Abstract
The carnivorous pitcher plants of the genus Nepenthes have long been known for their ethnobotanical applications. In this study, we prepared various extracts from the pitcher, stem, and leaf of Nepenthes miranda using 100% ethanol and assessed their inhibitory effects on key enzymes related to skin aging, including elastase, tyrosinase, and hyaluronidase. The cytotoxicity of the stem extract of N. miranda on H838 human lung carcinoma cells were also characterized by effects on cell survival, migration, proliferation, apoptosis induction, and DNA damage. The cytotoxic efficacy of the extract was enhanced when combined with the chemotherapeutic agent 5-fluorouracil (5-FU), indicating a synergistic effect. Flow cytometry analysis suggested that the stem extract might suppress H838 cell proliferation by inducing G2 cell cycle arrest, thereby inhibiting carcinoma cell proliferation. Gas chromatography–mass spectrometry (GC–MS) enabled the tentative identification of the 15 most abundant compounds in the stem extract of N. miranda. Notably, the extract showed a potent inhibition of the human RPA32 protein (huRPA32), critical for DNA replication, suggesting a novel mechanism for its anticancer action. Molecular docking studies further substantiated the interaction between the extract and huRPA32, highlighting bioactive compounds, especially the two most abundant constituents, stigmast-5-en-3-ol and plumbagin, as potential inhibitors of huRPA32’s DNA-binding activity, offering promising avenues for cancer therapy. Overall, our findings position the stem extract of N. miranda as a promising source of natural compounds for anticancer therapeutics and anti-skin-aging treatments, warranting further investigation into its molecular mechanisms and potential clinical applications.
1. Introduction
Nepenthes, a plant traditionally utilized in folk medicine, has been employed to remedy various ailments, including stomachaches and fevers [1]. Owing to its longstanding use in ethnomedicine, extracts from Nepenthes are considered relatively safe for pharmaceutical applications, potentially offering minimal side effects for human consumption. Consequently, the exploration and development of additional therapeutic uses are warranted.
Cancer, characterized by uncontrolled cell growth and the potential for tumors to spread locally or systemically, remains one of the world’s deadliest diseases [2,3,4]. WHO reports estimate around 20 million new cases and 10 million fatalities from cancer in 2023. Notably, lung cancer ranks among the primary causes of these fatalities. Non-small cell lung cancer (NSCLC), constituting approximately 85% of lung cancer cases, is commonly treated through surgery, chemotherapy, radiation, and targeted therapy. However, NSCLCs often show limited responsiveness to chemotherapy [5,6]. Recently, natural compounds are increasingly explored as potential anti-lung cancer agents and alternative treatment options [7,8,9,10]. These phytochemicals may also offer solutions to challenges like drug resistance, a common issue in lung cancer treatment [11]. Accordingly, this study assesses the cytotoxic effects of Nepenthes miranda extract on the viability, migration, proliferation, DNA damage, and apoptosis of human NSCLC H838 carcinoma cells. A notable advantage of using natural extracts against cancer cells lies in their multi-targeted modes of action [12]. Furthermore, we also investigate the combined effects of N. miranda extract with the clinical anticancer drug 5-fluorouracil (5-FU) to determine the presence of any synergistic effects in combating NSCLC.
The skin, the most extensive and intricate organ in the human body, serves as a crucial barrier protecting against external elements [13]. Its appearance significantly influences social interactions, with youthful and healthy skin often perceived positively in social contexts [14,15]. As we age, our skin undergoes changes that can profoundly affect our overall health and quality of life, manifesting as thinness, dryness, reduced elasticity, rough texture, wrinkles, and dark spots [16,17]. Accordingly, the continual discovery of anti-aging agents holds substantial value. Plant-based compounds, particularly secondary metabolites and whole plant extracts, have been extensively studied for their anti-aging properties [18]. Plants are rich in compounds like polyphenols, renowned for their potent antioxidant properties, combating aging and photodamage. Isolated polyphenols from green tea, such as catechin and epigallocatechin gallate (EGCG), are known to target aging-associated enzymes like elastase [19]. Other natural compounds, including quercetin (Que) and myricetin (Myr), have been identified as inhibitors of tyrosinase [20] and hyaluronidase [21]. Nepenthes thrives in extended exposure to bright sunlight, essential for its optimal growth. Given the association between UV light exposure and photodamage, skin aging, and skin cancer, exploring the anti-skin aging properties of Nepenthes extract is crucial. Accordingly, this study also aims to investigate this potential by examining the extract’s inhibitory effects on enzymes associated with skin aging, specifically elastase, tyrosinase, and hyaluronidase. Extracts from the Nepenthes plant, such as those from N. miranda [22,23], are noted for their high polyphenol content and significant antioxidant activity, suggesting their potential in anti-aging applications. Therefore, it is worth determining whether N. miranda extracts can specifically inhibit enzymes associated with the aging process.
Previously, the single-stranded DNA (ssDNA)-binding protein (SSB) [24], a pivotal player in DNA metabolism processes such as replication, repair, recombination, and replication fork restart in bacterial cells [25], was pinpointed as a molecular target inhibited by the N. miranda extract [26]. SSB, indispensable for bacterial DNA replication and cell reproduction, functions as a homotetramer where four oligonucleotide/oligosaccharide-binding (OB) folds create a DNA-binding domain [27,28,29]. The inhibition of SSB holds pharmacological promise for targeting bacterial pathogens [30,31,32]. Replication protein A (RPA) is a eukaryotic counterpart to bacterial SSB and exhibit similar mechanistic roles [33]. RPA, also an OB-fold protein, consists of a heterotrimer comprised of RPA70, RPA32, and RPA14 subunits, each capable of independent function. RPA32, in particular, binds ssDNA and interacts with various genome maintenance proteins, including RAD52, TIPIN, XPA, UNG2, and SMARCAL1 [34]. RPA32 shares structural and functional traits with alpha-accessory factor-44 (AAF-44), regulating DNA replication and enhancing DNA primase and polymerase-alpha activities [35]. Inhibition of RPA triggers replication stress, represses tumor growth [36], and is linked to cell death post-irradiation [37]. In addition, the decrease of RPA32 foci formation plays a role in suppressing NSCLC [38]. Considering that normal human cells do not require continuous DNA synthesis, an inhibitor targeting RPA32 could demonstrate selective cytotoxicity towards rapidly proliferating cells in need of genome replication. Similar to the case of inhibiting pathogenic SSB [26], N. miranda extract was investigated as a potential inhibitor of RPA32.
The Nepenthes plant, traditionally used in folk medicine, may offer novel therapeutic applications. This study broadens its medicinal scope to include cytotoxicity against H838 human NSCLC carcinoma cells, anti-skin aging, and RPA32 inhibition, employing a stem extract of N. miranda extracted with 100% ethanol. Ethanol, being a safer solvent for human use, is a preferable option for extractions. Using gas chromatography–mass spectrometry (GC–MS) and the docking program AutoDock Vina, we tentatively identified the chemical constituents of the extract and predicted the binding interactions of these compounds with RPA32. The results of this study collectively suggest that N. miranda holds potential pharmacological benefits, warranting further exploration for therapeutic applications.
2. Results
2.1. Anti-Elastase Activity of N. miranda Extract
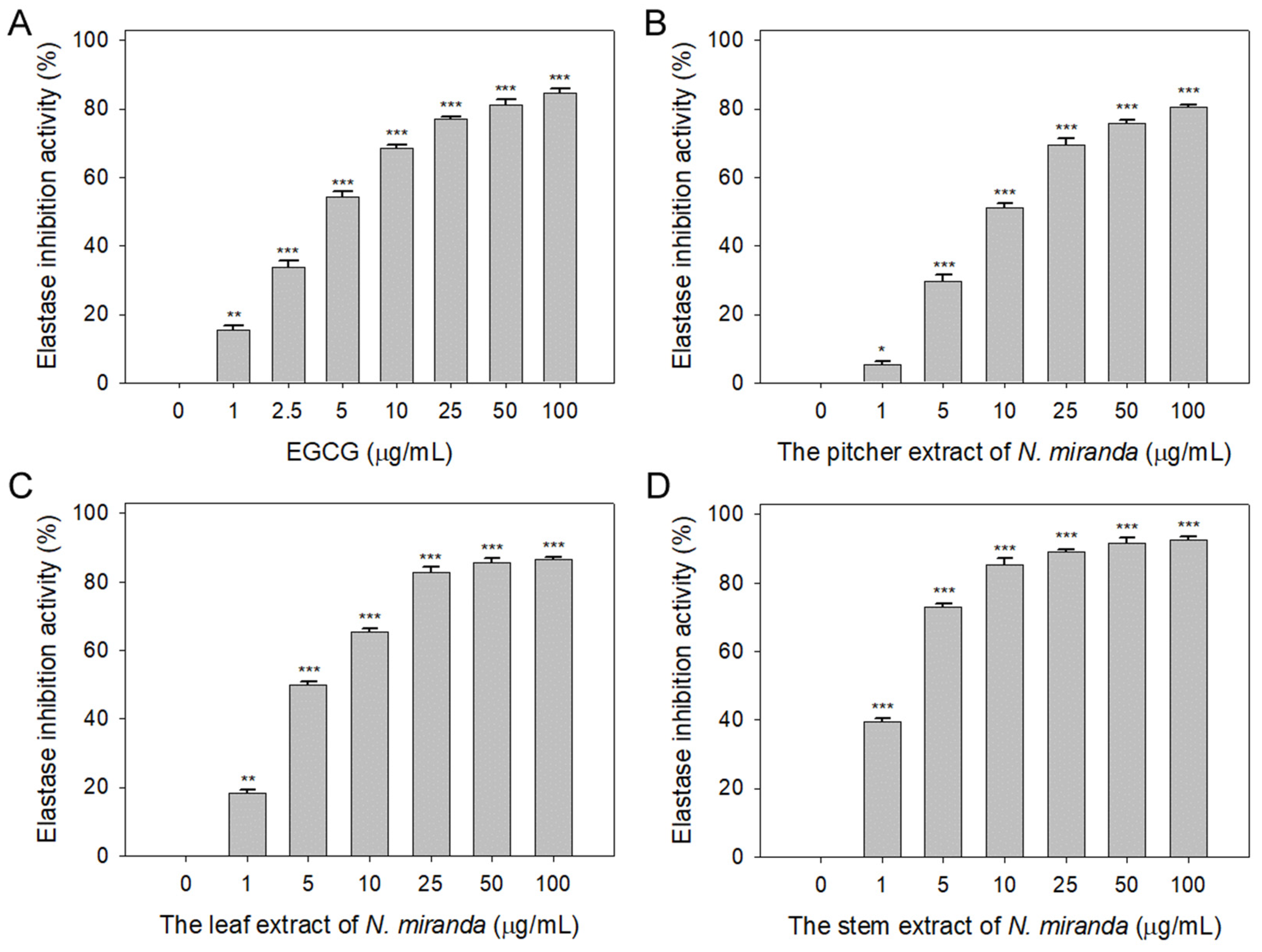
The role of elastin, which constitutes 2–4% of the extracellular matrix, is crucial in maintaining skin elasticity and hydration [39]. Elastase, a protease, is responsible for breaking protein bonds and degrading them into polypeptides or amino acids. Dysregulation in the extracellular release of elastase can lead to the degradation of elastin and other matrix components, causing damage to epithelial cells. The potential of Nepenthes plant extracts in inhibiting elastase activity has not been previously explored and warrants investigation. In this study, we assessed the elastase inhibitory activity of extracts from N. miranda’s pitcher, leaf, and stem (0–100 μg/mL) obtained using 100% ethanol. The assay employed N-Succinyl-Ala-Ala-Ala-p-nitroanilide (AAAPVN) as the substrate, with epigallocatechin gallate (EGCG) serving as a positive control (0–100 μg/mL). At a concentration of 5 μg/mL, EGCG (Figure 1A), and the extracts from the pitcher (Figure 1B), leaf (Figure 1C), and stem (Figure 1D) of N. miranda displayed inhibition levels of 54%, 29%, 50%, and 73%, respectively. The half-maximal inhibitory concentrations (IC50) were determined to be 4.41 ± 0.34 μg/mL for EGCG, 8.66 ± 1.15 μg/mL for the pitcher, 5.31 ± 0.31 μg/mL for the leaf, and 2.29 ± 0.68 μg/mL for the stem extract (Table 1). These findings reveal, for the first time, the significant inhibitory effect of N. miranda extract on elastase, suggesting its potential to prevent elastin degradation, loss of skin elasticity, and the formation of wrinkles.
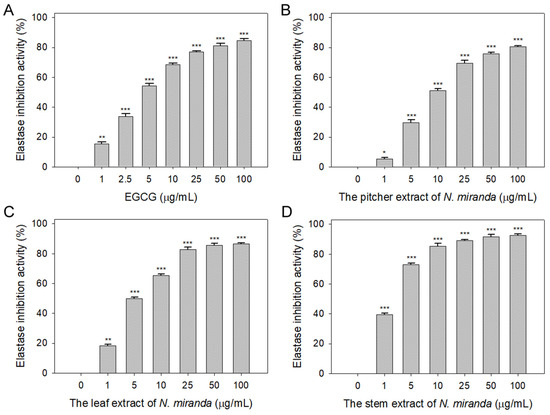
Figure 1.
Inhibition of elastase activity by N. miranda extract. The inhibitory effects were demonstrated on elastase activity by (A) EGCG and N. miranda extracts from the (B) pitcher, (C) leaf, and (D) stem. AAAPVN was utilized as the substrate. EGCG was employed as a positive control, while 10% DMSO was used as a negative control (indicating 0 μg/mL of N. miranda extract). Levels of statistical significance are denoted by * p < 0.05, ** p < 0.01, and *** p < 0.001 when compared to the control group.

Table 1.
Inhibition effects of N. miranda extract.
