Comparative Transcriptomics Analysis Reveals the Differences in Transcription between Resistant and Susceptible Pepper (Capsicum annuum L.) Varieties in Response to Anthracnose
Abstract
1. Introduction
2. Results
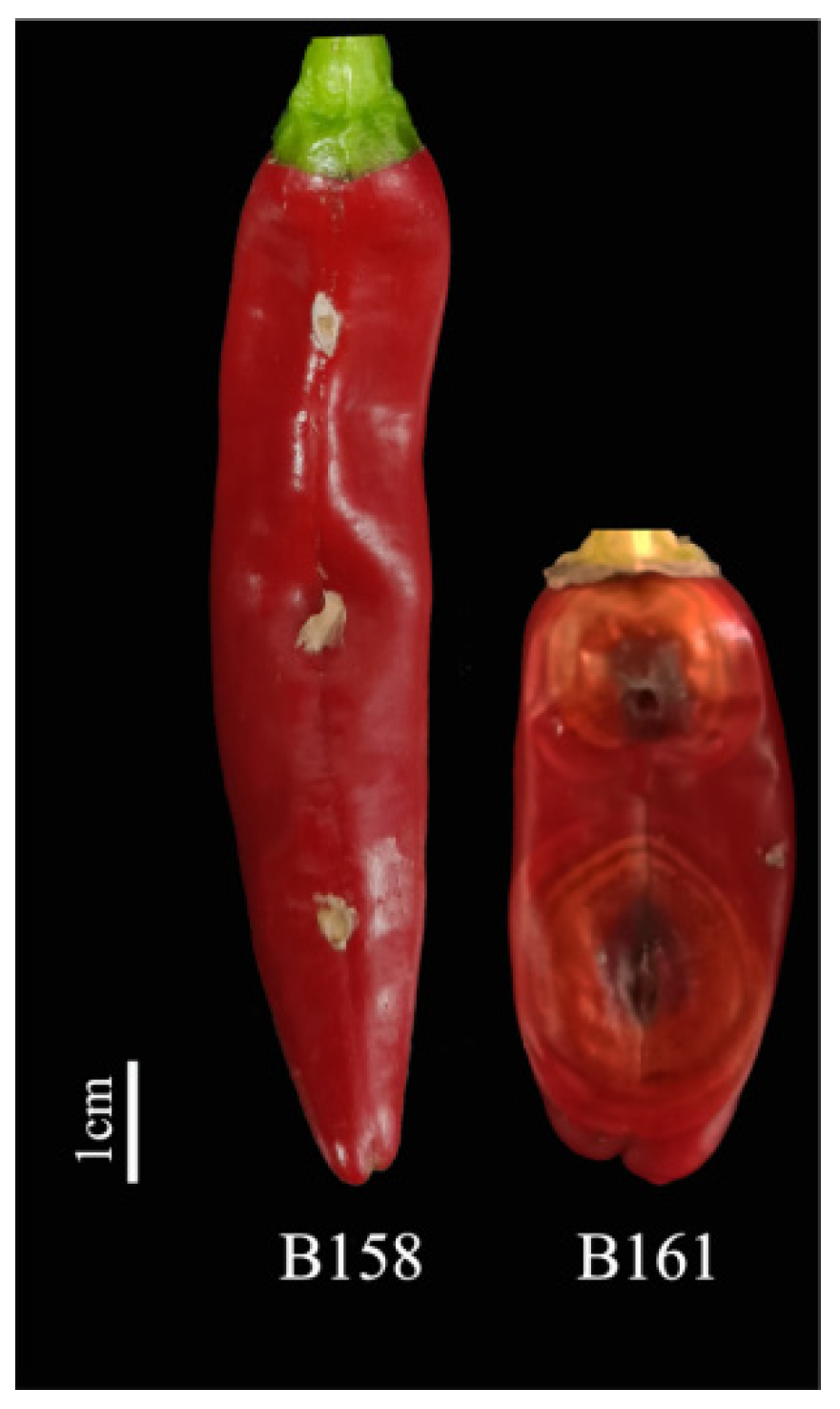
2.1. Screening of Pepper Materials for Disease Resistance
2.2. RNA-Seq and de Novo Transcription Assembly
