Differentially Expressed Genes Related to Isoflavone Biosynthesis in a Soybean Mutant Revealed by a Comparative Transcriptomic Analysis
Abstract
1. Introduction
2. Results
2.1. Variations in Isoflavone Contents among Soybean Seed Developmental Stages
2.2. Transcriptome Sequencing Data Analysis
2.3. Analysis of Differential Gene Expression in Soybean Seeds with Distinct Isoflavone Contents
2.4. GO and KEGG Enrichment Analyses of DEGs
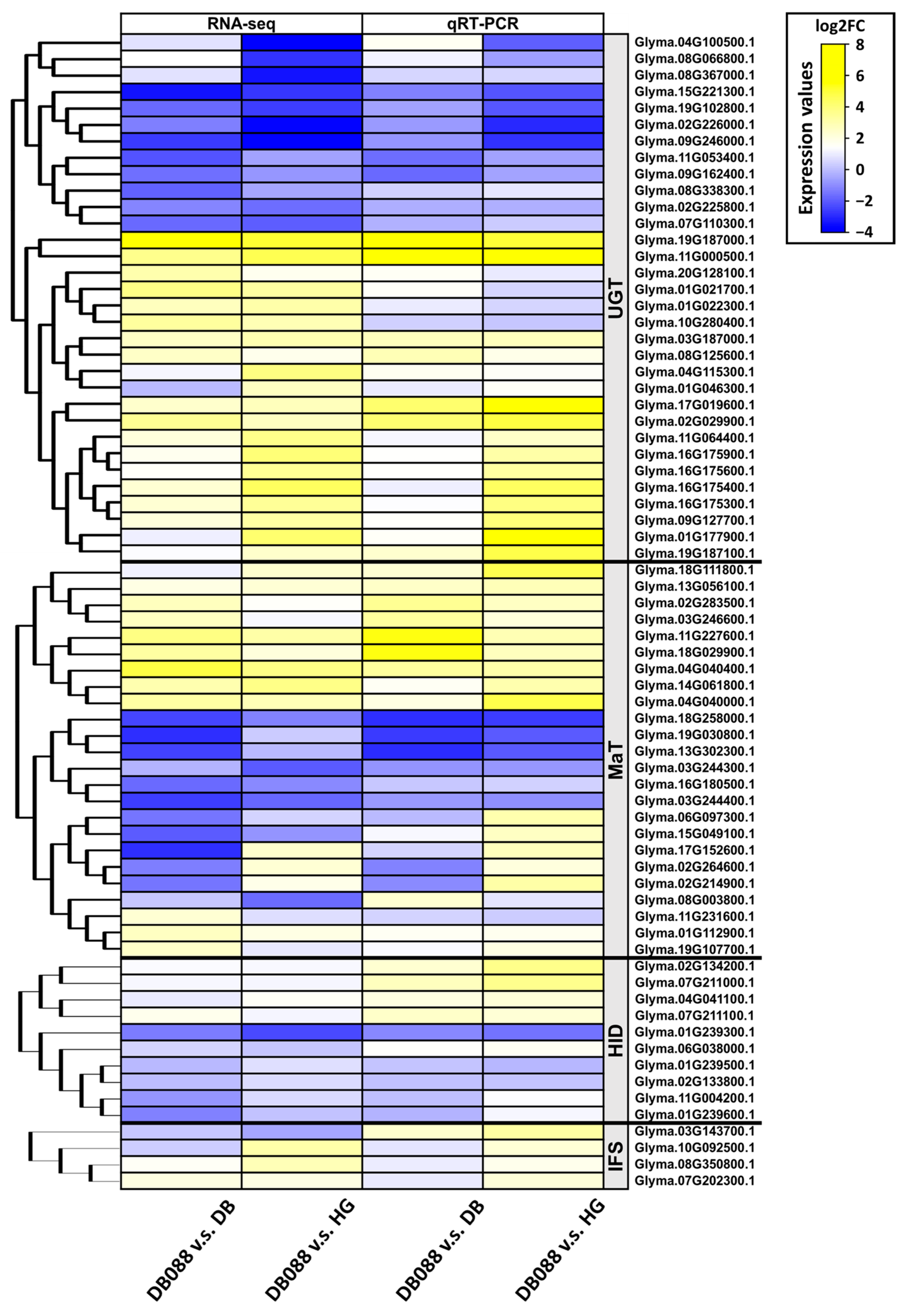
2.5. Expression Patterns of the Downstream Genes in the Isoflavone Biosynthetic Pathway
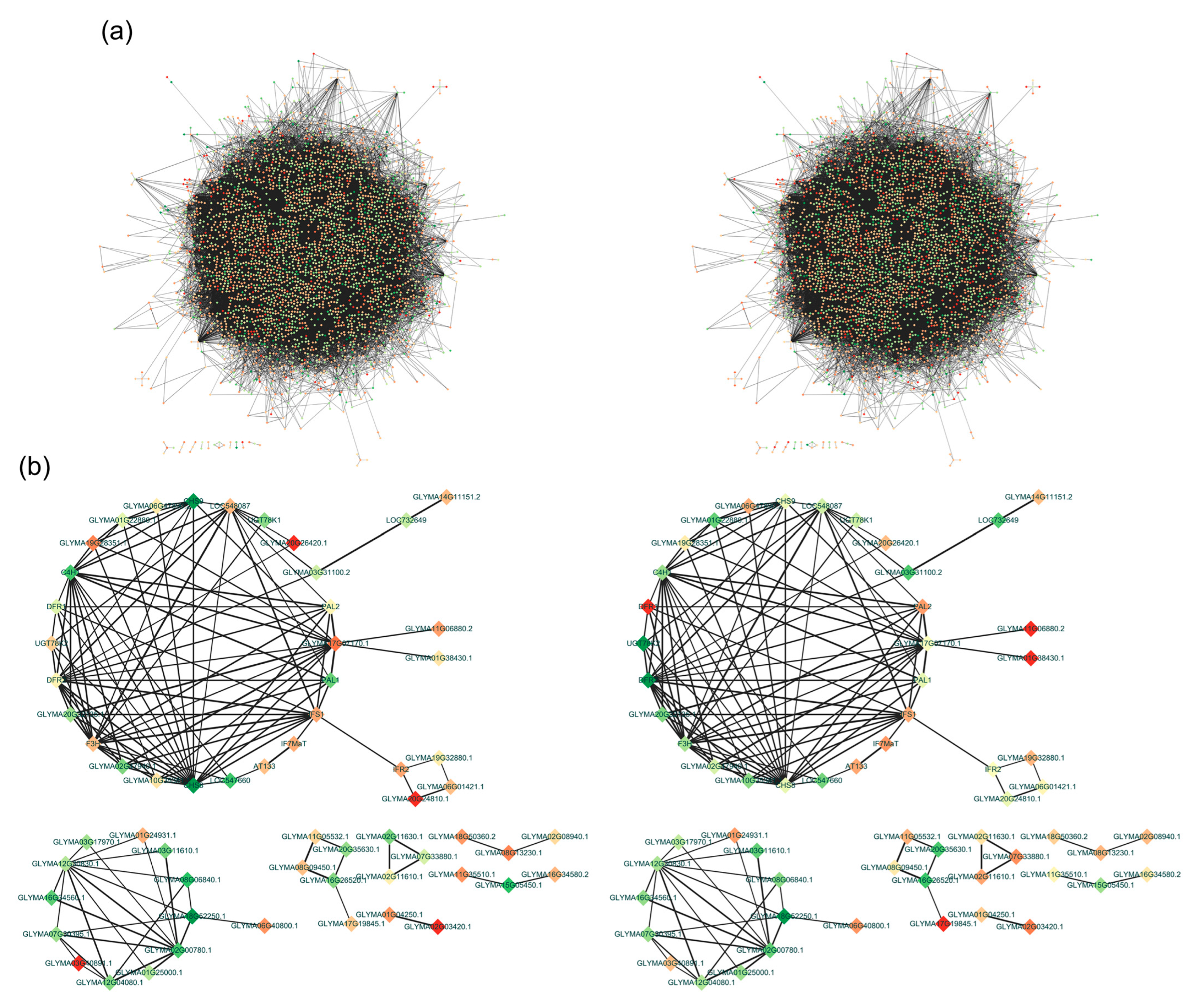
2.6. Co-Expression Network Analysis of the DEGs Related to Isoflavone Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Isoflavone Contents
4.3. RNA Extraction
4.4. cDNA Library Construction and RNA Sequencing
4.5. Data Preprocessing and Classification of Differentially Expressed Genes
4.6. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analyses and Protein–Protein Interaction Network Analysis
4.7. Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rochfort, S.; Panozzo, J. Phytochemicals for health, the role of pulses. J. Agric. Food Chem. 2007, 55, 7981–7994. [Google Scholar] [CrossRef]
- Sreevidya, V.; Srinivasa Rao, C.; Sullia, S.; Ladha, J.K.; Reddy, P.M. Metabolic engineering of rice with soybean isoflavone synthase for promoting nodulation gene expression in rhizobia. J. Exp. Bot. 2006, 57, 1957–1969. [Google Scholar] [CrossRef]
- Arjmandi, B.H.; Smith, B.J. Soy isoflavones’ osteoprotective role in postmenopausal women: Mechanism of action. J. Nutr. Biochem. 2002, 13, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Vitale, D.C.; Piazza, C.; Melilli, B.; Drago, F.; Salomone, S. Isoflavones: Estrogenic activity, biological effect and bioavailability. Eur. J. Drug Metab. Pharmacokinet. 2013, 38, 15–25. [Google Scholar] [CrossRef]
- Dixit, A.; Antony, J.; Sharma, N.K.; Tiwari, R.K. 12. Soybean constituents and their functional benefits. Res. Signpost 2011, 37, 2. [Google Scholar]
- Smallwood, C.J.; Nyinyi, C.N.; Kopsell, D.A.; Sams, C.E.; West, D.R.; Chen, P.; Kantartzi, S.K.; Cregan, P.B.; Hyten, D.L.; Pantalone, V.R. Detection and confirmation of quantitative trait loci for soybean seed isoflavones. Crop Sci. 2014, 54, 595–606. [Google Scholar] [CrossRef]