2.2. Anti-Tyrosinase Activity of N. miranda Extract
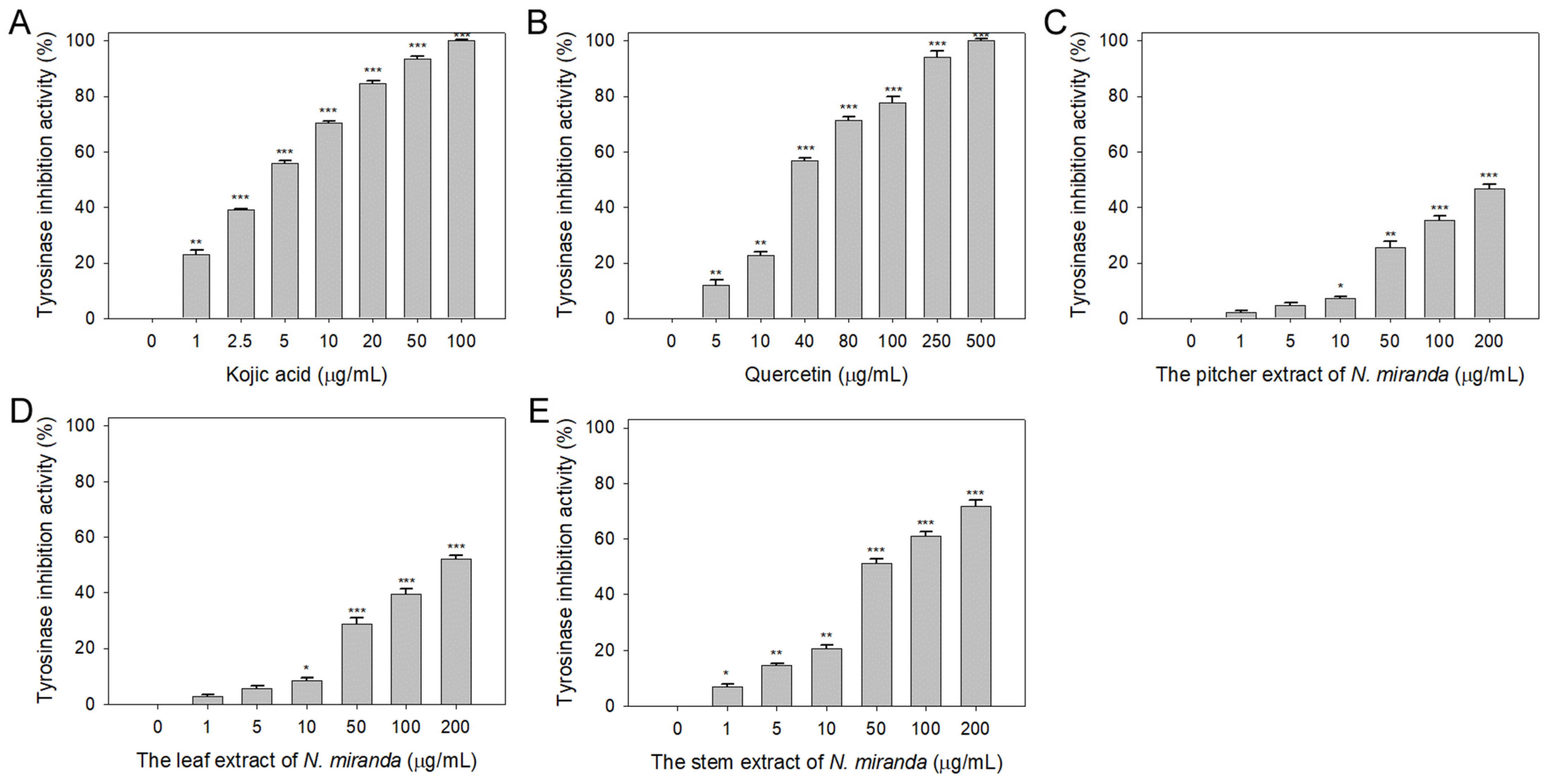
Beyond elastase, melanin synthesis is a key factor in addressing skin aging, with tyrosinase playing an essential role as the primary enzyme in melanogenesis [40,41]. Lowering tyrosinase activity can diminish melanin production, hence the potential inhibitory effect of N. miranda extracts on tyrosinase was explored in this study. The capability of Nepenthes plant extracts to curb tyrosinase activity had not been previously established. The tyrosinase-inhibiting properties of the extracts (concentrations 0–100 μg/mL) were assessed utilizing the modified dopachrome method, with 3,4-dihydroxy-L-phenylalanine (L-DOPA) as the substrate. Kojic acid (KA) and quercetin (Que) served as positive controls. At a concentration of 10 μg/mL, KA (Figure 2A), Que (Figure 2B), and N. miranda extracts from the pitcher (Figure 2C), leaf (Figure 2D), and stem (Figure 2E) exhibited inhibitory effects at rates of 70%, 23%, 7%, 9%, and 21%, respectively. The IC50 values were determined to be 3.94 ± 0.32 for KA, 33.64 ± 2.00 for Que, 180.57 ± 0.75 for the leaf, and 48.33 ± 2.92 μg/mL for the stem extract (Table 1). The IC50 for the pitcher extract was not ascertainable due to its minimal inhibitory action. These findings mark the first instance of demonstrating that N. miranda extracts, particularly the stem extract, can counteract tyrosinase activity, potentially offering protection against the formation of dark pigments and providing therapeutic avenues for treating skin hyperpigmentation and lightening the skin.
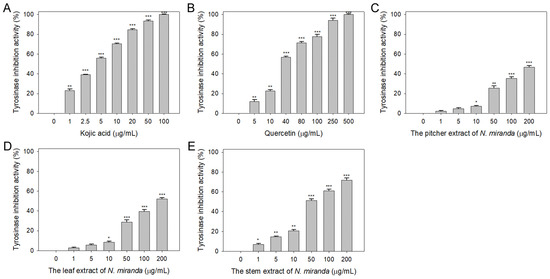
Figure 2.
Inhibition of tyrosinase activity by N. miranda extract. The inhibitory effects were investigated on tyrosinase activity by (A) KA, (B) Que, and N. miranda extracts from the (C) pitcher, (D) leaf, and (E) stem. L-DOPA was employed as the substrate. KA and Que were utilized as positive controls, while 10% DMSO was used as a negative control (representing 0 μg/mL of N. miranda extract). Levels of statistical significance are indicated by * p < 0.05, ** p < 0.01, and *** p < 0.001 in comparison to the control group.
2.3. Anti-Hyaluronidase Activity of N. miranda Extract
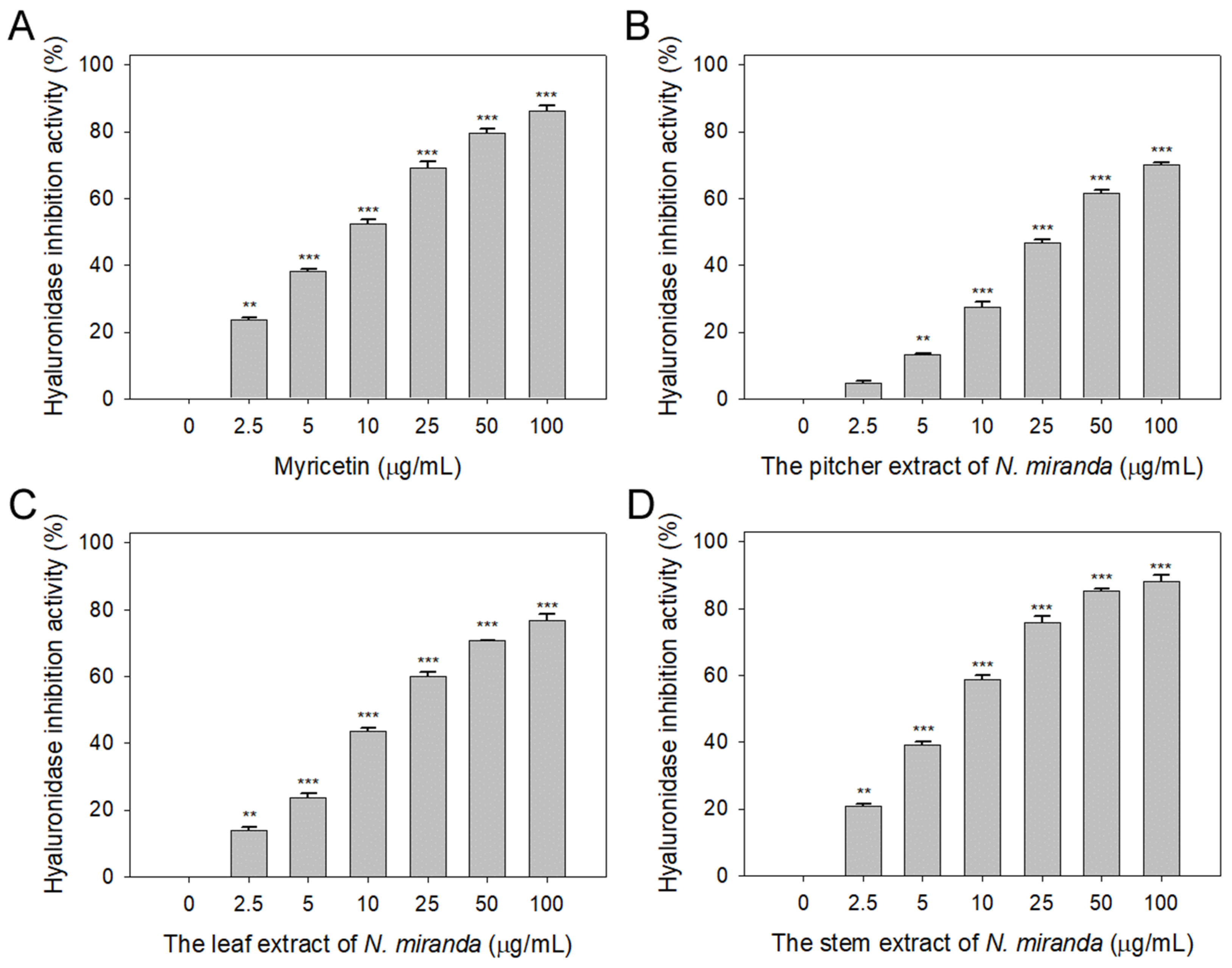
Prior to this study, the potential of Nepenthes plant extracts to inhibit hyaluronidase activity remained unexplored. Hyaluronic acid plays a pivotal role in maintaining moisture, providing a supple, hydrated texture to the body, and mitigating dryness, roughness, and wrinkles. Therefore, inhibitors of hyaluronidase may have significant implications in preserving skin moisture and texture [42]. Accordingly, we evaluated the hyaluronidase-inhibiting properties of extracts from N. miranda’s pitcher, leaf, and stem (concentrations ranging from 0–100 μg/mL), extracted using 100% ethanol. Hyaluronic acid was used as the substrate, with myricetin (Myr) as a positive control (0–100 μg/mL). At a concentration of 10 μg/mL, Myr (Figure 3A), and the extracts from the pitcher (Figure 3B), leaf (Figure 3C), and stem (Figure 3D) of N. miranda exhibited inhibitory effects at rates of 52%, 28%, 44%, and 59%, respectively. The IC50 values were calculated to be 9.52 ± 0.27 μg/mL for Myr, 31.67 ± 2.96 μg/mL for the pitcher, 16.18 ± 1.03 μg/mL for the leaf, and 7.89 ± 0.64 μg/mL for the stem extract (Table 1). These results, for the first time, highlight the considerable inhibitory potential of N. miranda extract against hyaluronidase, underscoring its potential role in maintaining hydration and preventing the signs of aging in the skin.
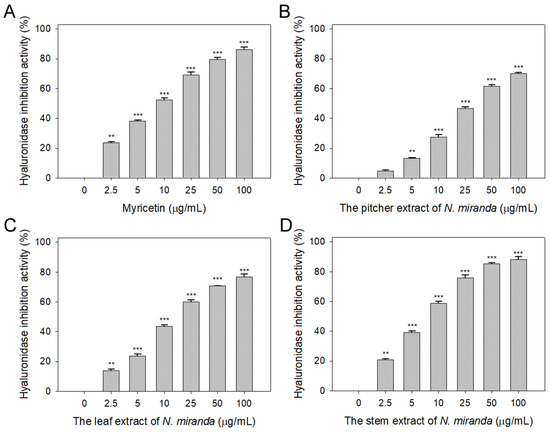
Figure 3.
Inhibition of hyaluronidase activity by N. miranda extract. The inhibitory effects were investigated on hyaluronidase activity by (A) Myr, and N. miranda extracts from the (B) pitcher, (C) leaf, and (D) stem. Hyaluronic acid was employed as the substrate. Myr was utilized as a positive control, while 10% DMSO was used as a negative control (representing 0 μg/mL of N. miranda extract). Levels of statistical significance are indicated by ** p < 0.01 and *** p < 0.001 in comparison to the control group.
