A New Record of Pogonatum tahitense (Polytrichaceae) from Tibet, China: Taxonomic Description, Range Expansion, and Biogeographic History
Abstract
1. Introduction
2. Results
2.1. Morphological Study
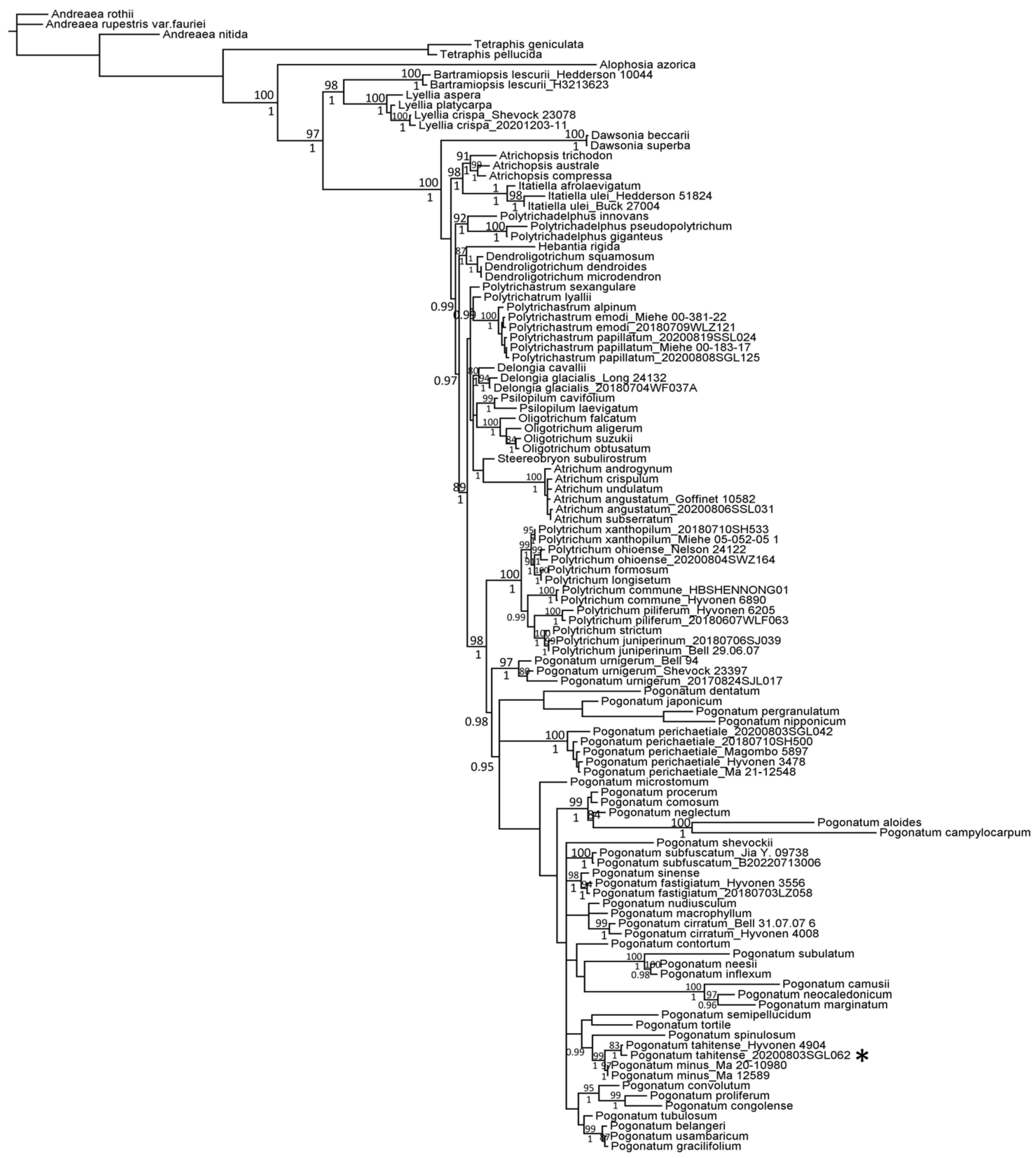
2.2. Phylogenetic Relationship
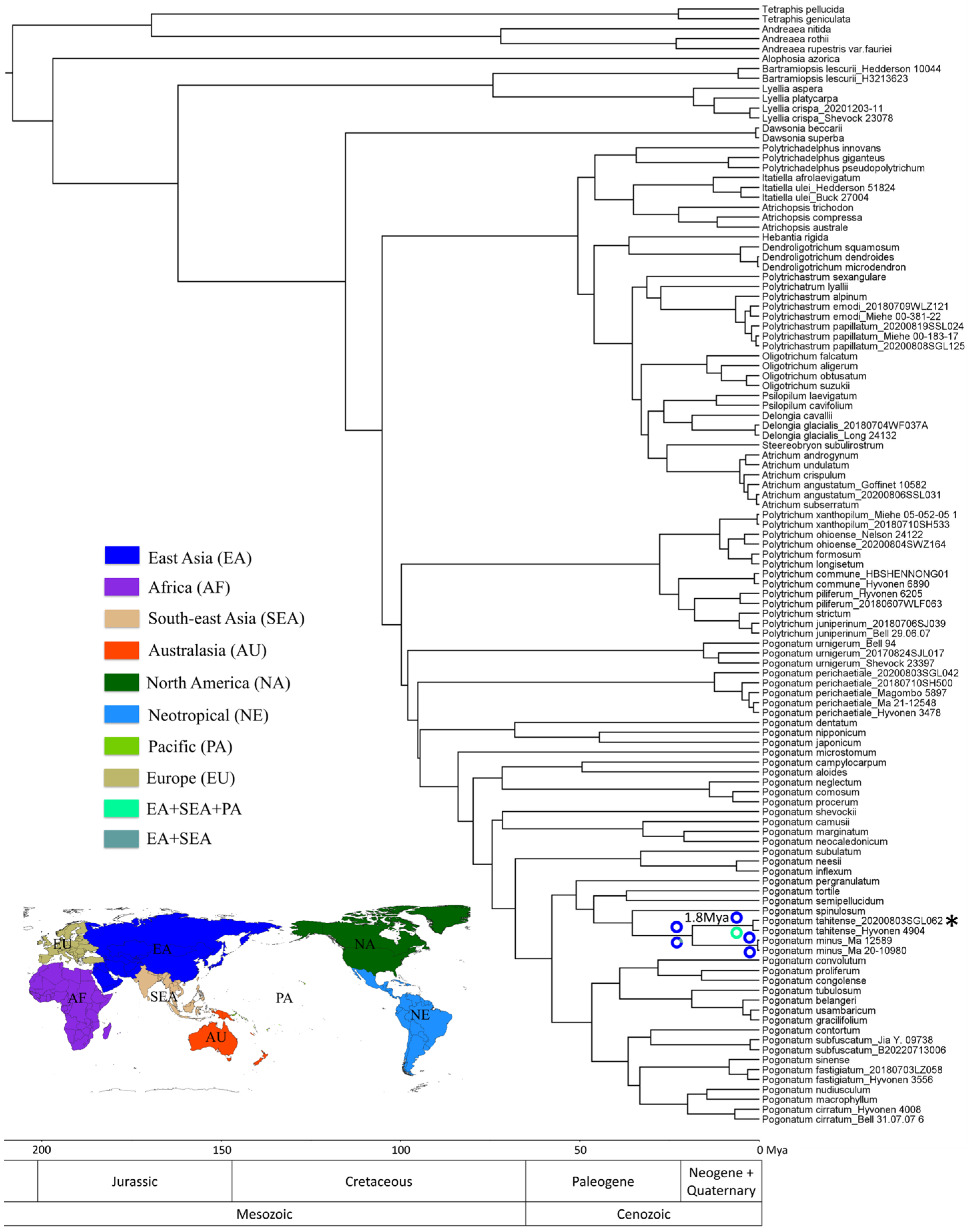
2.3. Divergence Time and Biogeographic History
3. Discussion
3.1. Phylogenetic Relationships
3.2. Pogonatum Tahitense and Its Related Species
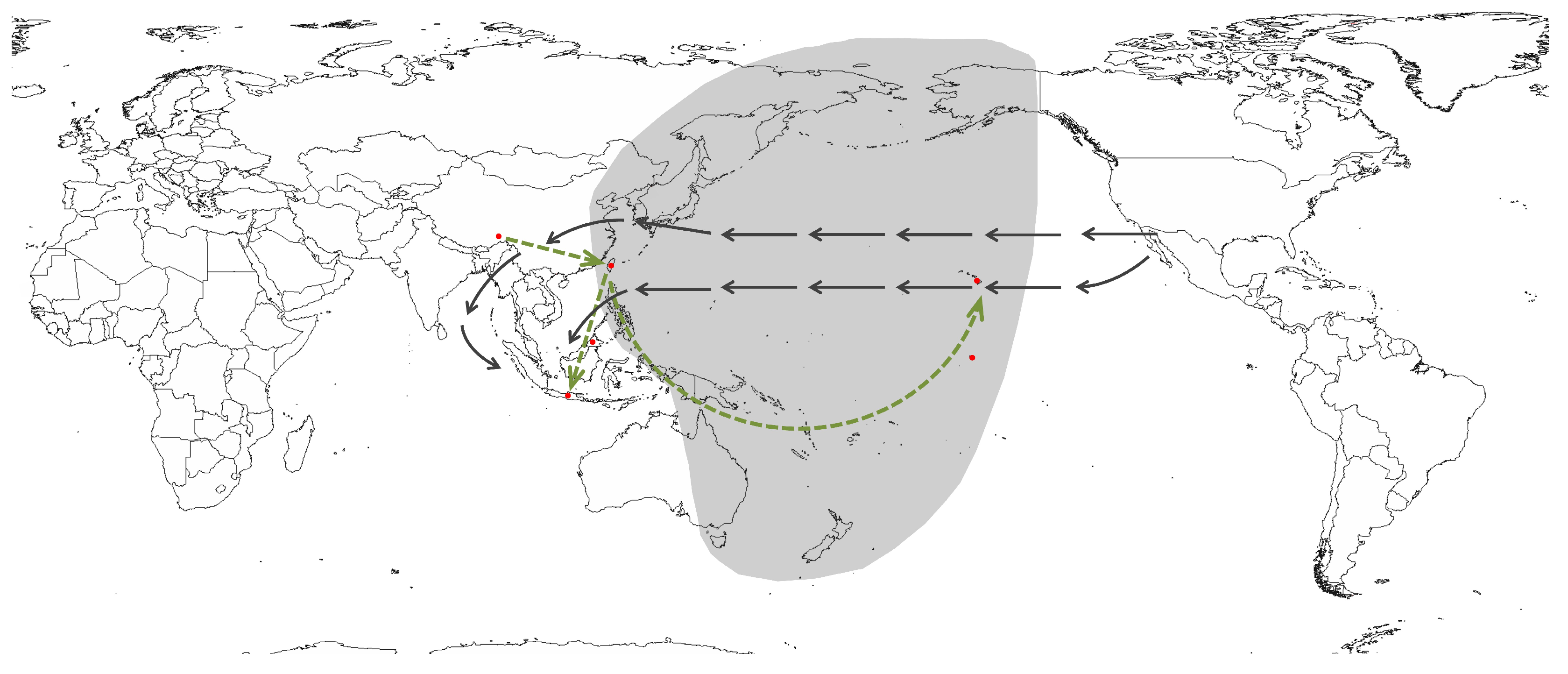
3.3. Biogeographic History of Distribution Patterns
4. Materials and Methods
4.1. Taxon Sampling
4.2. Morphological Study
4.3. DNA Isolation, PCR, and Sequencing
4.4. Phylogenetic Analyses
4.5. Divergence Time Estimates
4.6. Biogeographic Inference
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwägrichen, C.F. Species Muscorum Frondosorum Xiv; G.N. Nauck: Berlin, Germany, 1830. [Google Scholar]
- Bell, N.; Kariyawasam, I.; Flores, J.; Hyvnen, J. The diversity of the Polytrichopsida—A review. Bryophyt. Divers. Evol. 2021, 43, 98–111. [Google Scholar] [CrossRef]
- Palisot de Beauvois, A.M.F.J. Prodrome de l’aethéogamie our d’un traité sur les familles de plantes dont la fructification est extraordinaire. Mag. Encycl. 1804, 9, 289–330. [Google Scholar]
- Hyvönen, J. A synopsis of genus Pogonatum (Polytrichaceae, Musci). Acta Bot. Fenn. 1989, 138, 1–87. [Google Scholar]
- Hedwig, J. Species Muscorum Frondosorum vi; Joannis Ambrosii Barthii: Lipsiae, Germany, 1801. [Google Scholar]
- Koskinen, S.; Hyvönen, J. Pogonatum (Polytrichales, Bryophyta) revisited. In Molecular Systematics of Bryophytes; Goffinet, B., Hollowell, V., Magill, R., Eds.; Monographs in Systematic Botany from the Missouri Botanical Garden 98; Missouri Botanical Garden Press: St. Louis, MI, USA, 2004; pp. 255–269. [Google Scholar] [CrossRef]
- Bell, N.E.; Hyvönen, J. Phylogeny of the moss class Polytrichopsida (BRYOPHYTA): Generic-level structure and incongruent gene trees. Mol. Phylogenet. Evol. 2010, 55, 381–398. [Google Scholar] [CrossRef] [PubMed]
- Bescherelle, É. Florule bryologique de Tahiti et des iles de Nukahiva et Mangareva. Ann. Sci. Nat. Bot. 1895, 20, 1–62. [Google Scholar]