- Murphy, S.E.; Lee, E.A.; Woodrow, L.; Seguin, P.; Kumar, J.; Rajcan, I.; Ablett, G.R. Genotype× environment interaction and stability for isoflavone content in soybean. Crop Sci. 2009, 49, 1313–1321. [Google Scholar] [CrossRef]
- Eldridge, A.C.; Kwolek, W.F. Soybean isoflavones: Effect of environment and variety on composition. J. Agric. Food Chem. 1983, 31, 394–396. [Google Scholar] [CrossRef]
- Ng, M.-S.; Ku, Y.-S.; Yung, W.-S.; Cheng, S.-S.; Man, C.-K.; Yang, L.; Song, S.; Chung, G.; Lam, H.-M. MATE-type proteins are responsible for isoflavone transportation and accumulation in soybean seeds. Int. J. Mol. Sci. 2021, 22, 12017. [Google Scholar] [CrossRef] [PubMed]
- Barz, W.; Mackenbrock, U. Constitutive and elicitation induced metabolism of isoflavones and pterocarpans in chickpea (Cicer arietinum) cell suspension cultures. Plant Cell Tissue Organ Cult. 1994, 38, 199–211. [Google Scholar] [CrossRef]
- Welle, R.; Grisebach, H. Isolation of a novel NADPH-dependent reductase which coacts with chalcone synthase in the biosynthesis of 6′-deoxychalcone. FEBS Lett. 1988, 236, 221–225. [Google Scholar] [CrossRef]
- Jung, W.; Yu, O.; Lau, S.-M.C.; O’Keefe, D.P.; Odell, J.; Fader, G.; McGonigle, B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 2000, 18, 208–212. [Google Scholar] [CrossRef]
- Akashi, T.; Aoki, T.; Ayabe, S.-i. Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 2005, 137, 882–891. [Google Scholar] [CrossRef]
- Dhaubhadel, S.; Farhangkhoee, M.; Chapman, R. Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 2008, 59, 981–994. [Google Scholar] [CrossRef]
- Yu, X.H.; Chen, M.H.; Liu, C.J. Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso) flavones in Medicago truncatula. Plant J. 2008, 55, 382–396. [Google Scholar] [CrossRef]
- St-Pierre, B.; De Luca, V. Origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv. Phytochem. 2000, 34, 285–315. [Google Scholar]