2.4. Cytotoxic Activity of the Ethanol Extract of Nepenthes miranda on H838 Lung Adenocarcinoma Cells
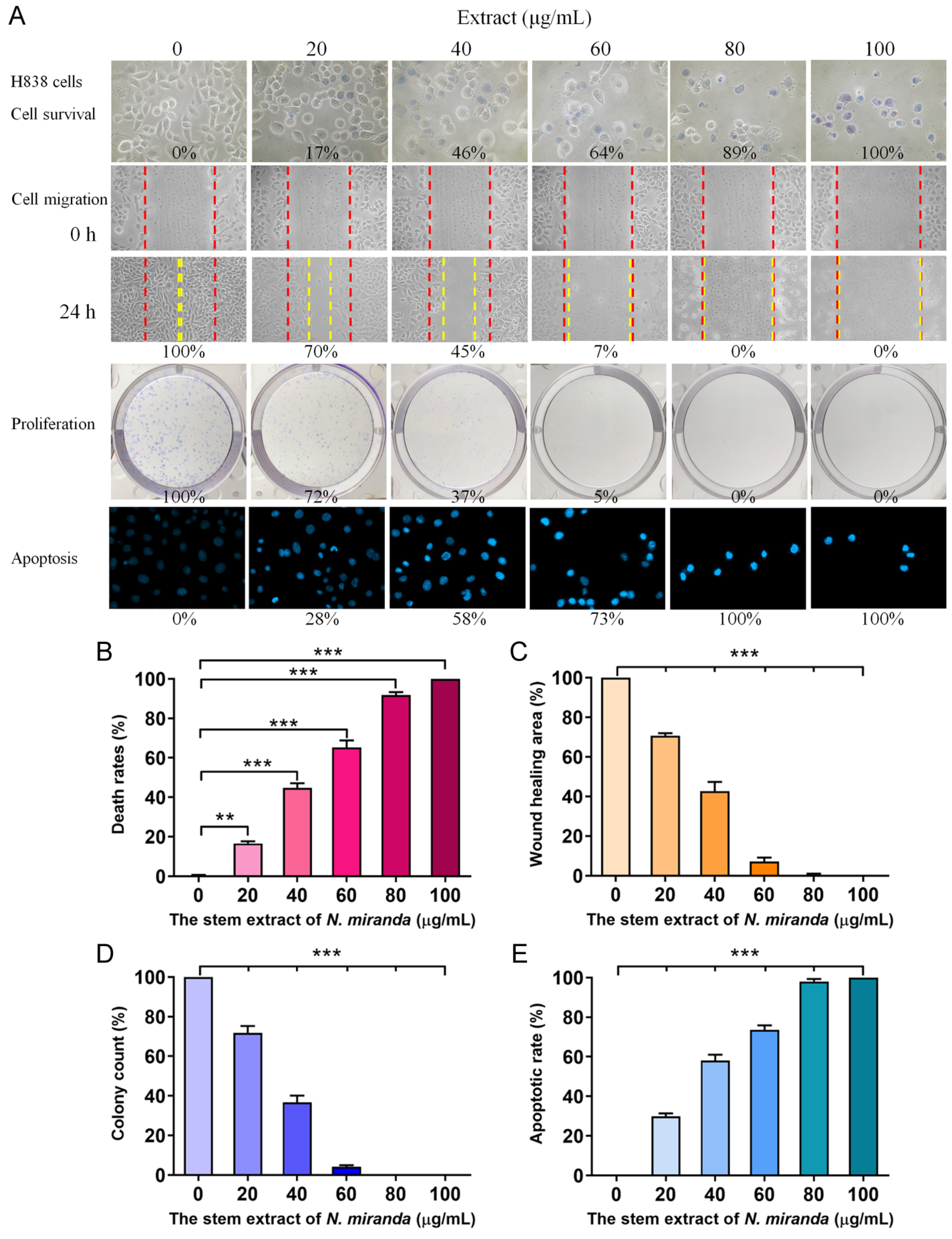
To delve deeper into the cytotoxic effects on cancer cells, particularly H838 human lung carcinoma cells, we utilized the stem extract of N. miranda prepared with 100% ethanol to explore its anticancer capabilities. These capabilities encompass cell viability, cell migration inhibition, suppression of cell proliferation, and DNA fragmentation indicative of apoptosis (Figure 4). The anti-H838 efficacy of N. miranda extracts was found to follow the order: stem > leaf > pitcher. Consequently, our research primarily focused on investigating the properties of the stem extract. The H838 cell line, derived from the lung tissue of a 59-year-old White male diagnosed with stage 3B adenocarcinoma and displaying epithelial morphology, served as the model for these investigations [43]. For a comprehensive overview, the collective results of these analyses are aggregated in Figure 4A. Initially, we focused on the impact of N. miranda extract on H838 cell viability (Figure 4B), utilizing the trypan blue staining assay to determine cytotoxicity. This method is predicated on the concept that viable cells will exclude trypan blue due to their intact membranes, whereas non-viable cells will absorb the dye, thus facilitating the distinction and counting of viable (clear) and non-viable (blue-stained) cells. H838 cell cultures in 96-well plates were subjected to varying concentrations of the extract. The extract stock (20 mg/mL) was diluted with culture medium to obtain the desired experimental concentrations, and cells were incubated with these dilutions or with culture medium containing 0.2% DMSO as the control. Our results demonstrated a pronounced cytotoxic effect of the N. miranda extract on H838 cells, with a dose-dependent increase in mortality rates at increasing extract concentrations. Specifically, exposure to 0, 20, 40, 60, 80, and 100 μg/mL of N. miranda extract led to 0%, 17%, 46%, 64%, 89%, and 100% cell mortality, respectively (Figure 4B). The sharp differentiation in cell survival between treated and control groups highlights the significant cytotoxic potential of N. miranda extract on this cancer cell line. The observed dose-dependent cytotoxicity suggests the presence of active components in the extract that are highly effective in promoting cell death.
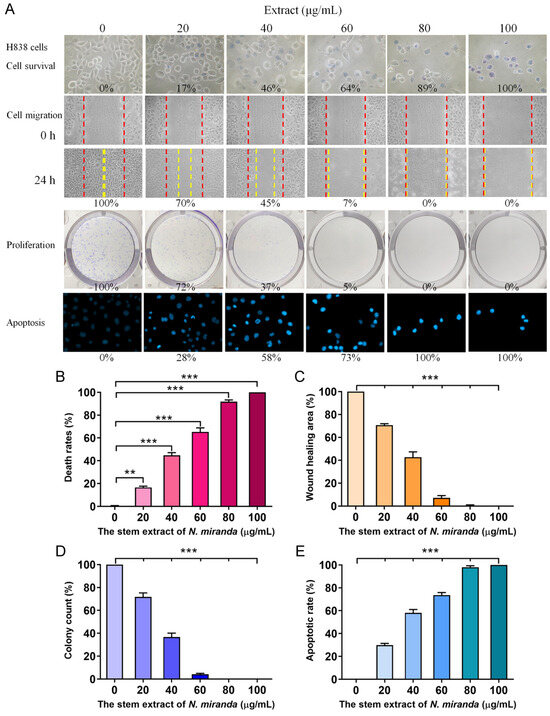
Figure 4.
Anticancer potential of N. miranda stem extract on H838 cells. (A) The effect of N. miranda stem extract on H838 cell survival, migration, proliferation, and nuclear condensation. (B) Trypan blue exclusion assay showing H838 cell viability following treatment with various concentrations of N. miranda extract. (C) Wound healing assay depicting the migration of H838 cells treated with different concentrations of N. miranda extract. Images were captured immediately and 24 h post-treatment. (D) Clonogenic assay assessing the proliferative and colony-forming potential of H838 cells pre-treated with varying concentrations of N. miranda extract. (E) Hoechst staining illustrating apoptosis and DNA fragmentation in H838 cells at varying concentrations of N. miranda extract. Statistical significance is denoted by ** p < 0.01 and *** p < 0.001 when compared to the control group.
2.5. The Extract of N. miranda Inhibited the Migration of H838 Cells
The N. miranda extract also displayed notable suppressive effects on the migration of H838 cells, which is a critical aspect of cancer metastasis. These effects were monitored using a wound healing assay, a reliable in vitro method to evaluate the migratory response of cells in a two-dimensional framework. For this assay, a wound was introduced into a dense layer of H838 cells, after which various concentrations of N. miranda extract were applied. The resultant gap acts as a stimulus for the adjacent cells to move and fill the void. Sequential images taken immediately and 24 h after the treatment of the extract showed a marked reduction in the migration rate as the extract’s concentration was stepped up. Specifically, incubation with 0, 20, 40, 60, 80, and 100 μg/mL of N. miranda extract resulted in a 0%, 30%, 55%, 93%, 100%, and 100% reduction in wound closure, respectively (Figure 4C). The decline in cell movement with increasing extract dosage was statistically significant, indicating a strong anti-metastatic effect. The halt in cell migration at concentrations of 80 μg/mL and above demonstrates the extract’s potential in curtailing cancer cell dissemination, a crucial aspect of cancer progression. Therefore, N. miranda extract may have valuable applications in curbing cancer metastasis, suggesting its therapeutic potential in cancer treatment strategies.
2.6. The Extract of N. miranda Inhibited the Proliferation of H838 Cells
The suppressive action of N. miranda extract on the proliferation of H838 cells was evaluated with the clonogenic assay. This assay is a cornerstone for assessing the ability of individual cells to grow and form colonies, thus serving as an indicator of cell survival and proliferation capabilities. The data from the clonogenic assay reveal a substantial reduction in both the quantity and size of colonies with increasing extract doses, indicating a significant impact on cell survival and proliferative potential. Specifically, the application of the extract at 0, 20, 40, 60, 80, and 100 μg/mL concentrations led to a graded inhibition of growth and colony formation in H838 cells by 0%, 28%, 63%, 95%, 100%, and 100%, respectively (Figure 4D). A concentration of 80 μg/mL achieved a complete cessation of cell proliferation, suggesting the extract’s efficacy in blocking the clonal expansion of H838 cells. The marked reduction in colony growth highlights its potential role of the N. miranda extract in impeding cancer cell proliferation. Such antiproliferative effects are vital for any agent intended to diminish cancer cell viability and prevent their dissemination. Consequently, the N. miranda extract may contain active compounds that may disrupt critical pathways for cell growth and replication, offering a viable direction for future investigations into its utility as a cancer therapeutic.
2.7. The Extract of N. miranda Induced Apoptosis of H838 Cells
Our findings indicate that N. miranda extract induces apoptosis in H838 cells, as visualized with Hoechst staining. Hoechst 33342, the stain employed in our assay, is a cell-permeant dye that stains the nuclei of all cells. It has a pronounced affinity for the condensed chromatin of apoptotic cells, resulting in a more intense fluorescence compared to non-apoptotic cells. Quantitative analysis of DNA fragmentation, a hallmark of apoptosis, showed that treatment with 0, 20, 40, 60, 80, and 100 μg/mL concentrations of the extract corresponded to 0%, 28%, 58%, 73%, 100%, and 100% increases in DNA fragmentation, respectively (Figure 4E). Morphological signs of apoptosis, such as chromatin condensation and nuclear fragmentation, became increasingly evident with higher concentrations of N. miranda extract, indicating a dose-responsive escalation in apoptotic cell death. These apoptotic features are detectable with the intensified blue fluorescence of Hoechst dye binding to the apoptotic chromatin. The induction of complete apoptosis at concentrations of 80 μg/mL and above highlights the strong pro-apoptotic capacity of the extract. Thus, the N. miranda extract appears to possess bioactive constituents that may trigger specific apoptotic pathways, offering insights into the extract’s mechanism of action and its potential for cancer therapy.
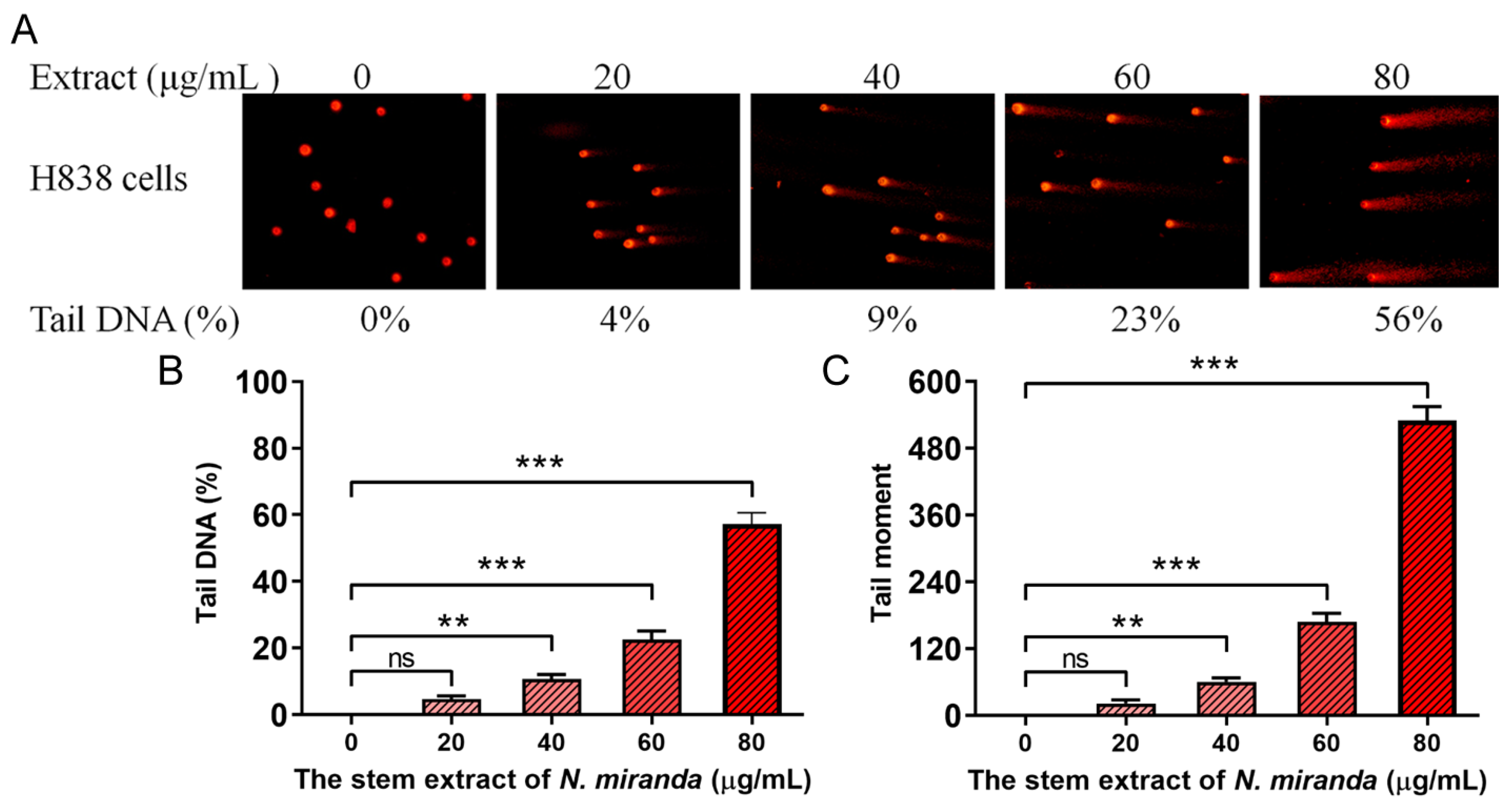
2.8. The Extract of N. miranda Caused DNA Damage in H838 Cells
Our research revealed that N. miranda extract induces DNA damage in H838 cells, a finding that was demonstrated using the comet assay. This assay, known for its sensitivity in detecting DNA strand breaks at the single-cell level (single-cell gel electrophoresis) [44], showed a distinct increase in DNA damage with higher concentrations of the extract. In the process, H838 cells encapsulated in agarose on a slide underwent lysis, and subsequent electrophoresis produced the characteristic ‘comet’ tails indicative of DNA fragmentation, which were more pronounced in cells treated with N. miranda extract compared to controls (Figure 5A). The assay images display a spectrum of DNA damage among the cells, from minimal, evidenced by short or absent comet tails, to severe, indicated by longer tails. These comet tails provide a visual and measurable indicator of the extent of DNA fragmentation. Specifically, treatment with 0, 20, 40, 60, and 80 μg/mL of the extract resulted in 0%, 4%, 9%, 23%, and 56% tail DNA, respectively (Figure 5B). The tail moments in H838 cells, induced by the extract, also exhibited a dose-dependent increase (Figure 5C). The extract’s treatment led to a noticeable increment in both the comet tails’ length and density, indicating a concentration-dependent increase in DNA damage. This kind of damage often leads to cell death, particularly via apoptotic mechanisms, and serves as an essential marker of the extract’s genotoxic capabilities. The induction of DNA strand breaks at elevated extract doses reveals potent cytotoxic and genotoxic activities, pointing to a mode of action that compromises the genetic material within the cancer cells. The ability of the extract to cause such genotoxicity is central to its anticancer effects and offers significant insights into its potential use in cancer therapy by promoting cell death via DNA damage.