- Hyvönen, J.; Lai, M.J. Polytrichaceae (Musci) in Taiwan (China). J. Hattori Bot. Lab. 1991, 70, 119–141. [Google Scholar] [CrossRef]
- Suleiman, M.; Edwards, S.R. Mosses of Mt. Trus Madi, Sabah, Malaysia. Bryophyt. Divers. Evol. 2002, 21, 57–64. [Google Scholar] [CrossRef]
- Bell, N.E.; Hyvönen, J.; Yao, K.Y.; Ma, W.Z. Description and phylogenetic investigation of Pogonatum shevockii N.E.Bell & Hyvönen (Polytrichaceae), a new East Asian species with a unique leaf morphology. J. Bryol. 2017, 39, 235–246. [Google Scholar] [CrossRef]
- Deng, T.; Zhang, J.; Meng, Y.; Volis, S.; Sun, H.; Nie, Z. Role of the Qinghai-Tibetan Plateau uplift in the Northern Hemisphere disjunction: Evidence from two herbaceous genera of Rubiaceae. Sci. Rep. 2017, 7, 13411. [Google Scholar] [CrossRef]
- Niu, Y.T.; Ye, J.F.; Zhang, J.L.; Wan, J.Z.; Yang, T.; Wei, X.X.; Lu, L.M.; Li, J.H.; Chen, Z.D. Long-distance dispersal or postglacial contraction? Insights into disjunction between Himalaya-Hengduan Mountains and Taiwan in a cold-adapted herbaceous genus, Triplostegia. Ecol. Evol. 2017, 8, 1131–1146. [Google Scholar] [CrossRef]
- Jin, W.Y.; Li, H.W.; Wei, R.; Huang, B.H.; Liu, B.; Sun, T.T.; Mabberly, D.J.; Liao, P.C.; Yang, Y. New insights into biogeographical disjunctions between Taiwan and the Eastern Himalayas: The case of Prinsepia (Rosaceae). Taxon 2020, 69, 278–289. [Google Scholar] [CrossRef]
- Anderson, F.E.; Swofford, D.L. Should we be worried about long-branch attraction in real data sets? Investigations using metazoan 18S rDNA. Mol. Phylogenet. Evol. 2004, 33, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Bergsten, J. A review of long-branch attraction. Cladistics 2005, 21, 163–193. [Google Scholar] [CrossRef] [PubMed]
- Derda, G.S.; Wyatt, R.; Hyvönen, J. Genetic similarities among the hair-cap mosses (Polytrichaceae) as revealed by enzyme electrophoresis. Bryologist 1999, 102, 352–365. [Google Scholar] [CrossRef]
- Derda, G.S.; Wyatt, R. Isozyme evidence regarding the origins of three allopolyploid species of Polytrichastrum (Polytrichaceae, Bryophyta). Plant Syst. Evol. 2000, 220, 37–53. [Google Scholar] [CrossRef]