- Ahmad, M.Z.; Li, P.; Wang, J.; Rehman, N.U.; Zhao, J. Isoflavone malonyltransferases GmIMaT1 and GmIMaT3 differently modify isoflavone glucosides in soybean (Glycine max) under various stresses. Front. Plant Sci. 2017, 8, 735. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Zhang, Y.; Zeng, X.; Li, P.; Wang, X.; Benedito, V.A.; Zhao, J. Isoflavone malonyl-CoA acyltransferase GmMaT2 is involved in nodulation of soybean by modifying synthesis and secretion of isoflavones. J. Exp. Bot. 2021, 72, 1349–1369. [Google Scholar] [CrossRef]
- Matsuda, H.; Nakayasu, M.; Aoki, Y.; Yamazaki, S.; Nagano, A.J.; Yazaki, K.; Sugiyama, A. Diurnal metabolic regulation of isoflavones and soyasaponins in soybean roots. Plant Direct 2020, 4, e00286. [Google Scholar] [CrossRef]
- Kudou, S.; Fleury, Y.; Welti, D.; Magnolato, D.; Uchida, T.; Kitamura, K.; Okubo, K. Malonyl isoflavone glycosides in soybean seeds (Glycine max Merrill). Agric. Biol. Chem. 1991, 55, 2227–2233. [Google Scholar] [CrossRef]
- Suzuki, H.; Nishino, T.; Nakayama, T. cDNA cloning of a BAHD acyltransferase from soybean (Glycine max): Isoflavone 7-O-glucoside-6 ″-O-malonyltransferase. Phytochemistry 2007, 68, 2035–2042. [Google Scholar] [CrossRef]
- Zhao, J.; Huhman, D.; Shadle, G.; He, X.-Z.; Sumner, L.W.; Tang, Y.; Dixon, R.A. MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 2011, 23, 1536–1555. [Google Scholar] [CrossRef]
- Patra, B.; Schluttenhofer, C.; Wu, Y.; Pattanaik, S.; Yuan, L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 1236–1247. [Google Scholar] [CrossRef]
- Weisshaar, B.; Jenkins, G.I. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1998, 1, 251–257. [Google Scholar] [CrossRef]
- Chu, S.; Wang, J.; Zhu, Y.; Liu, S.; Zhou, X.; Zhang, H.; Wang, C.-E.; Yang, W.; Tian, Z.; Cheng, H. An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PLoS Genet. 2017, 13, e1006770. [Google Scholar] [CrossRef]
- Joshi, A.; Jeena, G.S.; Kumar, R.S.; Pandey, A.; Shukla, R.K. Ocimum sanctum, OscWRKY1, regulates phenylpropanoid pathway genes and promotes resistance to pathogen infection in Arabidopsis. Plant Mol. Biol. 2022, 110, 235–251. [Google Scholar] [CrossRef]
- Spelt, C.; Quattrocchio, F.; Mol, J.; Koes, R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 2002, 14, 2121–2135. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Ramachandran, S. Natural and artificial mutants as valuable resources for functional genomics and molecular breeding. Int. J. Biol. Sci. 2010, 6, 228. [Google Scholar] [CrossRef]