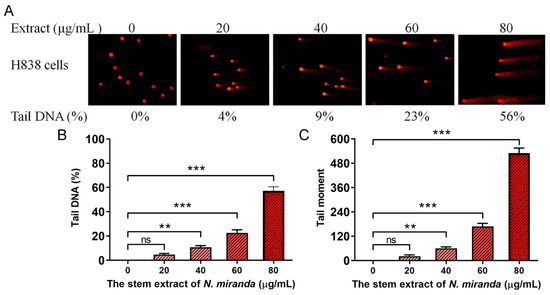
Figure 5.
DNA damage induced in H838 cells by N. miranda extract. (A) Comet assay results display a significant escalation in DNA damage as the concentration of N. miranda extract increases. (B) A marked increase in comet tail density and (C) an extension in comet tail length were observed, reflective of a concentration-dependent rise in DNA damage. Levels of statistical significance are indicated by ** p < 0.01 and *** p < 0.001 in comparison to the control group. ns—non-significant.
2.9. Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
In light of the anti-H838 activities and anti-skin-aging potential of N. miranda extract, we conducted a gas chromatography–mass spectrometry (GC–MS) analysis to identify the predominant compounds within the extract. The spectral data generated were compared with the NIST 2011 and Wiley 10th edition mass spectral libraries, allowing for the tentative identification of the compounds. Compounds with a similarity index (SI) greater than 800 were considered for identification. Accordingly, the top 15 compounds, each constituting more than 0.5% of the extract, were identified as follows: stigmast-5-en-3-ol (11.7%), plumbagin (10.9%), hexadecanoic acid (3.4%), hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester (2.4%), catechol (2.3%), oleic acid (2.0%), pyrogallol (2.0%), 13-docosenamide (1.7%), stigmasterol (1.7%), 3-methoxycatechol (1.5%), 1-eicosanol (1.2%), glyceryl monostearate (1.2%), linoleic acid (1.1%), stearic acid (0.8%), and heptadecanol (0.5%). It is conceivable that one or more compounds in the stem extract of N. miranda, extracted with 100% ethanol, may act as potential anticancer and anti-skin-aging agents, either individually or synergistically.
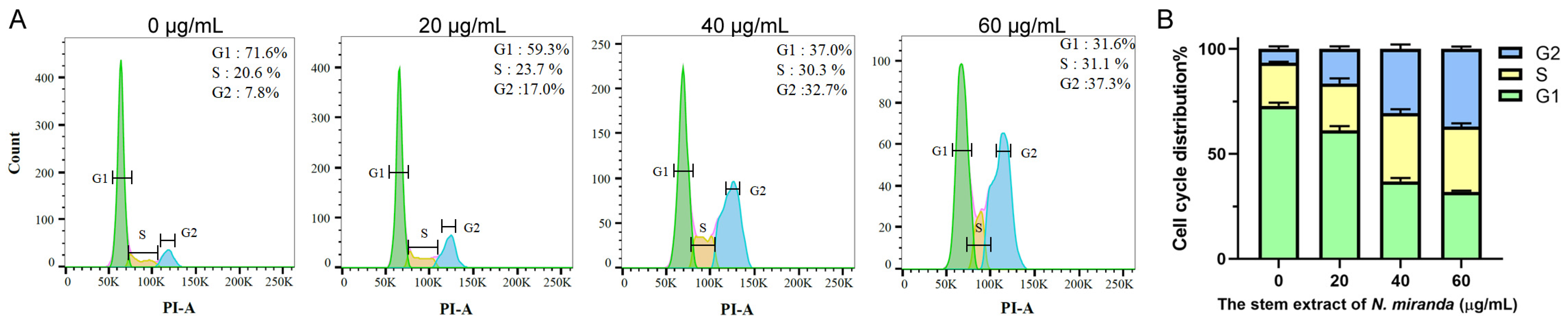
2.10. The Stem Extract of N. miranda Suppressed Carcinoma Cell Proliferation by Inducing G2 Cell-Cycle Arrest
The progression of the cell cycle in H838 cells, upon treatment with N. miranda stem extract, was assessed using flow cytometry (Figure 6A). Treatment with the extract at 20, 40, and 60 µg/mL concentrations resulted in an increase in the G2 phase cell population, from 7.8% in untreated cells to 17.0%, 32.7%, and 37.3% in treated cells, respectively, indicating a concentration-dependent accumulation in the G2 phase (Figure 6B). This shift implies a disruption in the normal progression of the cell cycle, which is crucial for cell division. The enrichment in the G2 phase suggests that the N. miranda stem extract may exert its anticarcinogenic effects by inducing G2 phase arrest, thereby inhibiting the proliferation of carcinoma cells.
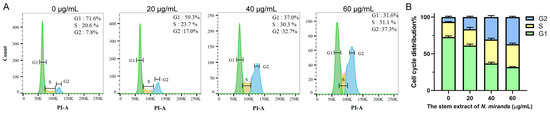
Figure 6.
(A) Alteration of cell cycle progression by N. miranda stem extract in H838 cells. H838 cells underwent treatment with a control solution (0.1% DMSO) or with N. miranda stem extract at specified concentrations for 24 h and were subsequently fixed in 70% ethanol overnight. The cells were then stained with propidium iodide (PI) for 30 min before analysis via flow cytometry. (B) The cell cycle distribution.
2.11. Co-Treatment of N. miranda Stem Extract with the Anticancer Drug 5-Fluorouracil (5-FU) against H838 Cells
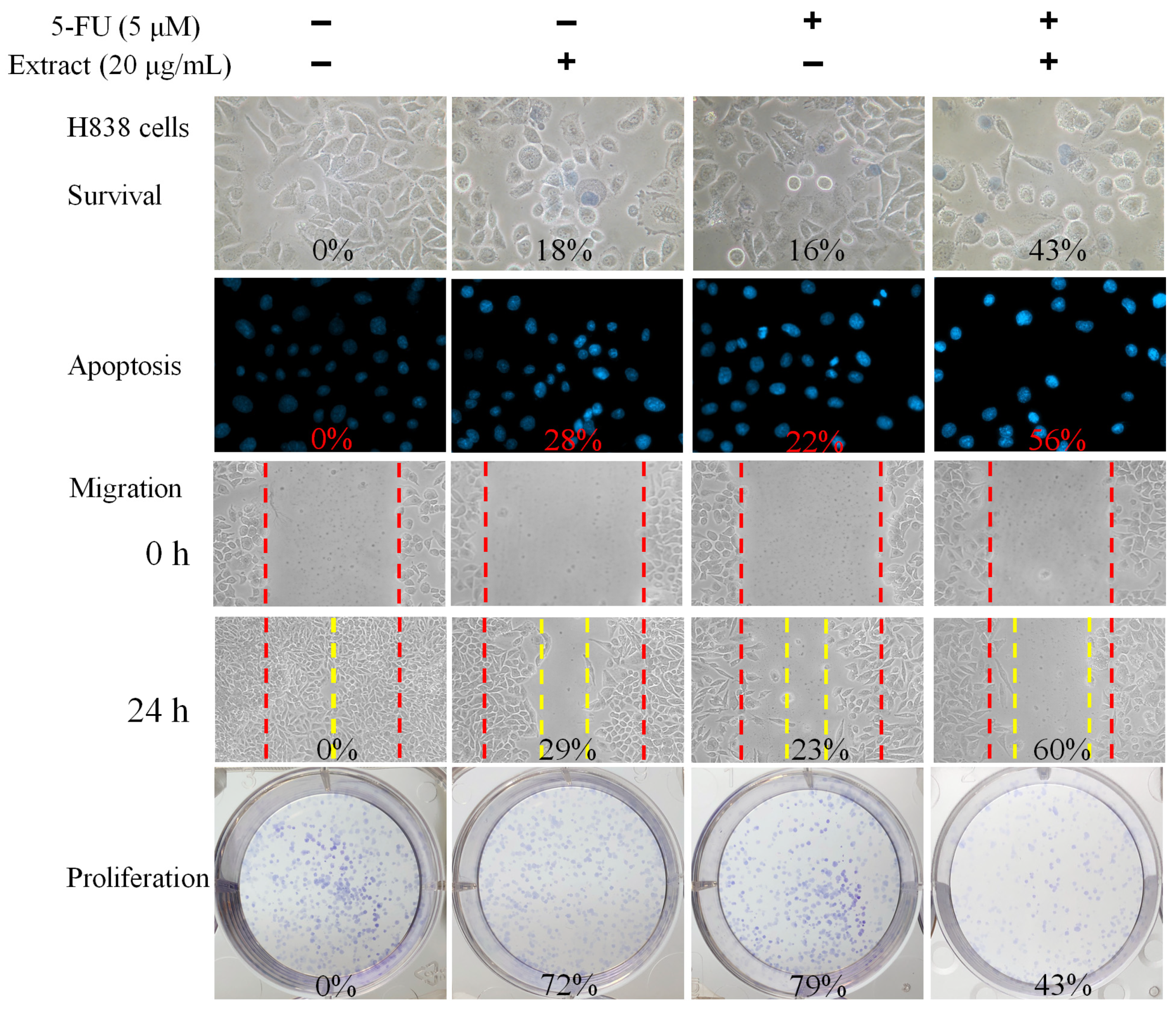
We also evaluated the combined effect of N. miranda stem extract and the chemotherapeutic agent 5-fluorouracil (5-FU) on H838 carcinoma cells (Figure 7). 5-FU, an FDA-approved drug [45], is known for its effectiveness in treating various cancers and has been shown to work synergistically with natural compounds such as myricetin [46], sinomenine [47], lapatinib [48], and plumbagin [22,23] in cancer treatment. Hence, we investigated the potential enhanced efficacy of 5-FU when used in combination with N. miranda stem extract against H838 cells. For these experiments, we used a 20 μg/mL concentration of N. miranda stem extract, which by itself induces minimal cytotoxicity. Treatment of H838 cells with N. miranda stem extract, 5-FU at 5 μM, and their combination resulted in cell death rates of 18%, 16%, and 43%, apoptosis rates of 28%, 22%, and 56%, migration reduction of 29%, 23%, and 60%, and proliferation and colony formation inhibition of 28%, 21%, and 57%, respectively. These findings suggest a synergistic cytotoxic effect, as the combination treatment led to increased cell death, enhanced inhibition of migration and proliferation, and greater induction of DNA fragmentation in H838 cells compared to either agent alone. These results propose that 5-FU could be effectively paired with N. miranda stem extract to potentially improve cancer treatment outcomes. However, such synergistic effects need to be validated through further experimental and clinical research.
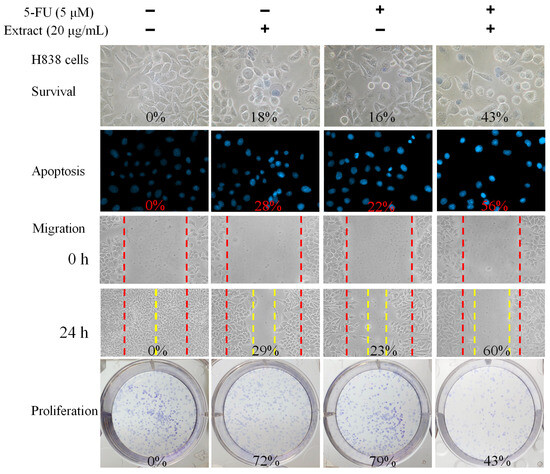
Figure 7.
Synergistic anticancer effects of N. miranda stem extract and 5-FU on H838 Cells. The combined impact of N. miranda stem extract (20 μg/mL) and 5-FU (5 μM) on the survival, migration, proliferation, and apoptosis of H838 cells was assessed. Evaluations were conducted using trypan blue dye exclusion staining for cell viability, Hoechst staining for apoptosis detection, a wound-healing assay for migration analysis, and a clonogenic assay for proliferative capacity. The outcomes of the combination treatment suggest that 5-FU, when used alongside N. miranda stem extract, may enhance therapeutic efficacy against cancer.