- Maciel-Silva, A.S.; Da Silva, F.C.L.; Válio, I.F.M. All green, but equal? Morphological traits and ecological implications on spores of three species of mosses in the Brazilian Atlantic Forest. An. Acad. Bras. Ciênc. 2014, 86, 1249–1262. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Zhao, L.; Song, X.; Wang, Q.; Kou, J.; Jiang, Y.; Shao, X. Morphological traits of Bryum argenteum and its response to environmental variation in arid and semi-arid areas of Tibet. Ecol. Eng. 2019, 136, 101–107. [Google Scholar] [CrossRef]
- Buerki, S.; Forest, F.; Alvarez, N.; Nylander, J.A.A.; Arrigo, N.; Sanmartín, I. An evaluation of new parsimony based versus parametric inference methods in biogeography: A case study using the globally distributed plant family Sapindaceae. J. Biogeogr. 2011, 38, 531–550. [Google Scholar] [CrossRef]
- Abdel-Monem, A.; Fernandez, L.; Boone, G. K-Ar ages from the eastern Azores group (Santa Maria, São Miguel and the Formigas islands). Lithos 1975, 8, 247–254. [Google Scholar] [CrossRef]
- Serralheiro, A.; Madeira, J.M. Stratigraphy and geochronology of Santa Maria Island (Azores). Açoreana 1993, 7, 575–592. [Google Scholar]
- Sibrant, A.L.R.; Hildenbrand, A.; Marques, F.O.; Costa, A.C.G. Volcano-tectonic evolution of the Santa Maria Island (Azores): Implications for palaeostress evolution at the western Eurasia-Nubia plate boundary. J. Volcanol. Geotherm. Res. 2015, 291, 49–62. [Google Scholar] [CrossRef]
- Sibuet, J.C.; Hsu, S.K. Geodynamics of the Taiwan arc-arc collision. Tectonophysics 1997, 274, 221–251. [Google Scholar] [CrossRef]
- Sibuet, J.C.; Hsu, S.K. How was Taiwan created? Tectonophysics 2004, 379, 159–181. [Google Scholar] [CrossRef]
- Harrison, T.M.; Copeland, P.; Kidd, W.S.F.; Yin, A. Raising Tibet. Science 1992, 255, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Molnar, P.; England, P.; Martiod, J. Mantle dynamics, uplift of the Tibetan Plateau and the Indian monsoon development. Rev. Geophys. 1993, 34, 357–396. [Google Scholar] [CrossRef]
- Olson, S. Evolution in Hawaii: A Supplement to ‘Teaching about Evolution and the Nature of Science’; The National Academies Press: Washington, DC, USA, 2004. [Google Scholar] [CrossRef]