- Beyaz, R.; Yildiz, M. The use of gamma irradiation in plant mutation breeding. Plant Eng. 2017, 34–46. [Google Scholar]
- Kim, S.H.; Ryu, J.; Kim, W.J.; Kang, R.; Seo, E.; Kim, G.; Kang, S.-Y.; Lee, J.-D.; Ha, B.-K. Identification of a new GmSACPD-C allele in high stearic acid mutant Hfa180 derived from gamma-ray irradiation. Mol. Breed. 2019, 39, 17. [Google Scholar] [CrossRef]
- Che, Z.; Liu, H.; Yi, F.; Cheng, H.; Yang, Y.; Wang, L.; Du, J.; Zhang, P.; Wang, J.; Yu, D. Genome-wide association study reveals novel loci for SC7 resistance in a soybean mutant panel. Front. Plant Sci. 2017, 8, 1771. [Google Scholar] [CrossRef]
- Hwang, S.-G.; Lee, S.C.; Lee, J.; Lee, J.W.; Kim, J.-H.; Choi, S.Y.; Kim, J.-B.; Choi, H.-I.; Jang, C.S. Resequencing of a core rice mutant population induced by gamma-ray irradiation and its application in a genome-wide association study. J. Plant Biol. 2020, 63, 463–472. [Google Scholar] [CrossRef]
- Kim, D.-G.; Lyu, J.I.; Kim, J.M.; Seo, J.S.; Choi, H.-I.; Jo, Y.D.; Kim, S.H.; Eom, S.H.; Ahn, J.-W.; Bae, C.-H. Identification of Loci Governing Agronomic Traits and Mutation Hotspots via a GBS-Based Genome-Wide Association Study in a Soybean Mutant Diversity Pool. Int. J. Mol. Sci. 2022, 23, 10441. [Google Scholar] [CrossRef]
- Hung, N.N.; Kim, D.-G.; Lyu, J.I.; Park, K.-C.; Kim, J.M.; Kim, J.-B.; Ha, B.-K.; Kwon, S.-J. Detecting genetic mobility using a transposon-based marker system in gamma-ray irradiated soybean mutants. Plants 2021, 10, 373. [Google Scholar] [CrossRef]
- Kim, J.M.; Lyu, J.I.; Kim, D.-G.; Hung, N.N.; Seo, J.S.; Ahn, J.-W.; Lim, Y.J.; Eom, S.H.; Ha, B.-K.; Kwon, S.-J. Genome wide association study to detect genetic regions related to isoflavone content in a mutant soybean population derived from radiation breeding. Front. Plant Sci. 2022, 13, 968466. [Google Scholar] [CrossRef]
- Kim, J.M.; Seo, J.S.; Lee, J.W.; Lyu, J.I.; Ryu, J.; Eom, S.H.; Ha, B.-K.; Kwon, S.-J. QTL mapping reveals key factors related to the isoflavone contents and agronomic traits of soybean (Glycine max). BMC Plant Biol. 2023, 23, 517. [Google Scholar] [CrossRef]