2.12. Inhibition of Human RPA32 by N. miranda Extracts
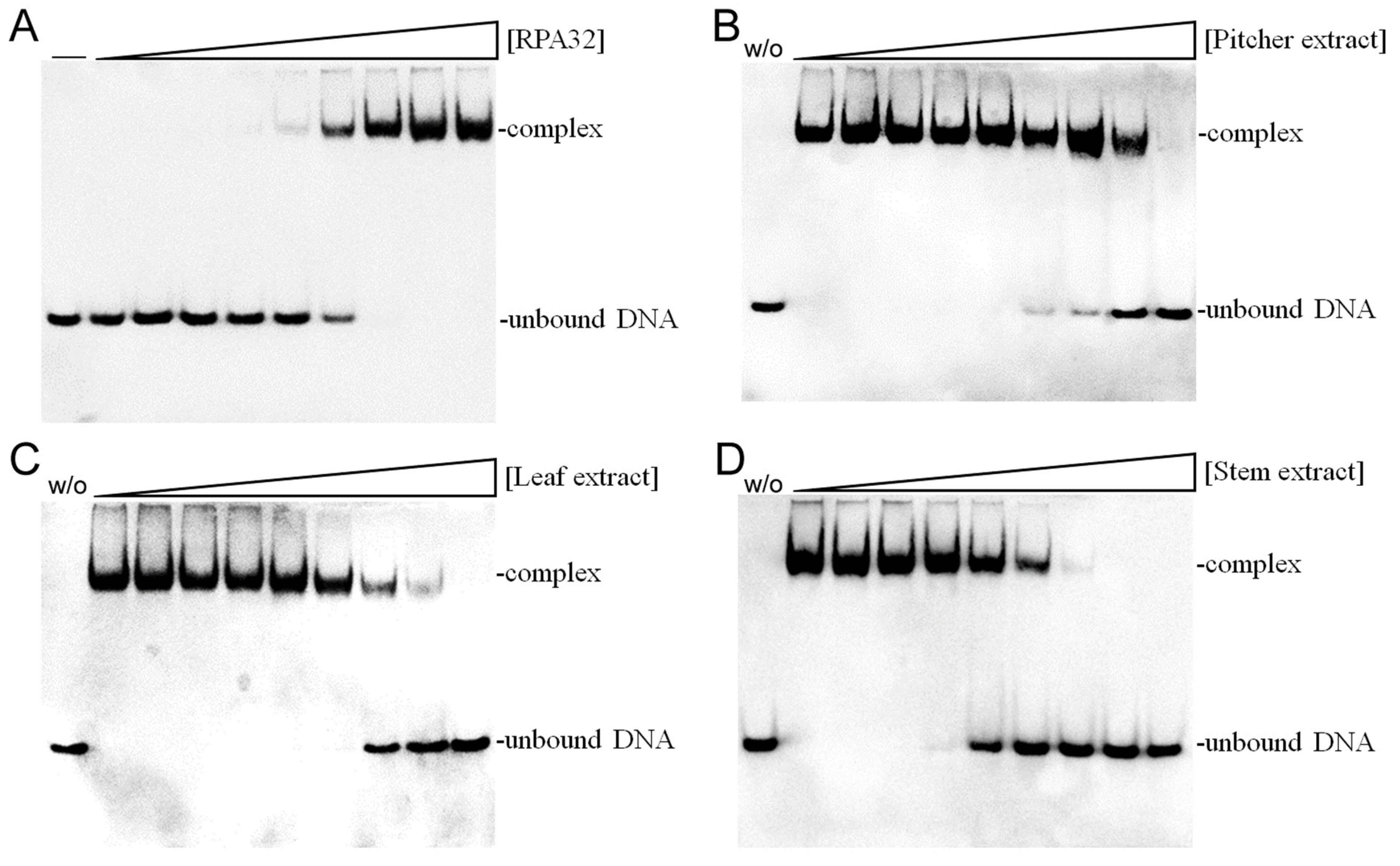
Given the notable anti-proliferative effects of N. miranda stem extract, it is possible that it contains bioactive compounds capable of disrupting key cellular growth and replication pathways. This potential positions it as a promising candidate for further exploration as a cancer treatment. Accordingly, we investigated the possibility of human RPA32 (huRPA32), a critical protein for DNA replication due to its role in binding single-stranded DNA, being a target for inhibition by N. miranda stem extract. Considering huRPA32’s necessity in cell division and the fact that normal cells do not perpetually undergo rapid replication, targeting huRPA32 could be an effective strategy for anticancer therapy. Electrophoretic mobility shift analysis (EMSA) was used to evaluate the binding activity of recombinant huRPA32 (concentrations 0–40 μM), which was expressed and purified from E. coli (Figure 8A). The principle behind EMSA is that the formation of stable protein–DNA complexes will result in reduced electrophoretic mobility compared to unbound DNA. In this study, the deoxythymidine homopolymer dT25 was used. Incubation with dT25 led to a notable band shift, indicative of stable complex formation. The EMSA titration curve allowed the determination of the midpoint value for ssDNA-binding ([Protein]50) of huRPA32 to dT25 as 6.3 ± 0.5 μM. Subsequently, a concentration of 10 μM huRPA32, which effectively binds to dT25, was selected to screen various concentrations (0–1000 μg/mL) of N. miranda plant extracts for inhibitory activity. At 250 μg/mL, the extracts from the pitcher (Figure 8B), leaf (Figure 8C), and stem (Figure 8D) of N. miranda inhibited huRPA32 activity by 14%, 56%, and 89%, respectively. The IC50 values were determined to be 706.6 ± 32.6 μg/mL for the pitcher, 231.4 ± 12.5 μg/mL for the leaf, and 101.6 ± 6.3 μg/mL for the stem extract (Table 1). Therefore, it appears that certain components within the stem extract of N. miranda, extracted using 100% ethanol, might act individually or synergistically as potent inhibitors of huRPA32.
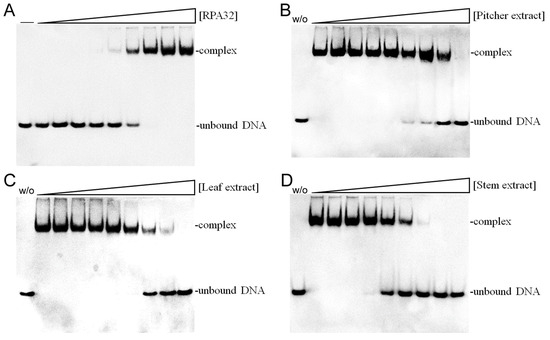
Figure 8.
Inhibition of the ssDNA-binding activity of huRPA32 by different extracts of N. miranda. (A) Binding of huRPA32 to ssDNA dT25. Purified huRPA32 (0, 0.32, 0.63, 1.25, 2.5, 5, 7.5, 10, 20, and 40 μM) was incubated with biotin-labeled ssDNA dT25 and the binding was analyzed with EMSA. The binding constant ([Protein]50) of huRPA32 was quantified through linear interpolation based on the protein concentrations. The inhibitory effects were investigated on the DNA-binding activity of huRPA32 by N. miranda extracts (0, 0, 7.6, 15.1, 31.3, 62.5, 125, 250, 500, 1000 μg/mL) from the (B) pitcher, (C) leaf, and (D) stem. An amount of 1% DMSO was used as a negative control (representing 0 μg/mL of N. miranda extract). The “w/o” denotes the absence of huRPA32 and the extract during the incubation with the DNA dT25.
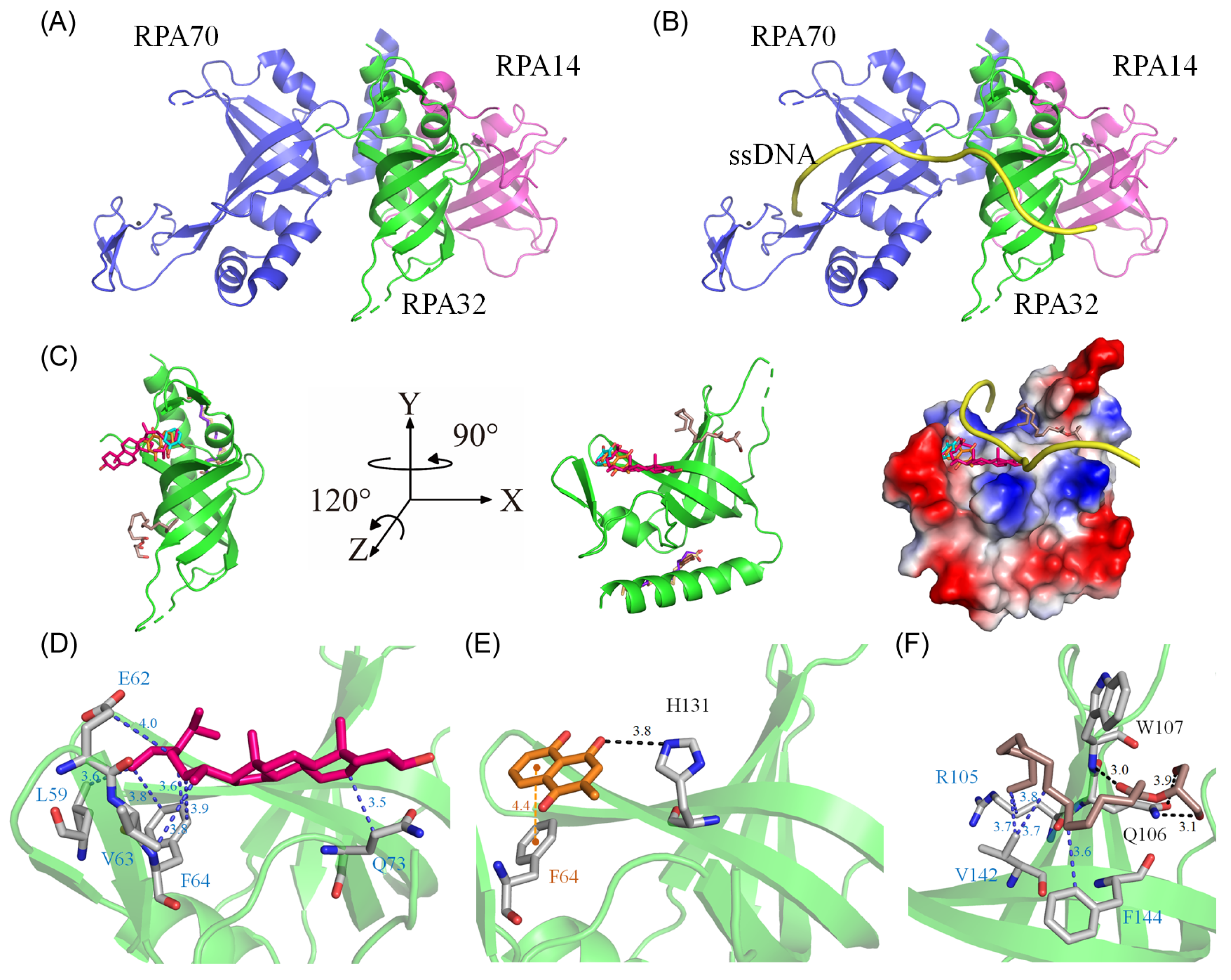
2.13. Molecular Docking of huRPA32
Our GC-MS analysis identified at least 15 compounds, each constituting over 0.5%, in the stem extract of N. miranda, suggesting that certain compounds might contribute to the inhibition of huRPA32. The crystal structure of huRPA32, in complex with huRPA70 and huRPA14 (PDB ID 1L1O) [49], is accessible in the Protein Data Bank (PDB) (Figure 9A), allowing molecular docking studies to predict potential binding sites and calculate binding energies using AutoDock Vina. Although the structure of the huRPA complex with ssDNA remains uncharacterized, the structure of the Pyrococcus abyssi RPA (PaRPA)–ssDNA complex has been recently resolved [50]. Consequently, we constructed a model of the huRPA–ssDNA complex (Figure 9B) by manually superimposing the apo-huRPA structure with the PaRPA complex (PDB ID 8AAS), under the assumption of similar ssDNA binding mechanisms across species. For docking analysis, the seven most abundant compounds from the stem extract were individually docked into huRPA32 (Figure 9C). Five of these seven compounds (stigmast-5-en-3-ol, plumbagin, hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester, catechol, and pyrogallol) targeted various ssDNA binding sites. However, oleic acid and hexadecanoic acid did not. These compounds, by occupying binding sites, may collectively hinder the binding of ssDNA to huRPA32. The binding efficiency of these compounds was ranked as follows (Table 2): stigmast-5-en-3-ol > plumbagin > hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester > oleic acid > hexadecanoic acid > pyrogallol > catechol. Notably, stigmast-5-en-3-ol and plumbagin, which are the two most prevalent compounds in N. miranda’s stem extract, demonstrated strong binding interactions as estimated with AutoDock Vina (Table 2). Stigmast-5-en-3-ol was situated within the cavity of the ssDNA-binding surface and engaged in extensive hydrophobic interactions with Leu59, Glu62, Val63, Phe64, and Gln73 of huRPA32 (Figure 9D). Plumbagin formed a hydrogen bond with His131 and engaged in π-stacking with Phe64 (Figure 9E). Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester, anchored within the groove for ssDNA binding of huRPA32, formed hydrogen bonds with Gln106 and Trp107, and also interacted hydrophobically with Arg105, Val142, and Phe144 (Figure 9F). Given the diverse interactions at different ssDNA-binding sites, the inhibitory potential of N. miranda’s stem extract against huRPA32 might be attributed to the combined effects of some of these compounds. Nonetheless, this hypothesis requires validation through biochemical and structural studies.
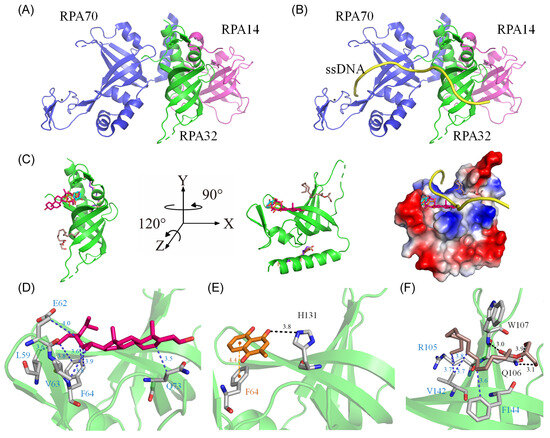
Figure 9.
Molecular docking analysis of huRPA32. (A) The crystal structure of huRPA32 in complex with huRPA70 and huRPA14 (PDB ID 1L1O), with huRPA70 colored blue, huRPA32 green, and huRPA14 light magenta. (B) The modeled huRPA–ssDNA complex, constructed by manually superimposing the apo-huRPA structure with the PaRPA complex (PDB ID 8AAS), assuming similar ssDNA binding mechanisms across species. The ssDNA from the PaRPA complex is colored yellow. (C) Docking analysis showing the seven most abundant compounds from the stem extract individually docked into huRPA32: stigmast-5-en-3-ol in hot pink, plumbagin in orange, hexadecanoic acid in purple-blue, hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester in dark salmon, catechol in cyan, oleic acid in wheat, and pyrogallol in chocolate. Five of these seven compounds targeted various ssDNA binding sites in huRPA32, potentially collectively hindering the binding of ssDNA to huRPA32. The charge distribution pattern is shown to indicate ssDNA binding sites for clarity. (D) The binding mode of stigmast-5-en-3-ol, situated within the cavity of the ssDNA-binding surface, engaging in extensive hydrophobic interactions with Leu59, Glu62, Val63, Phe64, and Gln73 of huRPA32. (E) The binding mode of plumbagin, which formed a hydrogen bond with His131 and engaged in π-stacking with Phe64. (F) The binding mode of hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester, anchored within the groove for ssDNA binding of huRPA32, formed hydrogen bonds with Gln106 and Trp107, and also interacted hydrophobically with Arg105, Val142, and Phe144.

Table 2.
Molecular docking analysis of huRPA32.