- Uto, K.; Yamamoto, Y.; Sudo, M.; Uchiumi, S.; Ishizuka, O.; Kogiso, T.; Tsunakawa, H. New K-Ar ages of the Society Islands, French Polynesia, and implications for the Society hotspot feature. Earth Planet. Space 2007, 59, 879–888. [Google Scholar] [CrossRef]
- Katili, J.A. Geochronology of West Indonesia and its implication on plate tectonics. Tectonophysics 1973, 19, 195–212. [Google Scholar] [CrossRef]
- Katili, J.A. Volcanism and plate tectonics in the Indonesian island arcs. Tectonophysics 1975, 26, 165–188. [Google Scholar] [CrossRef]
- Zeng, W.B. The passageway of the flora migration on both sides of the Taiwan Strait in Pleistocene epoch. Acta Bot. Yunnanica 1993, 16, 107–110. [Google Scholar]
- Chen, Z.D.; Ying, J.S.; Lu, A.M. Disjunct distribution of seed plants between southwestern China and Taiwan Island of China. Chin. Bull. Bot. 2012, 47, 551–570. [Google Scholar] [CrossRef]
- Ye, J.F.; Chen, Z.D.; Liu, B.; Qin, H.N.; Yang, Y. Disjunct distribution of vascular plants between southwestern area and Taiwan area in China. Biodivers. Sci. 2012, 20, 482–494. [Google Scholar] [CrossRef]
- Zhu, H. A biogeographical comparison between Yunnan, Southwest China, and Taiwan, Southeast China, with implications for the evolutionary history of the East Asian Flora. Ann. Mo. Bot. Gard. 2016, 101, 750–771. [Google Scholar] [CrossRef]
- Plášek, V.; Komínková, Z.; Ochyra, R.; Fialová, L.; Guo, S.; Sulayman, M. A Synopsis of Orthotrichum s. lato (Bryophyta, Orthotrichaceae) in China, with Distribution Maps and a Key to Determination. Plants 2021, 10, 499. [Google Scholar] [CrossRef] [PubMed]
- Longton, R.E. The Biology of Polar Bryophytes and Lichens; Cambridge Univ. Press: Cambridge, UK, 1988. [Google Scholar] [CrossRef]
- Price, J.P.; Wagner, W.L. Origins of the Hawaiian flora: Phylogenies and biogeography reveal patterns of long-distance dispersal. J. Syst. Evol. 2018, 56, 600–620. [Google Scholar] [CrossRef]
- Raine, A.F.; Driskill, S.; Raine, H.; Rothe, J.; Rossiter, S.; Anderson, T.; Bache, M. Post-fledging distribution of ’ua’u (Hawaiian petrel Pterodroma sandwichensis) from Kaua’i, Hawai’I and effectiveness of rehabilitation. Endanger. Species Res. 2023, 52, 27–40. [Google Scholar] [CrossRef]