- Hu, W.; Liu, X.; Xiong, Y.; Liu, T.; Li, Z.; Song, J.; Wang, J.; Wang, X.; Li, X. Nutritional evaluation and transcriptome analyses of short-time germinated seeds in soybean (Glycine max L. Merri.). Sci. Rep. 2021, 11, 22714. [Google Scholar] [CrossRef]
- Konzak, C.F. III. Genetic effects of radiation on higher plants. Q. Rev. Biol. 1957, 32, 27–45. [Google Scholar] [CrossRef]
- Alothman, M.; Bhat, R.; Karim, A. Effects of radiation processing on phytochemicals and antioxidants in plant produce. Trends Food Sci. Technol. 2009, 20, 201–212. [Google Scholar] [CrossRef]
- Hajika, M.; Igita, K.; Kitamura, K. A line lacking all the seed lipoxygenase isozymes in soybean [Glycine max (L.) Merrill] induced by gamma-ray irradiation. Jpn. J. Breed. 1991, 41, 507–509. [Google Scholar] [CrossRef]
- Lee, K.J.; Hwang, J.E.; Velusamy, V.; Ha, B.-K.; Kim, J.-B.; Kim, S.H.; Ahn, J.-W.; Kang, S.-Y.; Kim, D.S. Selection and molecular characterization of a lipoxygenase-free soybean mutant line induced by gamma irradiation. Theor. Appl. Genet. 2014, 127, 2405–2413. [Google Scholar] [CrossRef]
- Hwang, W.J.; Kim, M.Y.; Kang, Y.J.; Shim, S.; Stacey, M.G.; Stacey, G.; Lee, S.-H. Genome-wide analysis of mutations in a dwarf soybean mutant induced by fast neutron bombardment. Euphytica 2015, 203, 399–408. [Google Scholar] [CrossRef]
- Kim, D.-G.; Lyu, J.I.; Lee, M.-K.; Kim, J.M.; Hung, N.N.; Hong, M.J.; Kim, J.-B.; Bae, C.-H.; Kwon, S.-J. Construction of soybean mutant diversity pool (MDP) lines and an analysis of their genetic relationships and associations using TRAP markers. Agronomy 2020, 10, 253. [Google Scholar] [CrossRef]
- Wang, Q.; Ge, X.; Tian, X.; Zhang, Y.; Zhang, J.; Zhang, P. Soy isoflavone: The multipurpose phytochemical. Biomed. Rep. 2013, 1, 697–701. [Google Scholar] [CrossRef]
- Zeng, G.; Li, D.; Han, Y.; Teng, W.; Wang, J.; Qiu, L.; Li, W. Identification of QTL underlying isoflavone contents in soybean seeds among multiple environments. Theor. Appl. Genet. 2009, 118, 1455–1463. [Google Scholar] [CrossRef]
- Hoeck, J.A.; Fehr, W.R.; Murphy, P.A.; Welke, G.A. Influence of genotype and environment on isoflavone contents of soybean. Crop Sci. 2000, 40, 48–51. [Google Scholar] [CrossRef]