3. Discussion
Natural products have attracted significant interest as viable alternatives for treating various diseases due to their multifaceted modes of action on problematic cells [51,52,53,54]. Before this study, the potential of Nepenthes plant extracts for anti-skin-aging had not been investigated. Nepenthes, recognized for their ethnobotanical uses such as alleviating stomachaches and fevers, show minimal side effects and have demonstrated notable in vitro anticancer and antibacterial properties [55]. This underscores the potential for new therapeutic discoveries and the importance of understanding how Nepenthes extracts, such as that from N. miranda [56]—a distinctive hybrid of N. maxima and N. northiana with unique physiological characteristics—act and inhibit specific targets for medical advancement. The goal of this study was to identify natural sources for developing treatments against skin aging (Figure 1, Figure 2 and Figure 3) and cancer (Figure 4, Figure 5, Figure 6 and Figure 7). This research represents the first exploration into the anti-skin-aging potential of Nepenthes plants by targeting crucial enzymes involved in skin degradation. The strong inhibitory effects on skin-aging enzymes identified in this study (Table 1), coupled with high polyphenol and flavonoid contents and significant antioxidant activities [23], suggest that N. miranda extracts could potentially serve as a natural alternative or complementary therapy. This includes applications such as adjuvants or cosmeceutical ointments for treating symptoms of skin aging. Furthermore, this study identifies RPA32 [36], an essential protein for DNA replication and cell development, as a novel target of Nepenthes extract, offering insight into a potential mechanism for inhibiting cancer cell growth by targeting RPA32. The identification of additional plant-derived compounds for anti-aging and anticancer therapies is crucial for further pharmaceutical development.
Skin aging is an inevitable process that affects everyone, yet lifestyle and environmental factors can expedite its onset, leading to prematurely aged skin [57,58]. This research illuminates the comprehensive capability of N. miranda extract in mitigating the effects of skin aging. Notably, the extract displays robust inhibitory activity against elastase (Figure 1), tyrosinase (Figure 2), and hyaluronidase (Figure 3), highlighting its effectiveness in preserving skin’s elasticity, minimizing hyperpigmentation, and maintaining moisture and softness. Particularly impressive are its anti-elastase and anti-hyaluronidase properties (Table 1), where the IC50 values for N. miranda’s stem extract surpassed those of known positive controls EGCG and Myr. Our GC–MS analysis identified at least 15 compounds, each constituting over 0.5%, in the stem extract of N. miranda, suggesting that certain compounds might contribute to the strong inhibition of elastase and hyaluronidase. These results underscore N. miranda extract’s potential in anti-aging skin care products and open avenues for further investigation into its bioactive constituents as innovative enzyme inhibitors relevant to the skin aging process.
Elastase, a protease enzyme, plays a critical role in breaking down elastin, which is crucial for skin elasticity due to its ability to snap back into shape. Inhibiting elastase can thus prevent the loss of skin elasticity and the formation of wrinkles. Our findings indicate that N. miranda’s stem extract is a strong inhibitor of elastase. Although the significant presence of compounds like stigmast-5-en-3-ol (11.7%) and plumbagin (10.9%) in the extract has yet to be explored for their inhibitory effects, hexadecanoic acid (also known as palmitic acid; 3.4%), identified in the extract, is already recognized as a moderate elastase inhibitor [59,60]. The marked anti-elastase activity observed in N. miranda’s stem extract could be attributed to a synergistic interaction among the compounds present rather than solely to the action of hexadecanoic acid. Nonetheless, this hypothesis regarding the collective activity of the identified compounds within the stem extract requires further experimental validation.
Hyaluronic acid is crucial for maintaining the body’s hydration, providing smoothness, and ensuring lubrication by binding water molecules. Inhibiting hyaluronidase, therefore, helps to prevent the loss of this moisture. Key components of N. miranda’s stem extract, particularly plumbagin [61] and stigmasterol [62], have been identified as potential inhibitors of hyaluronidase. Consequently, the notable anti-hyaluronidase effect of the stem extract may stem from the action of these compounds, possibly in conjunction with other constituents. It merits further investigation to ascertain if N. miranda’s stem extract contains any new substances that inhibit hyaluronidase, which could be beneficial for cosmeceutical uses. Our laboratory is currently exploring the hyaluronidase inhibitory effects of these and other active ingredients found in the stem extract of N. miranda.
The significant cytotoxic impact of N. miranda stem extract on H838 lung carcinoma cells underscores its promise as an anticancer treatment. The dose-dependent increase in cell mortality, suppression of cell migration and proliferation, and induction of apoptosis (Figure 4) and DNA damage (Figure 5) in H838 cells indicates the presence of bioactive compounds in N. miranda stem extract capable of targeting multiple pathways involved in cancer cell growth and survival. Our GC–MS analysis revealed that the primary components of the N. miranda stem extract, such as stigmast-5-en-3-ol [63], plumbagin [64], and hexadecanoic acid [65], are known for their anticancer properties, contributing to the extract’s potent cytotoxic effects. Exploring whether the various active components in N. miranda stem extract can offer significant polypharmacological and synergistic effects, and determining the optimal ratios for cancer treatment, requires further elucidation. Furthermore, flow cytometry findings suggest that the extract may halt the proliferation of NSCLC H838 cells by causing G2 phase cell-cycle arrest (Figure 6). The G2 cell-cycle checkpoint serves as an essential guardian of the genome in tumor cells and, therefore, targeting the cell cycle proteins and abrogating this checkpoint has emerged as a promising therapeutic strategy against cancer [66]. Accordingly, our laboratory is delving into the specific cellular pathways that lead to G2 arrest in H838 cells, aiming to advance the development of new NSCLC therapies.
Leveraging the anti-proliferative effects of N. miranda extract, this study suggests that the extract could influence key pathways involved in cellular growth and replication. Additionally, given its capacity to inhibit the ssDNA-binding activity of bacterial SSB [26], we explored and confirmed the extract’s ability to inhibit the crucial DNA replication protein huRPA32 (Figure 8). This indicates that huRPA32 could be a significant target for the anticancer activity of N. miranda extract. Given the structural and functional similarities between huRPA32 and SSB, inhibitors of SSB might also effectively target huRPA32. Our recent discoveries include the identification of natural products such as myricetin, taxifolin, dihydrokaempferol, and rutin as SSB inhibitors [30,31,67]. It remains to be seen whether these flavonoids, along with active compounds found in the stem extract of N. miranda, can also inhibit huRPA32, potentially contributing to the development of new chemotherapy agents. Docking studies (Figure 9) have indicated that five compounds in the N. miranda extract (stigmast-5-en-3-ol, plumbagin, hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester, catechol, and pyrogallol) target various ssDNA binding sites, suggesting a collective mechanism that may impede ssDNA’s binding to huRPA32. Therefore, the effectiveness of N. miranda’s stem extract against huRPA32 could result from the synergistic action of these identified compounds. Nonetheless, this speculation regarding the collective activity of the identified compounds within the stem extract requires further experimental validation.
Despite the advent of new drug development, 5-FU continues to be a fundamental component in cancer treatment, particularly for cancers of the gastrointestinal tract, breast, head, and neck [45]. Our previous research determined the complexed crystal structures of SSB [68], dihydropyrimidinase (DHPase) [69], and dihydroorotase (DHOase) [70] with 5-FU, suggesting that 5-FU’s mechanism of action extends beyond its primary target, thymidylate synthase, to potentially regulate the activity of these proteins. In this study, we discovered that combining the stem extract of N. miranda with 5-FU significantly amplifies its cytotoxic impact (Figure 7). 5-FU disrupts RNA function and DNA synthesis [71], while plumbagin—a prominent compound in N. miranda’s stem extract—promotes anticancer effects by elevating intracellular reactive oxygen species (ROS) levels and triggering apoptosis, with ROS activation leading to cell death through a p53-dependent pathway [64]. Moreover, plumbagin’s inhibition of DHPase [23] and DHOase [22] affects cellular DNA base metabolism [72]. Our structural analysis showed plumbagin and 5-FU binding at distinct sites within DHPase’s active site [23,72], indicating that plumbagin’s role in enhancing ROS may significantly boost 5-FU’s chemotherapeutic efficacy by concurrently affecting DHPase and DHOase functions. This synergy between N. miranda’s stem extract and 5-FU in cytotoxicity suggests a potential pathway for enhancing treatment effectiveness. Further exploration of this synergistic relationship could reveal optimal treatment combinations and dosages, leading to more precise and potent cancer therapies.
While our study provides novel insights into the anti-aging and anticancer potential of N. miranda stem extract, we recognize several limitations. The study is predominantly based on in vitro assays, which, although valuable for initial screening, cannot fully replicate the complexity of living organisms. Future studies involving in vivo models are crucial to validate our findings and to assess the pharmacokinetics, pharmacodynamics, and potential toxicity of the extract in a physiological context.
4. Materials and Methods
4.1. Materials
All solvents and chemicals were of the highest grade and commercially obtained from Sigma-Aldrich (St. Louis, MO, USA). The Escherichia coli strain BL21(DE3) pLysS (Novagen, Cambridge, UK) was used for recombinant protein expression. The H838 cell line was kindly provided by Dr. Kuo-Ting Chang (Tao Yuan General Hospital). H838 cells were maintained in RPMI 1640 supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were incubated at 37 °C in a 95% air and 5% CO2 atmosphere.
4.2. Expression and Purification of the Recombinant Protein
The synthesized gene encoding huRPA32 (anticipated to express 270 amino acid residues) was cloned using forward and reverse primers (with underlining indicating the restriction site) huRPA32-EcoRI-N (5′-GGGGAATTCATGTGGAACAGTGGATTCGAAAG-3′) and huRPA32-HindIII-C (5′-GGGAAGCTTTTCTGCATCTGTGGATTTAAA-3′). These primers facilitated the insertion of the gene into the pET21e vector, resulting in the construction of the expression plasmid pET21e-huRPA32. This plasmid was designed to express full-length recombinant huRPA32 in E. coli. The pET21e vector, a derivative of the pET21b vector (Novagen Inc., Madison, WI, USA), was modified to avoid the fusion of the N-terminal T7 tag with the gene product. The expressed gene product from pET21e-huRPA32 is expected to have two additional artificial residues, EF, introduced by the EcoRI site at the N-terminus, and a C-terminal His tag (KLAAALEHHHHHH), facilitating the purification of the recombinant protein. This resulting plasmid was introduced into E. coli BL21 (DE3) cells, which were cultured in LB medium with 250 μg/mL ampicillin until reaching an OD600 of 0.9, under vigorous shaking at 37 °C. Recombinant huRPA32 expression was induced with 1 mM isopropyl thiogalactopyranoside (IPTG) for 12 h at 25 °C. The induced cells were harvested, resuspended in sonication buffer (20 mM Tris–HCl, 0.5 M NaCl, pH 8.0), and lysed via sonication. The pellet was then collected, washed with sonication buffer containing 2 M urea, and harvested again. The resulting pellet was solubilized in sonication buffer with 8 M urea for 12 h at 4 °C. The solution was dialyzed twice daily against a refolding buffer (5 mM imidazole, 20 mM Tris–HCl, 0.5 M NaCl, and 1 mM DTT, pH 7.4). The resulting solution was dialyzed again against a purification buffer (5 mM imidazole, 20 mM Tris–HCl, and 0.5 M NaCl, pH 7.9) for 4 h. The recombinant protein was purified from the soluble supernatant using Ni2+-affinity chromatography (HisTrap HP; GE Healthcare Bio-Sciences, Piscataway, NJ, USA). Elution was carried out using an elution buffer (20 mM Tris-HCl, 250 mM imidazole, and 0.5 M NaCl, pH 7.9). The purified protein was then dialyzed against a storage buffer (40 mM Tris-HCl, 50 mM NaCl, pH 7.5) and concentrated to 15 mg/mL for future use. Protein purity was verified by electrophoresis on a 12% SDS-PAGE.
4.3. Electrophoretic Mobility Shift Analysis (EMSA)
For the analysis of huRPA32 binding, a biotinylated deoxythymidine (dT) homopolymer dT25 was employed as a standard assay substrate. The ssDNA dT25 was biotinylated at its 5′ end. Purified huRPA32 was incubated with this labeled ssDNA (30 fmol/μL) at various concentrations (0, 0.32, 0.63, 1.25, 2.5, 5, 7.5, 10, 20, and 40 μM). The electrophoretic mobility shift analysis (EMSA) was performed using the LightShift Chemiluminescent EMSA Kit. Briefly, huRPA32 and the DNA substrate dT25 were mixed and incubated for 60 min at 37 °C in a buffer containing 40 mM Tris–HCl (pH 7.5) and 50 mM NaCl. After adding the dye mixture, the reaction samples were resolved on an 8% native polyacrylamide gel using EMSA. Electrophoresis was conducted at 100 V for 1 h in TBE running buffer. The protein–DNA complexes were then electroblotted onto a positively charged nylon membrane (GE, Boston, MA, USA). Cross-linking of the transferred DNA to the membrane was achieved using a UV-light instrument equipped with 312 nm bulbs, with an exposure time of 10 min. Detection of the DNA was carried out using the streptavidin–horseradish peroxidase conjugate and chemiluminescent substrate (Pierce Biotechnology, Waltham, MA, USA). The [Protein]50, representing the concentration of protein that bound 50% of the input DNA, was estimated from the EMSA results.
4.4. huRPA32 Inhibition
An inhibition assay was conducted with huRPA32 at a concentration of 10 μM, incubated with both dT25 and various concentrations of N. miranda extract (0, 7.6, 15.1, 31.3, 62.5, 125, 250, 500, 1000 μg/mL). Following EMSA, a titration curve was generated based on the experimental data. The concentration of N. miranda extract required to achieve 50% inhibition (IC50) of huRPA32 activity was directly determined from this curve using graphical analysis.
4.5. Plant Materials and Extract Preparations
The pitcher, leaf, and stem extracts of N. miranda were obtained using 100% ethanol [26]. The plant material, procured from Guoguang Flower Market and Taiwan Provincial Flower Marketing Cooperative, was identified by Dr. Zhong-Bao Zhang in December 2020. The collected samples were dried, cut into small pieces, and then pulverized into a powder. For the extraction process, 1 g of the plant powder was placed into a 250 mL conical flask, to which 100 mL of ethanol was added. The mixture was then shaken on an orbital shaker for 5 h. The resulting extract was filtered through a 0.45 μm filter, and the ethanol was subsequently removed using a hot air circulation oven set at 40 °C. The extracts were stored at −80 °C until needed. The extract powder was dissolved in 20% DMSO to prepare a stock solution at a concentration of 20 mg/mL. For anticancer cell assays, the stock solution was diluted with supplemented culture medium to reach the desired assay concentrations. Cancer cells were then incubated with these extract solutions or with the culture medium containing 0.2% DMSO, serving as the treatment and control groups, respectively. For EMSA, the stock solution was diluted with protein storage buffer (40 mM Tris-HCl, 50 mM NaCl, pH 7.5) to the indicated assay concentrations. The protein was then incubated with these extract solutions or with the storage buffer containing 1% DMSO, also serving as treatment and control groups, respectively.