- Bamford, M.; Watkins, D.; Bancroft, W.; Tischler, G.; Wahl, J. Migratory Shorebirds of the East Asian—Australasian Flyway Population Estimates and Internationally Important Sites; Wetlands International—Oceania: Canberra, Australia, 2008. [Google Scholar]
- Shaffer, S.A.; Tremblay, Y.; Weimerskirch, H.; Scott, D.; Thompson, D.R.; Sagar, P.M.; Moller, H.; Taylor, G.A.; Foley, D.G.; Block, B.A.; et al. Migratory shearwaters integrate oceanic resources across the Pacific Ocean in an endless summer. Proc. Natl. Acad. Sci. USA 2006, 103, 12799–12802. [Google Scholar] [CrossRef]
- Tropicos v3.4.2. Tropicos.org. Missouri Botanical Garden. Available online: https://tropicos.org (accessed on 8 January 2024).
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Ahonen, I.; Muona, J.; Piippo, S. Inferring the phylogeny of the Lejeuneaceae (Jungermanniopsida): A first appraisal of molecular data. Bryol 2003, 106, 297–308. [Google Scholar] [CrossRef]
- Cox, C.J.; Hedderson, T.A.J. Phylogenetic relationships among the ciliate arthrodontous mosses: Evidence from chloroplast and nuclear DNA sequences. Plant Syst. Evol. 1999, 215, 119–139. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Beckert, S.; Steinhauser, S.; Muhle, H.; Knoop, V. A molecular phylogeny of bryophytes based on nucleotide sequences of the mitochondrial nad5 gene. Plant Syst. Evol. 1999, 218, 179–192. [Google Scholar] [CrossRef]
- Shaw, A.J. Molecular phylogeography and cryptic speciation in the mosses, Mielichhoferia elongata and M. mielichhoferiana (Bryaceae). Mol. Ecol. 2000, 9, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Troshin, P.V.; Procter, J.B.; Barton, G.J. Java bioinformatics analysis web services for multiple sequence alignment–JABAWS:MSA. Bioinformatics 2011, 27, 2001–2002. [Google Scholar] [CrossRef] [PubMed]
- Troshin, P.V.; Procter, J.B.; Sherstnev, A.; Barton, D.L.; Madeira, F.; Barton, G.J. JABAWS 2.2 Distributed Web Services for Bioinformatics: Protein Disorder, Conservation and RNA Secondary Structure. Bioinformatics 2018, 34, 1939–1940. [Google Scholar] [CrossRef]