- Kim, J.A.; Chung, I.M. Change in isoflavone concentration of soybean (Glycine max L.) seeds at different growth stages. J. Sci. Food Agric. 2007, 87, 496–503. [Google Scholar] [CrossRef]
- Yu, O.; Shi, J.; Hession, A.O.; Maxwell, C.A.; McGonigle, B.; Odell, J.T. Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 2003, 63, 753–763. [Google Scholar] [CrossRef]
- Liu, R.; Hu, Y.; Li, J.; Lin, Z. Production of soybean isoflavone genistein in non-legume plants via genetically modified secondary metabolism pathway. Metab. Eng. 2007, 9, 1–7. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Le, T.H.T.; Nguyen, T.N.L.; Nguyen, T.G.; Sy, D.T.; Tu, Q.T.; Vu, T.T.T.; Le, V.S.; Chu, H.M.; Vu, T.K.L. Overexpressing GmCHI1A increases the isoflavone content of transgenic soybean (Glycine max (L.) Merr.) seeds. Vitro Cell. Dev. Biol. Plant 2020, 56, 842–850. [Google Scholar] [CrossRef]
- Azam, M.; Zhang, S.; Abdelghany, A.M.; Shaibu, A.S.; Feng, Y.; Li, Y.; Tian, Y.; Hong, H.; Li, B.; Sun, J. Seed isoflavone profiling of 1168 soybean accessions from major growing ecoregions in China. Food Res. Int. 2020, 130, 108957. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Ha, J.; Kang, Y.-G.; Lee, T.; Kim, M.; Yoon, M.Y.; Lee, E.; Yang, X.; Kim, D.; Kim, Y.-J.; Lee, T.R. Comprehensive RNA sequencing and co-expression network analysis to complete the biosynthetic pathway of coumestrol, a phytoestrogen. Sci. Rep. 2019, 9, 1934. [Google Scholar] [CrossRef]
- Livingstone, J.M.; Zolotarov, Y.; Strömvik, M.V. Transcripts of soybean isoflavone 7-O-glucosyltransferase and hydroxyisoflavanone dehydratase gene homologues are at least as abundant as transcripts of their well known counterparts. Plant Physiol. Biochem. 2011, 49, 1071–1075. [Google Scholar] [CrossRef]
- Wang, J.; Hossain, M.S.; Lyu, Z.; Schmutz, J.; Stacey, G.; Xu, D.; Joshi, T. SoyCSN: Soybean context-specific network analysis and prediction based on tissue-specific transcriptome data. Plant Direct 2019, 3, e00167. [Google Scholar] [CrossRef]