4.6. GC-MS Analysis
The composition of the stem extract of N. miranda, obtained through ethanol extraction, was tentatively identified using established methodologies [23]. The analysis was performed using a Thermo Scientific TRACE 1300 Gas Chromatograph coupled with a Thermo Scientific ISQ Single Quadrupole Mass Spectrometer System. The chromatographic separation was achieved on a Rxi-5ms column (30 m × 0.25 mm i.d. × 0.25 μm film). Helium, serving as the carrier gas, was maintained at a constant flow rate of 1 mL/min. The oven temperature program started at 40 °C, held for 3 min, followed by a ramp of 10 °C/min to a final temperature of 300 °C, which was then sustained for 1 min. The injection port temperature was set at 300 °C. The eluted compounds were detected by a quadrupole mass detector, with ionization achieved via the electron ionization method. Operational settings included a quadrupole temperature of 150 °C, a source temperature of 300 °C, electron energy of 70 eV, a detector temperature of 300 °C, an emission current multiplier voltage of 1624 V, and an interface temperature of 300 °C. The mass range scanned spanned from 29 to 650 amu. Relative mass fractions of the individual chemical components were calculated employing the peak area normalization method. Compounds were tentatively identified by comparing the generated spectra with the NIST 2011 and Wiley 10th edition mass spectral libraries. Those with a similarity index (SI) exceeding 800 were considered positively identified and are reported in this study.
4.7. Elastase Inhibition
The elastase inhibition assay was conducted in accordance with modified methods from previous studies [19]. The assay was carried out in 200 mM Tris buffer (pH 8.0). Porcine pancreatic elastase was prepared as a 3.33 mg/mL stock solution in the same buffer. The substrate N-Succinyl-Ala-Ala-Ala-p-nitroanilide (AAAPVN) was dissolved in 200 mM Tris buffer at a concentration of 1.6 mM. Prior to initiating the reaction with the substrate, the test extracts were incubated with the enzyme for 15 min. The final reaction mixture, with a total volume of 250 μL, comprised the buffer, 0.8 mM AAAPVN, 2 μg/mL elastase, and the test extract at concentrations ranging from 0 to 100 μg/mL. Epigallocatechin gallate (EGCG) at concentrations of 0 to 100 μg/mL served as a positive control. Negative controls utilized 10% DMSO. Absorbance was measured at 405 nm immediately after substrate addition and then continuously over a 20 min period using 96-well plates. The percentage of elastase inhibition was calculated using the formula: Elastase inhibition % = [(A_control − A_sample)/A_control] × 100%.
4.8. Tyrosinase Inhibition
The inhibitory activity of tyrosinase was assessed using a modified dopachrome method with L-DOPA as the substrate, based on a spectrophotometric approach [73]. The extracts were initially dissolved in 20% DMSO and then diluted to various concentrations using 0.1 M phosphate buffer (pH 6.8). In each well of a 96-well microtitre plate, 5 μL of the extract was combined with 115 μL of 0.1 M phosphate buffer (pH 6.8), 40 μL of mushroom tyrosinase (200 units/mL), and 40 μL of L-DOPA (2.5 mM). The reaction mixture was incubated for 30 min at 37 °C. Alongside each sample, a blank was included containing all the components except L-DOPA to account for background absorbance. The absorbance of the samples was measured at 475 nm. Kojic acid and quercetin were utilized as positive controls for inhibition, while the negative control comprised 10% DMSO in place of the extracts. The percentage of tyrosinase inhibition was calculated using the formula: Tyrosinase inhibition % = [(A_control − A_sample)/A_control] × 100%.
4.9. Hyaluronidase Inhibition
The hyaluronidase inhibitory activity was assessed using a method modified from previous studies [74]. Initially, 25 μL of the test extract (concentration range: 0–100 μg/mL) was pre-incubated with 3 μL of hyaluronidase from bovine testes type I-S (H3506, Sigma-Aldrich, USA) at a concentration of 0.4 U/mL in 20 mM phosphate buffer (pH 7), containing 77 mM sodium chloride and 0.01% bovine serum albumin (BSA), for 10 min at 37 °C. Subsequently, 12 μL of phosphate buffer (300 mM, pH 5.35) was added, and the mixture was incubated for an additional 10 min at 37 °C. Then, 10 μL of hyaluronic acid substrate (0.03% in 300 mM phosphate buffer, pH 5.35) was introduced and incubated for 45 min at 37 °C. The reaction, which involves the decomposition of hyaluronic acid, was halted by adding 100 μL of acidic albumin solution (24 mM sodium acetate, 79 mM acetic acid, and 0.1% BSA, pH 3.75). The mixture was then allowed to stand at room temperature for 10 min, after which the absorbance was measured at a wavelength of 600 nm. Myricetin, at concentrations ranging from 0 to 100 μg/mL, served as a positive control, while 10% DMSO was used for the negative controls. The percentage of hyaluronidase inhibition was quantified using the formula: Hyaluronidase inhibition % = [(A_control − A_sample)/A_control] × 100%.
4.10. Trypan Blue Cytotoxicity Assay
Cell death was evaluated using the trypan blue cytotoxicity assay [75]. H838 cells (1 × 104) were incubated with N. miranda extract in a 100 μL volume for 24 h to assess the cytotoxic activity of the extract via trypan blue dye exclusion.
4.11. Chromatin Condensation Assay
Apoptosis in cancer cells was examined using the Hoechst 33342 staining method [76]. H838 cells were seeded in 96-well plates at a density of 5 × 103 cells per well and allowed to adhere for 16 h before incubation with N. miranda extract for 24 h. Afterward, cells were washed with PBS and stained with Hoechst dye (1 μg/mL) in the dark for 10 min. Stained cells were imaged using ImageXpress Pico (Molecular Devices, San Jose, CA, USA) with DAPI filter cubes. Image acquisition and analysis were performed using CellReporterXpress Version 2 software.
4.12. Clonogenic Formation Assay
The inhibition of H838 cell proliferation was evaluated using a clonogenic formation assay [77]. H838 cells were seeded at a density of 1 × 103 cells per well in 6-well plates and incubated overnight for attachment. Post-treatment with N. miranda extract for 5–7 days, cells were washed with PBS, fixed with methanol, and stained with 0.5% crystal violet for 20 min. The number of colonies was counted under a light microscope to assess proliferation inhibition.
4.13. Wound-Healing Assay
H838 cell migration inhibition was investigated using a wound healing assay [78], a technique to assess collective cell migration in two dimensions. A cell-free area was created in a confluent cell monolayer, and the presence of this gap induced cell migration to close the wound. H838 cells were grown in serum-reduced medium for 6 h before a linear wound was created using a pipette tip. After washing with serum-reduced medium, cells treated with N. miranda extract for 24 h were assessed for migration ability.
4.14. Flow Analysis
Flow cytometry was utilized for cell cycle analysis. H838 cells, treated with DMSO or N. miranda extract for 24 h, were harvested with trypsin. These cells were then washed, resuspended in PBS containing 1% FBS, and fixed with cold ethanol (70%). After another wash, cells were incubated in PBS for 5 min, then stained with a PI/RNase solution (PBS, RNase, and 50 μg/mL PI) for 30 min at 37 °C in the dark. Cell cycle distribution was analyzed using a BD FACSCanto II (BD Biosciences, San Jose, CA, USA) and visualized with FlowJo v10 software (Tree Star, Inc., Ashland, OR, USA).
4.15. Comet Assay
The comet assay was conducted following a modified method from previous research [44]. Cell pellets were resuspended in PBS and combined with 1% low melting point (LMP) agarose at a 1:10 ratio. Seventy-five μL of this mixture was then spread onto a slide pre-treated to enhance LMP agarose adherence. The slides were placed at 4 °C in the dark for 20 min. After the agarose solidified, slides were immersed in a lysis solution (10 mM Tris, 100 mM EDTA, 2.5 M NaCl, 1% Triton X-100, pH 10) for at least 90 min at 4 °C in a dark room. The excess lysis buffer from the slides was tapped off and the slides were washed twice with neutralization buffer (0.4 M Tris–HCl, pH 7.5) for 5 min each. The slides were placed in a horizontal electrophoresis chamber and covered with the electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13). Electrophoresis was conducted at 1.25 V/cm for 25 min. After electrophoresis, slides were washed with deionized water, dehydrated in 95% ethanol, air-dried, stained with PI (2.5 μg/mL; 50 μL per slide), and covered with cover slips in the dark for 30 min. Comet images were captured using a fluorescence microscope (ZEISS Axio Imager A2) and analyzed with Comet Score™ (TriTek Corp., Somerville, NJ, USA), evaluating at least 50 comets per slide for tail intensity percentage and tail moments.
4.16. Binding Analysis Using AutoDock Vina
The interaction between compounds and RPA32 was analyzed using AutoDock Vina [79,80,81]. RPA32’s atomic coordinates (PDB ID: 1L1O) were obtained from the RCSB PDB database. Pre-docking preparations, including charge assignments and volume measurements, were performed using AutoDockTools. The compounds’ 2D structures were sourced from PubChem and translated to .sdf format. Both ligands and the protein target were then prepared as PDBQT files for docking with AutoDock Vina through the PyRx Virtual Screening Tool. Docking results were visualized using PyMOL v2.2.0 software.
4.17. Statistical Analysis
Experiments were conducted in triplicate, with results presented as mean ± standard deviation (SD). IC50 values and statistical significance (p < 0.05) were determined using SigmaPlot version 12.0 and GraphPad Prism5 (GraphPad Software Inc., San Diego, CA, USA), respectively, with one-way ANOVA employed to assess differences between means.
5. Conclusions
We explored the anti-skin-aging and anti-RPA32 capabilities of various extracts from N. miranda, including the stem, leaf, and pitcher, all extracted using ethanol, a solvent deemed safer for human application. We investigated the cytotoxic effect of the stem extract on H838 cancer cells, focusing on cell survival, apoptosis, migration, proliferation, and DNA damage. The major components of this extract were identified through GC–MS analysis to evaluate their potential for multi-targeted therapeutic actions and synergistic benefits. The findings collectively suggest the promising pharmacological value of N. miranda, offering a foundation for its potential inclusion in future drug development.