- Flouri, T.; Izquierdo-Carrasco, F.; Darriba, D.; Aberer, A.J.; Nguyen, L.T.; Minh, B.Q.; von Haeseler, A.; Stamatakis, A. The Phylogenetic Likelihood Library. Syst. Biol. 2014, 64, 356–362. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinform. Appl. Note 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F. Bayesian inference of character evolution. Trends Ecol. Evol. 2004, 19, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis andpost-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Palmer, J.D. Plastid chromosome: Structure and evolution. In The Molecular Biology of Plastids; Bogorad, L., Vasil, I.K., Eds.; Academic Press: San Diego, CA, USA, 1991; pp. 5–53. [Google Scholar] [CrossRef]
- Schnabel, A.; Wendel, J.F. Cladistic biogeography of Gleditsia (Leguminosae) based on ndhf and rpl16 chloroplast gene sequences. Am. J. Bot. 1998, 85, 1753–1765. [Google Scholar] [CrossRef]
- Gaut, B.S. Molecular clocks and nucleotide substitution rates in higher plants. Evol. Biol. 1998, 30, 93–120. [Google Scholar] [CrossRef]
- Bakker, F.T.; Olsen, J.L.; Stam, W.T. Evolution of nuclear rDNA its sequences in the Cladophora albida/sericea clade (Chlorophyta). J. Mol. Evol. 1995, 40, 640–651. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, syy032. [Google Scholar] [CrossRef] [PubMed]
- Bechteler, J.; Peñaloza-Bojacá, G.; Bell, D.; Burleigh, J.G.; McDaniel, S.F.; Davis, E.C.; Sessa, E.B.; Bippus, A.; Cargill, D.C.; Chantanoarrapint, S.; et al. Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. Am. J. Bot. 2023, 110, e16249. [Google Scholar] [CrossRef] [PubMed]
- Bippus, A.C.; Stockey, R.A.; Rothwell, G.W.; Tomescu, A.M. Extending the fossil record of Polytrichaceae: Early Cretaceous Meantoinea alophosioides gen. et sp. nov., permineralized gametophytes with gemma cups from Vancouver Island. Am. J. Bot. 2017, 104, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A.S.; Herendeen, P.S.; Merrill, G.L.S.; Crane, P.R. Sporophytes and gametophytes of Polytrichaceae from the Campanian (Late Cretaceous) of Georgia, USA. Int. J. Plant Sci. 1997, 158, 489–499. [Google Scholar] [CrossRef]