- Livingstone, J.M.; Seguin, P.; Strömvik, M.V. An in silico study of the genes for the isoflavonoid pathway enzymes in soybean reveals novel expressed homologues. Can. J. Plant Sci. 2010, 90, 453–469. [Google Scholar] [CrossRef]
- Funaki, A.; Waki, T.; Noguchi, A.; Kawai, Y.; Yamashita, S.; Takahashi, S.; Nakayama, T. Identification of a highly specific isoflavone 7-O-glucosyltransferase in the soybean (Glycine max (L.) Merr.). Plant Cell Physiol. 2015, 56, 1512–1520. [Google Scholar] [CrossRef]
- Yin, Q.; Shen, G.; Di, S.; Fan, C.; Chang, Z.; Pang, Y. Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in Glycine max. Plant Cell Physiol. 2017, 58, 1558–1572. [Google Scholar] [CrossRef]
- Knizia, D.; Yuan, J.; Bellaloui, N.; Vuong, T.; Usovsky, M.; Song, Q.; Betts, F.; Register, T.; Williams, E.; Lakhssassi, N. The Soybean High Density ‘Forrest’by ‘Williams 82′SNP-Based Genetic Linkage Map Identifies QTL and Candidate Genes for Seed Isoflavone Content. Plants 2021, 10, 2029. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Shu, W.; Zhou, Q.; Xian, P.; Cheng, Y.; Lian, T.; Ma, Q.; Zhou, Y.; Li, H.; Nian, H.; Cai, Z. GmWRKY81 Encoding a WRKY Transcription Factor Enhances Aluminum Tolerance in Soybean. Int. J. Mol. Sci. 2022, 23, 6518. [Google Scholar] [CrossRef]
- Wei, W.; Liang, D.W.; Bian, X.H.; Shen, M.; Xiao, J.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Lv, J.; Chen, X. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 2019, 100, 384–398. [Google Scholar] [CrossRef]
- Wang, F.; Chen, H.W.; Li, Q.T.; Wei, W.; Li, W.; Zhang, W.K.; Ma, B.; Bi, Y.D.; Lai, Y.C.; Liu, X.L. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015, 83, 224–236. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef]
- Nam, J.; DePamphilis, C.W.; Ma, H.; Nei, M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 2003, 20, 1435–1447. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Anguraj Vadivel, A.K.; McDowell, T.; Renaud, J.B.; Dhaubhadel, S. A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max). Commun. Biol. 2021, 4, 356. [Google Scholar] [CrossRef]
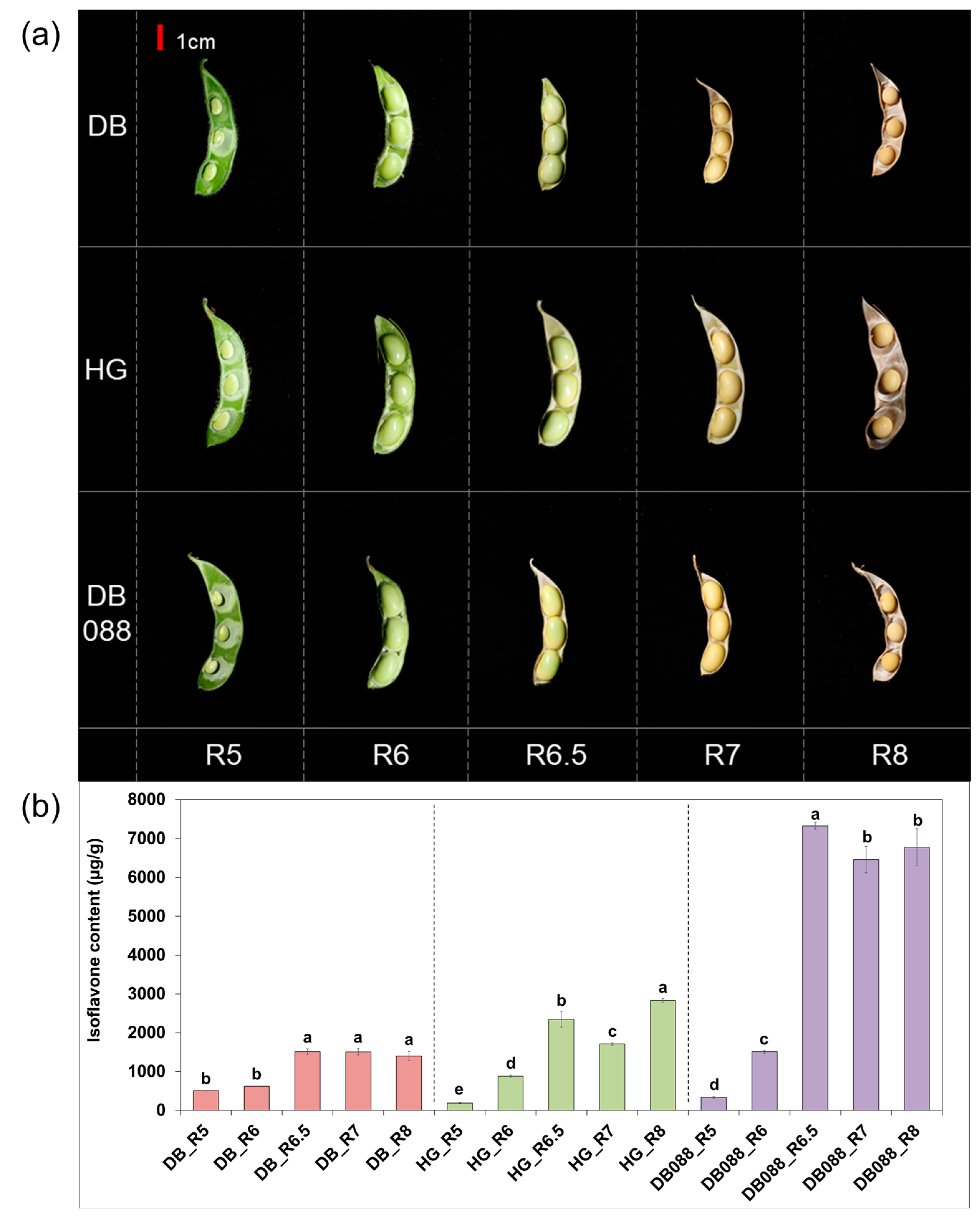
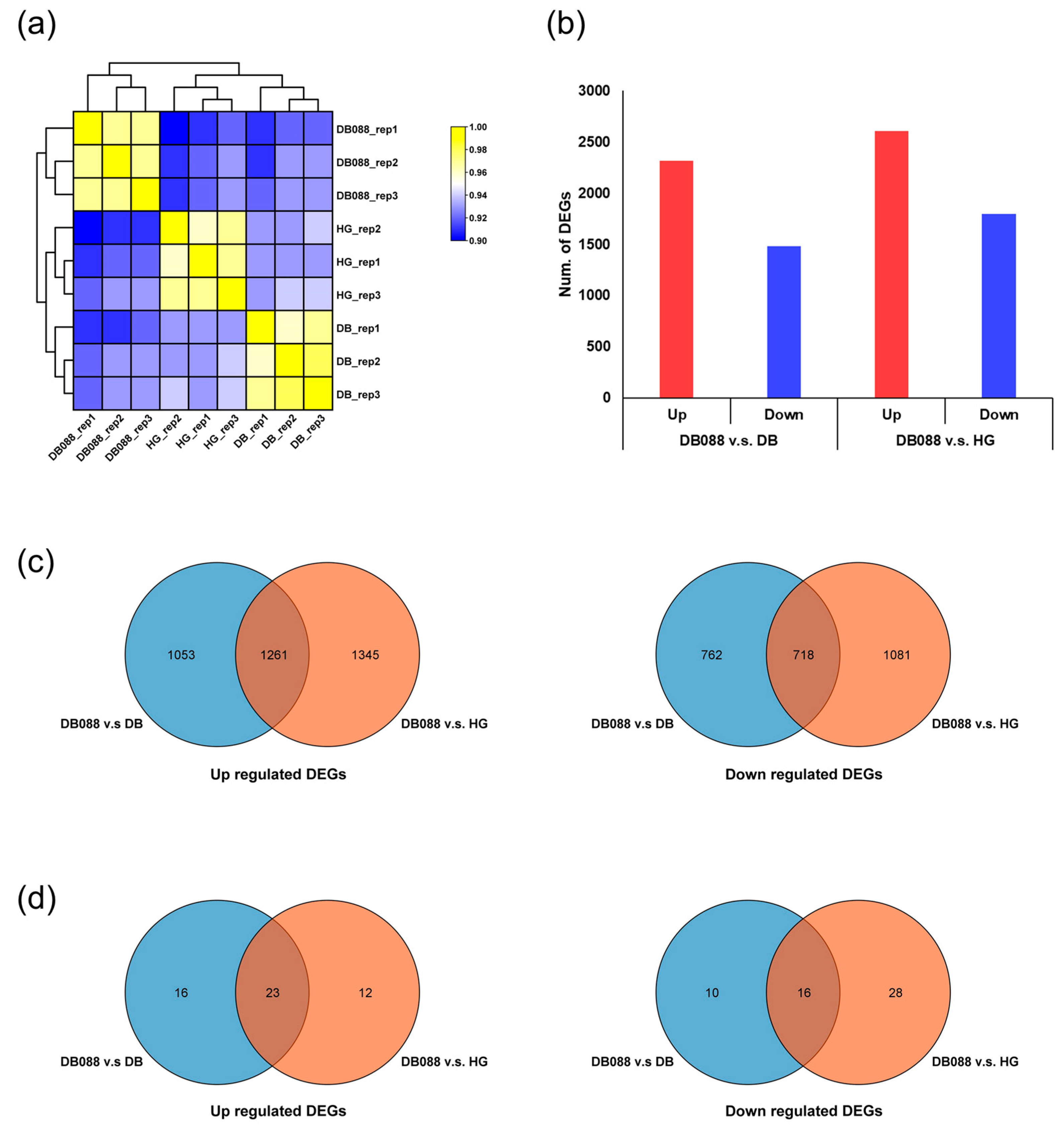
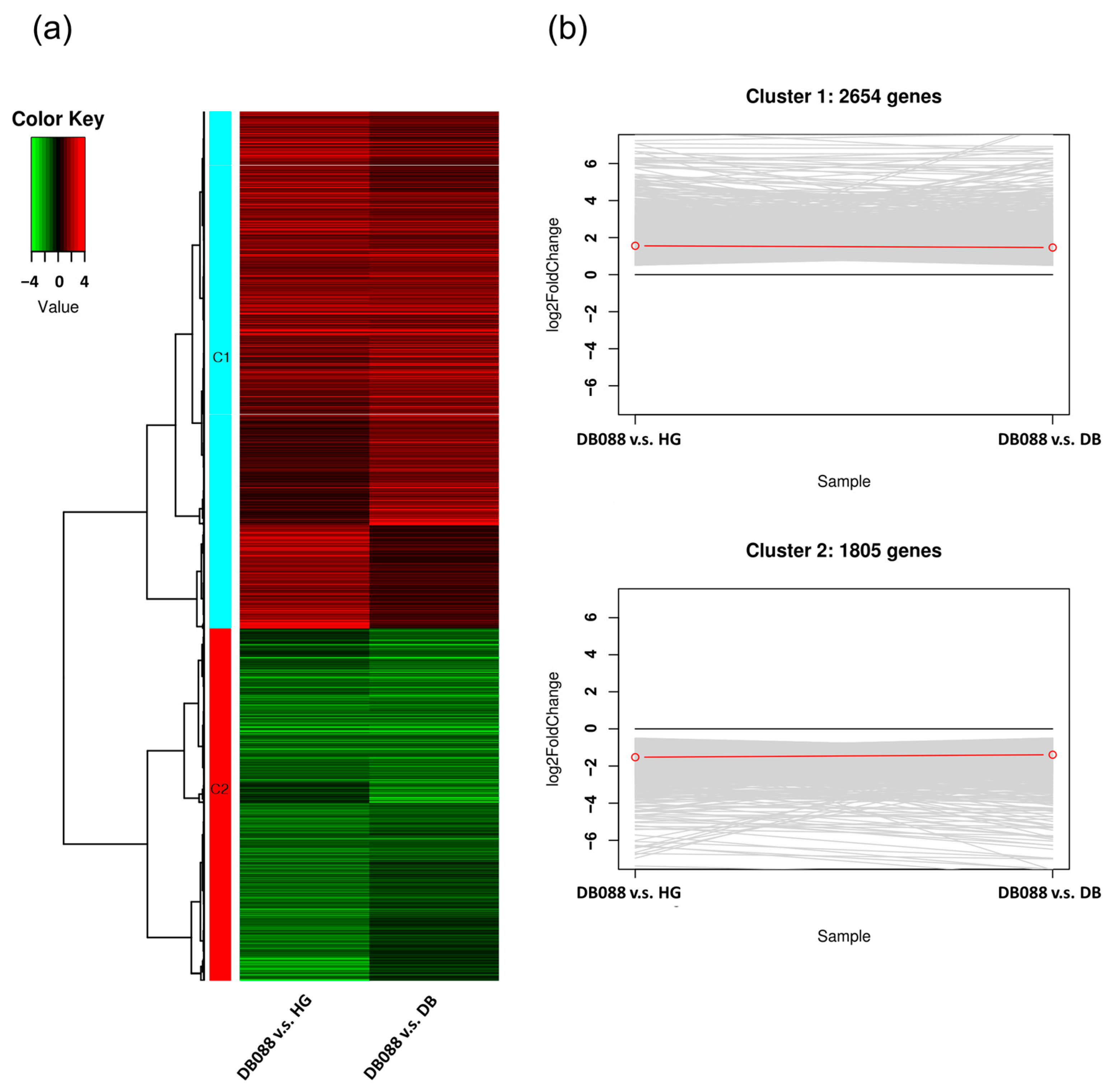

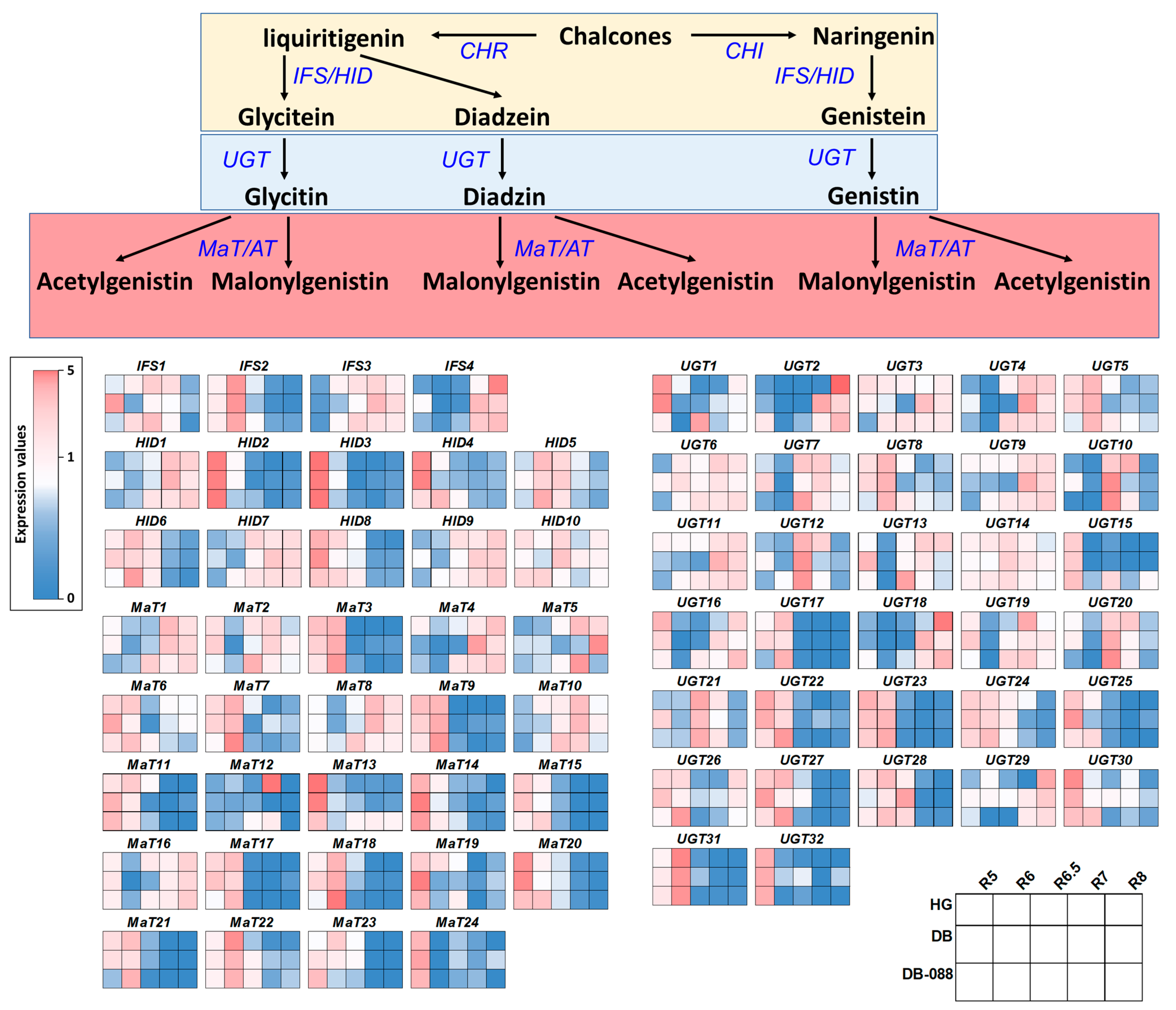