Author Contributions
C.-Y.L., Y.-C.C., Y.-H.H. and Y.L. performed the experiments; C.-Y.L. and Y.-C.C. analyzed the data; C.-Y.L., Y.-C.C. and C.-Y.H. contributed to the study design and manuscript writing. All authors reviewed the results and contributed to the data interpretation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the National Science and Technology Council, Taiwan (NSTC 112-2320-B-040-018 to C.-Y.H.) and Tao Yuan General Hospital, Taiwan (PTH112036 to C.-Y.L.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Miguel, S.; Hehn, A.; Bourgaud, F. Nepenthes: State of the art of an inspiring plant for biotechnologists. J. Biotechnol. 2018, 265, 109–115. [Google Scholar] [CrossRef]
- Rodney, S.; Banerji, U. Optimizing the FDA’s Project Optimus: Opportunities and challenges. Nat. Rev. Clin. Oncol. 2023, 21, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Sekine, S.; Sato, T. Decoding the basis of histological variation in human cancer. Nat. Rev. Cancer 2023, 24, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Dumontet, C.; Reichert, J.M.; Senter, P.D.; Lambert, J.M.; Beck, A. Antibody-drug conjugates come of age in oncology. Nat. Rev. Drug Discov. 2023, 22, 641–661. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Sun, H.; Wang, S.; Yang, H.; Kong, F.S. Treatment-Related Pneumonitis of EGFR Tyrosine Kinase Inhibitors Plus Thoracic Radiation Therapy in Patients with Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Int. J. Radiat. Oncol. Biol. Phys. 2024, 118, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Ghidini, A.; Luciani, A. Topotecan or other agents as second-line therapy for relapsed small-cell lung cancer: A meta-analysis of randomized studies. Mol. Clin. Oncol. 2021, 15, 218. [Google Scholar] [CrossRef]
- Peng, H.; Huang, Z.; Li, P.; Sun, Z.; Hou, X.; Li, Z.; Sang, R.; Guo, Z.; Wu, S.; Cao, Y. Investigating the efficacy and mechanisms of Jinfu’an decoction in treating non-small cell lung cancer using network pharmacology and in vitro and in vivo experiments. J. Ethnopharmacol. 2024, 321, 117518. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Chen, M.; Qin, Y.; Li, J.; Li, S.; Xu, X. Treatment of non-small cell lung cancer with Yiqi Buxue prescriptions combined with adjuvant chemotherapy on the cancer therapy-related cardiovascular toxicity: A systematic review and meta-analysis. J. Ethnopharmacol. 2024, 323, 117665. [Google Scholar] [CrossRef]
- González, A.S.C.; Valencia, M.G.; Cervantes-Villagrana, R.D.; Zapata, A.B.; Cervantes-Villagrana, A.R. Cytotoxic and Antitumor Effects of the Hydroalcoholic Extract of Tagetes erecta in Lung Cancer Cells. Molecules 2023, 28, 7055. [Google Scholar] [CrossRef]
- Dushkov, A.; Vosáhlová, Z.; Tzintzarov, A.; Kalíková, K.; Křížek, T.; Ugrinova, I. Analysis of the Ibotenic Acid, Muscimol, and Ergosterol Content of an Amanita Muscaria Hydroalcoholic Extract with an Evaluation of Its Cytotoxic Effect against a Panel of Lung Cell Lines In Vitro. Molecules 2023, 28, 6824. [Google Scholar] [CrossRef]
- Park, W.; Han, J.H.; Wei, S.; Yang, E.S.; Cheon, S.Y.; Bae, S.J.; Ryu, D.; Chung, H.S.; Ha, K.T. Natural Product-Based Glycolysis Inhibitors as a Therapeutic Strategy for Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitor-Resistant Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2024, 25, 807. [Google Scholar] [CrossRef]
- Mia, M.A.R.; Dey, D.; Sakib, M.R.; Biswas, M.Y.; Prottay, A.A.S.; Paul, N.; Rimti, F.H.; Abdullah, Y.; Biswas, P.; Iftehimul, M.; et al. The efficacy of natural bioactive compounds against prostate cancer: Molecular targets and synergistic activities. Phytother. Res. 2023, 37, 5724–5754. [Google Scholar] [CrossRef]
- Dańczak-Pazdrowska, A.; Gornowicz-Porowska, J.; Polańska, A.; Krajka-Kuźniak, V.; Stawny, M.; Gostyńska, A.; Rubiś, B.; Nourredine, S.; Ashiqueali, S.; Schneider, A.; et al. Cellular senescence in skin-related research: Targeted signaling pathways and naturally occurring therapeutic agents. Aging Cell 2023, 22, e13845. [Google Scholar] [CrossRef]
- Zhang, S.; Duan, E. Fighting against Skin Aging: The Way from Bench to Bedside. Cell Transplant. 2018, 27, 729–738. [Google Scholar] [CrossRef]
- Macia, E.; Duboz, P.; Chevé, D. The paradox of impossible beauty: Body changes and beauty practices in aging women. J. Women Aging 2015, 27, 174–187. [Google Scholar] [CrossRef]
- Rathore, G.; Das, K.; Landau, M.; Verner, I.; Kassir, M.; Galadari, H.I.; Gold, M.H.; Babaei, M.; Goldust, M. Clinical Assessment, Diagnosis, and Management of Infraorbital Wrinkles and Pigmentation. Dermatol. Clin. 2024, 42, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.F. Diversity of Asian skin: A review on skin biophysical properties. Exp. Dermatol. 2024, 33, e14959. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M. Plant Extracts as Skin Care and Therapeutic Agents. Int. J. Mol. Sci. 2023, 24, 15444. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Uyama, H.; Kobayashi, S. Inhibition effects of (+)-catechin-aldehyde polycondensates on proteinases causing proteolytic degradation of extracellular matrix. Biochem. Biophys. Res. Commun. 2004, 320, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.X.; Kubo, I. Kinetics of mushroom tyrosinase inhibition by quercetin. J. Agric. Food Chem. 2002, 50, 4108–4112. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, G.H. Evaluation of antioxidant and inhibitory activities for different subclasses flavonoids on enzymes for rheumatoid arthritis. J. Food Sci. 2010, 75, H212–H217. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Plumbagin, a Natural Product with Potent Anticancer Activities, Binds to and Inhibits Dihydroorotase, a Key Enzyme in Pyrimidine Biosynthesis. Int. J. Mol. Sci. 2021, 22, 6861. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lien, Y.; Chen, J.H.; Lin, E.S.; Huang, C.Y. Identification and characterization of dihydropyrimidinase inhibited by plumbagin isolated from Nepenthes miranda extract. Biochimie 2020, 171–172, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef] [PubMed]
- Antony, E.; Lohman, T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, Y.H.; Chung, J.C.; Su, H.H.; Huang, C.Y. The Inhibitory Effects and Cytotoxic Activities of the Stem Extract of Nepenthes miranda against Single-Stranded DNA-Binding Protein and Oral Carcinoma Cells. Plants 2023, 12, 2188. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lin, E.S.; Huang, C.Y. Complexed crystal structure of SSB reveals a novel single-stranded DNA binding mode (SSB)3:1: Phe60 is not crucial for defining binding paths. Biochem. Biophys. Res. Commun. 2019, 520, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Chen, I.C.; Huang, C.Y. Characterization of an SSB-dT25 complex: Structural insights into the S-shaped ssDNA binding conformation. RSC Adv. 2019, 9, 40388–40396. [Google Scholar] [CrossRef]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000, 7, 648–652. [Google Scholar] [CrossRef]
- Lin, E.S.; Luo, R.H.; Huang, C.Y. A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity. Int. J. Mol. Sci. 2022, 23, 588. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, Y.H.; Luo, R.H.; Basharat, Z.; Huang, C.Y. Crystal Structure of an SSB Protein from Salmonella enterica and Its Inhibition by Flavanonol Taxifolin. Int. J. Mol. Sci. 2022, 23, 4399. [Google Scholar] [CrossRef]
- Glanzer, J.G.; Endres, J.L.; Byrne, B.M.; Liu, S.; Bayles, K.W.; Oakley, G.G. Identification of inhibitors for single-stranded DNA-binding proteins in eubacteria. J. Antimicrob. Chemother. 2016, 71, 3432–3440. [Google Scholar] [CrossRef]
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef]
- Casteel, D.E.; Zhuang, S.; Zeng, Y.; Perrino, F.W.; Boss, G.R.; Goulian, M.; Pilz, R.B. A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 2009, 284, 5807–5818. [Google Scholar] [CrossRef]
- Glanzer, J.G.; Liu, S.; Wang, L.; Mosel, A.; Peng, A.; Oakley, G.G. RPA inhibition increases replication stress and suppresses tumor growth. Cancer Res. 2014, 74, 5165–5172. [Google Scholar] [CrossRef] [PubMed]
- MacPhail, S.H.; Olive, P.L. RPA foci are associated with cell death after irradiation. Radiat. Res. 2001, 155, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Almasan, A. USP14 Regulates DNA Damage Response and Is a Target for Radiosensitization in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2020, 21, 6383. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G. Molecular mechanisms of skin ageing. Mech. Ageing Dev. 2002, 123, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Logesh, R.; Prasad, S.R.; Chipurupalli, S.; Robinson, N.; Mohankumar, S.K. Natural tyrosinase enzyme inhibitors: A path from melanin to melanoma and its reported pharmacological activities. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188968. [Google Scholar] [CrossRef] [PubMed]
- Baber, M.A.; Crist, C.M.; Devolve, N.L.; Patrone, J.D. Tyrosinase Inhibitors: A Perspective. Molecules 2023, 28, 5762. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanouil, C.D.; Peña-García, J.; Banegas-Luna, A.J.; Kostagianni, A.D.; Gerothanassis, I.P.; Pérez-Sánchez, H.; Tzakos, A.G. ANTIAGE-DB: A Database and Server for the Prediction of Anti-Aging Compounds Targeting Elastase, Hyaluronidase, and Tyrosinase. Antioxidants 2022, 11, 2268. [Google Scholar] [CrossRef] [PubMed]
- Moody, T.W.; Zia, F.; Draoui, M.; Brenneman, D.E.; Fridkin, M.; Davidson, A.; Gozes, I. A vasoactive intestinal peptide antagonist inhibits non-small cell lung cancer growth. Proc. Natl. Acad. Sci. USA 1993, 90, 4345–4349. [Google Scholar] [CrossRef]
- Speit, G.; Hartmann, A. The comet assay: A sensitive genotoxicity test for the detection of DNA damage and repair. Methods Mol. Biol. 2006, 314, 275–286. [Google Scholar]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, J.; Chen, X.; Guo, W.; Du, Y.; Wang, Y.; Zang, W.; Zhang, S.; Zhao, G. Myricetin enhance chemosensitivity of 5-fluorouracil on esophageal carcinoma in vitro and in vivo. Cancer Cell Int. 2014, 14, 71. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Z.R.; Dong, W.G.; Zhang, J.X.; Guo, X.F.; Song, J.; Qiu, S. Cooperative inhibitory effect of sinomenine combined with 5-fluorouracil on esophageal carcinoma. World J. Gastroenterol. 2013, 19, 8292–8300. [Google Scholar] [CrossRef]
- Guo, X.F.; Zhu, X.F.; Zhong, G.S.; Deng, B.G.; Gao, Z.T.; Wang, H. Lapatinib, a dual inhibitor of EGFR and HER2, has synergistic effects with 5-fluorouracil on esophageal carcinoma. Oncol. Rep. 2012, 27, 1639–1645. [Google Scholar] [CrossRef]
- Bochkareva, E.; Korolev, S.; Lees-Miller, S.P.; Bochkarev, A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002, 21, 1855–1863. [Google Scholar] [CrossRef]
- Madru, C.; Martínez-Carranza, M.; Laurent, S.; Alberti, A.C.; Chevreuil, M.; Raynal, B.; Haouz, A.; Le Meur, R.A.; Delarue, M.; Henneke, G.; et al. DNA-binding mechanism and evolution of replication protein A. Nat. Commun. 2023, 14, 2326. [Google Scholar] [CrossRef]
- Hemmerling, F.; Piel, J. Strategies to access biosynthetic novelty in bacterial genomes for drug discovery. Nat. Rev. Drug Discov. 2022, 21, 359–378. [Google Scholar] [CrossRef]
- Huang, C.Y.; Ju, D.T.; Chang, C.F.; Muralidhar Reddy, P.; Velmurugan, B.K. A review on the effects of current chemotherapy drugs and natural agents in treating non-small cell lung cancer. Biomedicine 2017, 7, 23. [Google Scholar] [CrossRef]
- Dzialo, M.; Mierziak, J.; Korzun, U.; Preisner, M.; Szopa, J.; Kulma, A. The Potential of Plant Phenolics in Prevention and Therapy of Skin Disorders. Int. J. Mol. Sci. 2016, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Luo, Y.; Tian, M.; Zhang, S.Y.; Lu, R.; Wang, J.H.; Kasimu, R.; Li, X. Plant natural products: From traditional compounds to new emerging drugs in cancer therapy. Cell Prolif. 2014, 47, 506–515. [Google Scholar] [CrossRef]
- Wójciak, M.; Feldo, M.; Stolarczyk, P.; Płachno, B.J. Carnivorous Plants from Nepenthaceae and Droseraceae as a Source of Secondary Metabolites. Molecules 2023, 28, 2155. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, Q. Surface hydrophobicity of slippery zones in the pitchers of two Nepenthes species and a hybrid. Sci. Rep. 2016, 6, 19907. [Google Scholar] [CrossRef]
- Hajialiasgary Najafabadi, A.; Soheilifar, M.H.; Masoudi-Khoram, N. Exosomes in skin photoaging: Biological functions and therapeutic opportunity. Cell Commun. Signal. 2024, 22, 32. [Google Scholar] [CrossRef]
- Ahmed, I.A.; Mikail, M.A. Diet and skin health: The good and the bad. Nutrition 2024, 119, 112350. [Google Scholar] [CrossRef] [PubMed]
- Song, S.Y.; Lee, S.H.; Bae, M.S.; Park, D.H.; Cho, S.S. Strong inhibition of xanthine oxidase and elastase of Baccharis trimera (Less.) DC stem extract and analysis of biologically active constituents. Front. Pharmacol. 2023, 14, 1160330. [Google Scholar] [CrossRef]
- Altyar, A.E.; Ashour, M.L.; Youssef, F.S. Premna odorata: Seasonal Metabolic Variation in the Essential Oil Composition of Its Leaf and Verification of Its Anti-Ageing Potential via In Vitro Assays and Molecular Modelling. Biomolecules 2020, 10, 879. [Google Scholar] [CrossRef]
- Pinho, B.R.; Sousa, C.; Valentão, P.; Oliveira, J.M.; Andrade, P.B. Modulation of basophils’ degranulation and allergy-related enzymes by monomeric and dimeric naphthoquinones. PLoS ONE 2014, 9, e90122. [Google Scholar] [CrossRef]
- Xiong, Y.; Li, B.; Huang, D.; He, Q.; Yu, X. Anti-Deinagkistrodon acutus venom properties of ethanolic root extract from Cynanchum paniculatum (Bunge) kitag and its GC-MS analysis. J. Ethnopharmacol. 2018, 225, 189–197. [Google Scholar] [CrossRef]
- Fernando, I.P.S.; Sanjeewa, K.K.A.; Ann, Y.S.; Ko, C.I.; Lee, S.H.; Lee, W.W.; Jeon, Y.J. Apoptotic and antiproliferative effects of Stigmast-5-en-3-ol from Dendronephthya gigantea on human leukemia HL-60 and human breast cancer MCF-7 cells. Toxicol. In Vitro 2018, 52, 297–305. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Panda, M.; Biswal, B.K. Emerging role of plumbagin: Cytotoxic potential and pharmaceutical relevance towards cancer therapy. Food Chem. Toxicol. 2019, 125, 566–582. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jiao, W.; Xu, Y.; Hou, L.; Li, H.; Shao, J.; Zhang, X.; Wang, R.; Kong, D. Palmitic acid inhibits prostate cancer cell proliferation and metastasis by suppressing the PI3K/Akt pathway. Life Sci. 2021, 286, 120046. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y. Crystal structure of SSB complexed with inhibitor myricetin. Biochem. Biophys. Res. Commun. 2018, 504, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, C.Y. Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein. Biochem. Biophys. Res. Commun. 2021, 534, 41–46. [Google Scholar] [CrossRef]
- Huang, Y.H.; Ning, Z.J.; Huang, C.Y. Crystal structure of dihydropyrimidinase in complex with anticancer drug 5-fluorouracil. Biochem. Biophys. Res. Commun. 2019, 519, 160–165. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, Y.H.; Yang, P.C.; Peng, W.F.; Huang, C.Y. Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil. Biomolecules 2023, 13, 149. [Google Scholar] [CrossRef]
- Sethy, C.; Kundu, C.N. 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed. Pharmacother. 2021, 137, 111285. [Google Scholar] [CrossRef]
- Huang, C.Y. Structure, catalytic mechanism, posttranslational lysine carbamylation, and inhibition of dihydropyrimidinases. Adv. Protein Chem. Struct. Biol. 2020, 122, 63–96. [Google Scholar]
- Shimizu, K.; Kondo, R.; Sakai, K.; Lee, S.H.; Sato, H. The inhibitory components from Artocarpus incisus on melanin biosynthesis. Planta Med. 1998, 64, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Tu, P.T.; Tawata, S. Anti-Oxidant, Anti-Aging, and Anti-Melanogenic Properties of the Essential Oils from Two Varieties of Alpinia zerumbet. Molecules 2015, 20, 16723–16740. [Google Scholar] [CrossRef] [PubMed]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001, 21, A-3B. [Google Scholar]
- Larsson, R.; Nygren, P. A rapid fluorometric method for semiautomated determination of cytotoxicity and cellular proliferation of human tumor cell lines in microculture. Anticancer Res. 1989, 9, 1111–1119. [Google Scholar]
- Chen, M.H.; Yang, W.L.; Lin, K.T.; Liu, C.H.; Liu, Y.W.; Huang, K.W.; Chang, P.M.; Lai, J.M.; Hsu, C.N.; Chao, K.M.; et al. Gene expression-based chemical genomics identifies potential therapeutic drugs in hepatocellular carcinoma. PLoS ONE 2011, 6, e27186. [Google Scholar] [CrossRef]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).