- Bippus, A.C.; Escapa, I.E.; Tomescu, A.M.F. Wanted dead or alive (probably dead): Stem group Polytrichaceae. Am. J. Bot. 2018, 105, 1243–1263. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, X.; Dimitrov, D.; Pellissier, L.; Borregaard, M.K.; Shrestha, N.; Su, X.; Luo, A.; Zimmermann, N.E.; Rahbek, C.; et al. An updated floristic map of the world. Nat. Commun. 2023, 14, 2990. [Google Scholar] [CrossRef]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef]
- Ronquist, F. Dispersal-vicariance analysis: A new approach to the quantification of historical biogeography. Syst. Biol. 1997, 46, 195–203. [Google Scholar] [CrossRef]
- Yu, Y.; Harris, A.J.; He, X. S-DIVA (Statistical Dispersal-Vicariance Analysis): A tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 2010, 56, 848–850. [Google Scholar] [CrossRef]
- Ree, R.; Moore, B.R.; Webb, C.O.; Donoghue, M.T. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 2005, 59, 2299–2311. [Google Scholar] [CrossRef]
- Ree, R.; Smith, S. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Bot. 2008, 57, 4–14. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Song, X.; Chen, C.; Li, S.; Gu, J.; Shao, X. A New Record of Pogonatum tahitense (Polytrichaceae) from Tibet, China: Taxonomic Description, Range Expansion, and Biogeographic History. Plants 2024, 13, 846. https://doi.org/10.3390/plants13060846
Sun Y, Song X, Chen C, Li S, Gu J, Shao X. A New Record of Pogonatum tahitense (Polytrichaceae) from Tibet, China: Taxonomic Description, Range Expansion, and Biogeographic History. Plants. 2024; 13(6):846. https://doi.org/10.3390/plants13060846
Chicago/Turabian StyleSun, Yu, Xiaotong Song, Chunfa Chen, Shuang Li, Jiqi Gu, and Xiaoming Shao. 2024. "A New Record of Pogonatum tahitense (Polytrichaceae) from Tibet, China: Taxonomic Description, Range Expansion, and Biogeographic History" Plants 13, no. 6: 846. https://doi.org/10.3390/plants13060846
APA StyleSun, Y., Song, X., Chen, C., Li, S., Gu, J., & Shao, X. (2024). A New Record of Pogonatum tahitense (Polytrichaceae) from Tibet, China: Taxonomic Description, Range Expansion, and Biogeographic History. Plants, 13(6), 846. https://doi.org/10.3390/plants13060846