| Sample ID | Raw Reads | Avg. Length (bp) | Total Length (bp) | GC (%) | Q30 (%) | Clean Reads | Mapping Rate | |
|---|---|---|---|---|---|---|---|---|
| NO. Reads | Percent (%) | |||||||
| Danbaek_1 | 33,164,566 | 151 | 5,007,849,466 | 45.03 | 94.01 | 31,428,746 | 14,831,715 | 94% |
| Danbaek_2 | 33,036,170 | 151 | 4,988,461,670 | 46.73 | 92.70 | 30,724,592 | 15,034,644 | 98% |
| Danbaek_3 | 32,927,962 | 151 | 4,972,122,262 | 46.1 | 92.74 | 30,650,778 | 15,029,361 | 98% |
| DB088_1 | 33,359,878 | 151 | 5,037,341,578 | 46.79 | 92.93 | 31,149,264 | 14,982,713 | 96% |
| DB088_2 | 32,669,030 | 151 | 4,933,023,530 | 47.20 | 92.69 | 30,444,128 | 14,799,324 | 97% |
| DB088_3 | 33,099,718 | 151 | 4,998,057,418 | 46.62 | 92.57 | 30,661,986 | 14,957,058 | 98% |
| Hwanggeum_2 | 33,033,674 | 151 | 4,988,084,774 | 46.99 | 92.95 | 30,862,092 | 14,801,354 | 96% |
| Hwanggeum_3 | 33,363,984 | 151 | 5,037,961,584 | 46.35 | 92.90 | 31,142,916 | 15,097,658 | 97% |
| Hwanggeum_5 | 33,329,700 | 151 | 5,032,784,700 | 46.74 | 92.67 | 31,005,740 | 15,057,759 | 97% |
| Total | 297,984,682 | 44,995,686,982 | 278,070,242 | 134,591,586 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.M.; Lee, J.W.; Seo, J.S.; Ha, B.-K.; Kwon, S.-J. Differentially Expressed Genes Related to Isoflavone Biosynthesis in a Soybean Mutant Revealed by a Comparative Transcriptomic Analysis. Plants 2024, 13, 584. https://doi.org/10.3390/plants13050584
Kim JM, Lee JW, Seo JS, Ha B-K, Kwon S-J. Differentially Expressed Genes Related to Isoflavone Biosynthesis in a Soybean Mutant Revealed by a Comparative Transcriptomic Analysis. Plants. 2024; 13(5):584. https://doi.org/10.3390/plants13050584
Chicago/Turabian StyleKim, Jung Min, Jeong Woo Lee, Ji Su Seo, Bo-Keun Ha, and Soon-Jae Kwon. 2024. "Differentially Expressed Genes Related to Isoflavone Biosynthesis in a Soybean Mutant Revealed by a Comparative Transcriptomic Analysis" Plants 13, no. 5: 584. https://doi.org/10.3390/plants13050584
APA StyleKim, J. M., Lee, J. W., Seo, J. S., Ha, B.-K., & Kwon, S.-J. (2024). Differentially Expressed Genes Related to Isoflavone Biosynthesis in a Soybean Mutant Revealed by a Comparative Transcriptomic Analysis. Plants, 13(5), 584. https://doi.org/10.3390/plants13050584








