in Responding to Adverse Stresses and TaLIM8-4D Function as a Susceptible Gene. Plant Genome 2022, 15, e20246. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-Wide Survey and Expression Analysis of the OSCA Gene Family in Rice. BMC Plant Biol. 2015, 15, 261. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Kurt, F.; Filiz, E.; Aydın, A. Genome-Wide Identification of Serine Acetyltransferase (SAT) Gene Family in Rice (Oryza sativa) and Their Expressions under Salt Stress. Mol. Biol. Rep. 2021, 48, 6277–6290. [Google Scholar] [CrossRef] [PubMed]
- Cen, Q.; Kang, L.; Zhou, D.; Zhang, X.; Tian, Q.; Zhang, X.; Mou, W.; Dang, C.; Fang, Y.; Xue, D. Genome-Wide Identification and Expression Analysis of RCC1 Gene Family under Abiotic Stresses in Rice (Oryza sativa L.). Agronomy 2023, 13, 703. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
| Gene Name | Accession Number | Chromosome | Genomic Location | CDS Length (bp) | Protein | Subcellular Localization | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| RAP-ID | MSU-ID | Size (aa) | MV (Da) | pI | GRAVY | |||||
| OsFAR1 | Os09g0567500 | LOC_Os09g39410 | 9 | 22660725–22665351 | 1518 | 505 | 56,713.1 | 8.79 | 0.044 | Nucleus |
| OsFAR2 | Os03g0167600 | LOC_Os03g07140 | 3 | 3653709–3657069 | 1827 | 608 | 65,228.14 | 7.03 | −0.105 | Chloroplast |
| OsFAR3 | Os04g0354600 | LOC_Os04g28620 | 4 | 16945210–16951596 | 1500 | 499 | 56,729.88 | 8.53 | −0.083 | Cytoplasm |
| OsFAR4 | Os08g0557800 | LOC_Os08g44360 | 8 | 27916313–27922609 | 1530 | 509 | 57,435.52 | 8.1 | −0.082 | Cytoplasm |
| OsFAR5 | Os04g0353600 | LOC_Os04g28520 | 4 | 16882610–16890074 | 1494 | 497 | 56,404.22 | 7.12 | −0.08 | Cytoplasm |
| OsFAR6 | NA | LOC_Os07g23340 | 7 | 13149738–13153339 | 1242 | 413 | 47,928.54 | 8.98 | −0.249 | Cytoplasm |
| OsFAR7 | Os07g0489100 | LOC_Os07g30600 | 7 | 18106225–18111251 | 1428 | 475 | 53,384.97 | 8.94 | 0.017 | Nucleus |
| OsFAR8 | Os08g0298700 | LOC_Os08g20200 | 8 | 12115583–12121337 | 1491 | 496 | 56,266.4 | 8.67 | −0.114 | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, D.; Ding, M.; Wen, S.; Tian, Q.; Zhang, X.; Fang, Y.; Xue, D. Characterization of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Its Response to Abiotic Stress in Rice (Oryza sativa L.). Plants 2024, 13, 1010. https://doi.org/10.3390/plants13071010
Zhou D, Ding M, Wen S, Tian Q, Zhang X, Fang Y, Xue D. Characterization of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Its Response to Abiotic Stress in Rice (Oryza sativa L.). Plants. 2024; 13(7):1010. https://doi.org/10.3390/plants13071010
Chicago/Turabian StyleZhou, Danni, Mingyu Ding, Shuting Wen, Quanxiang Tian, Xiaoqin Zhang, Yunxia Fang, and Dawei Xue. 2024. "Characterization of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Its Response to Abiotic Stress in Rice (Oryza sativa L.)" Plants 13, no. 7: 1010. https://doi.org/10.3390/plants13071010
APA StyleZhou, D., Ding, M., Wen, S., Tian, Q., Zhang, X., Fang, Y., & Xue, D. (2024). Characterization of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Its Response to Abiotic Stress in Rice (Oryza sativa L.). Plants, 13(7), 1010. https://doi.org/10.3390/plants13071010