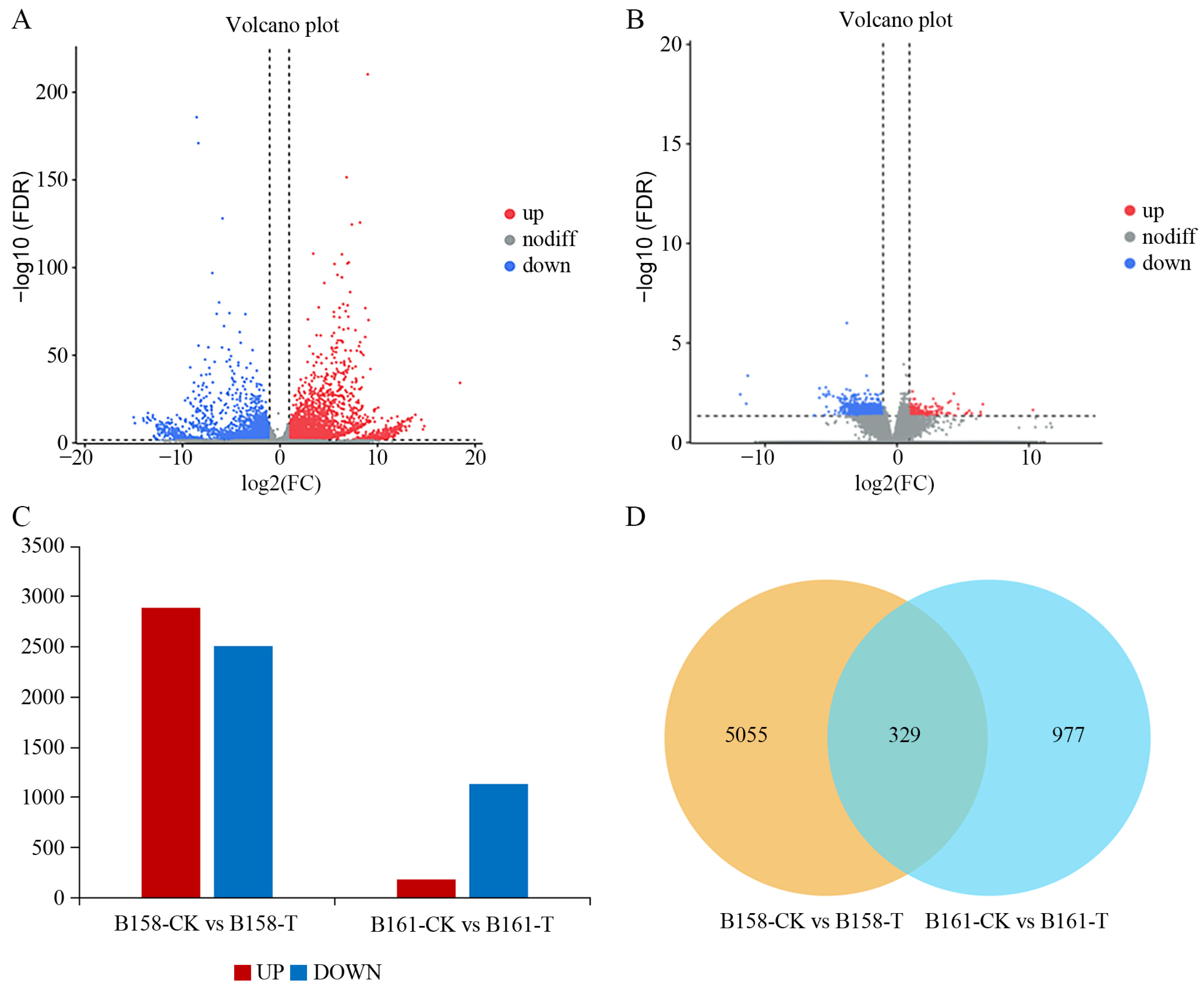
2.3. DEGs Analysis of Pepper in Response to C. capsici Infection
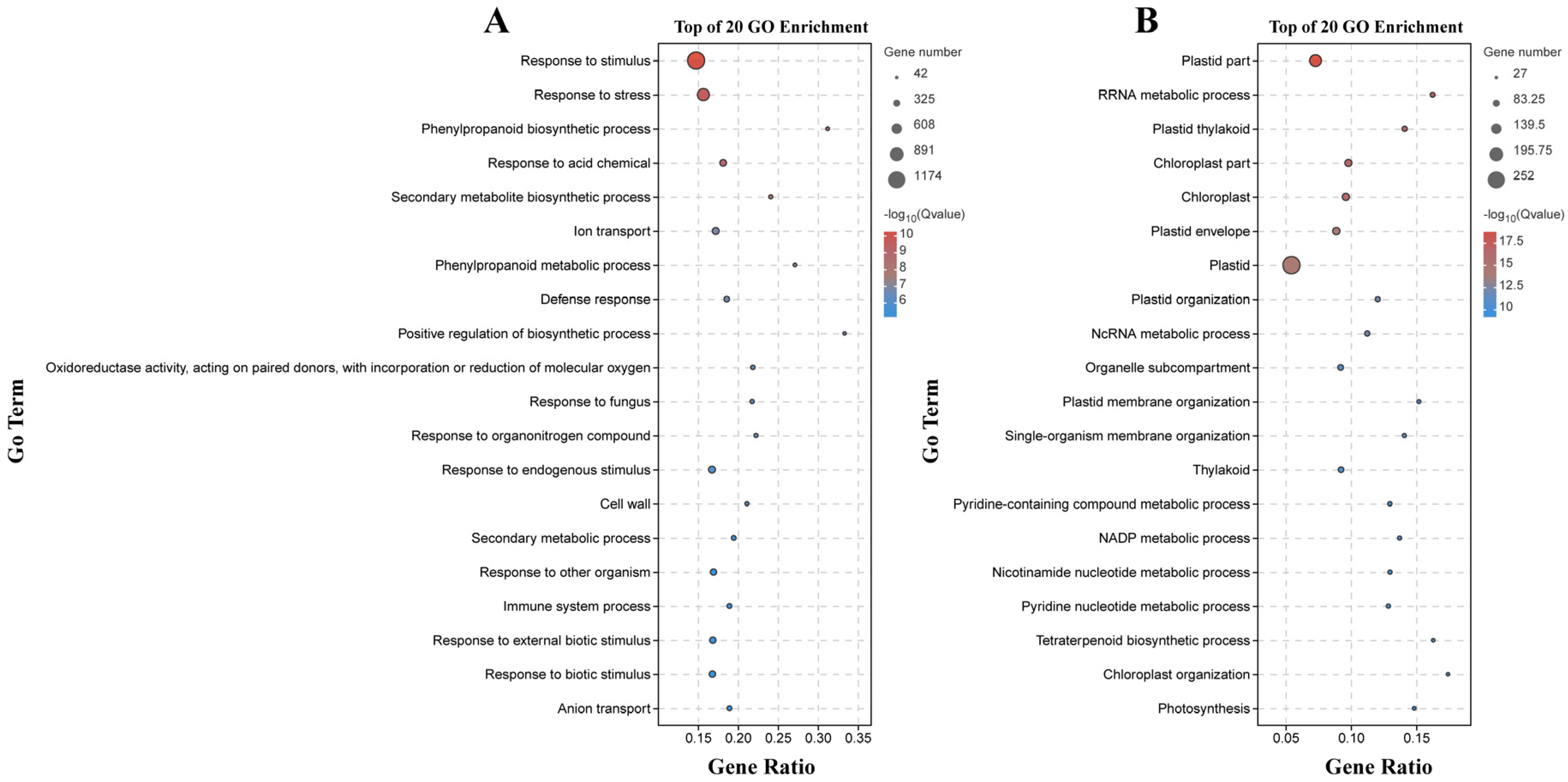
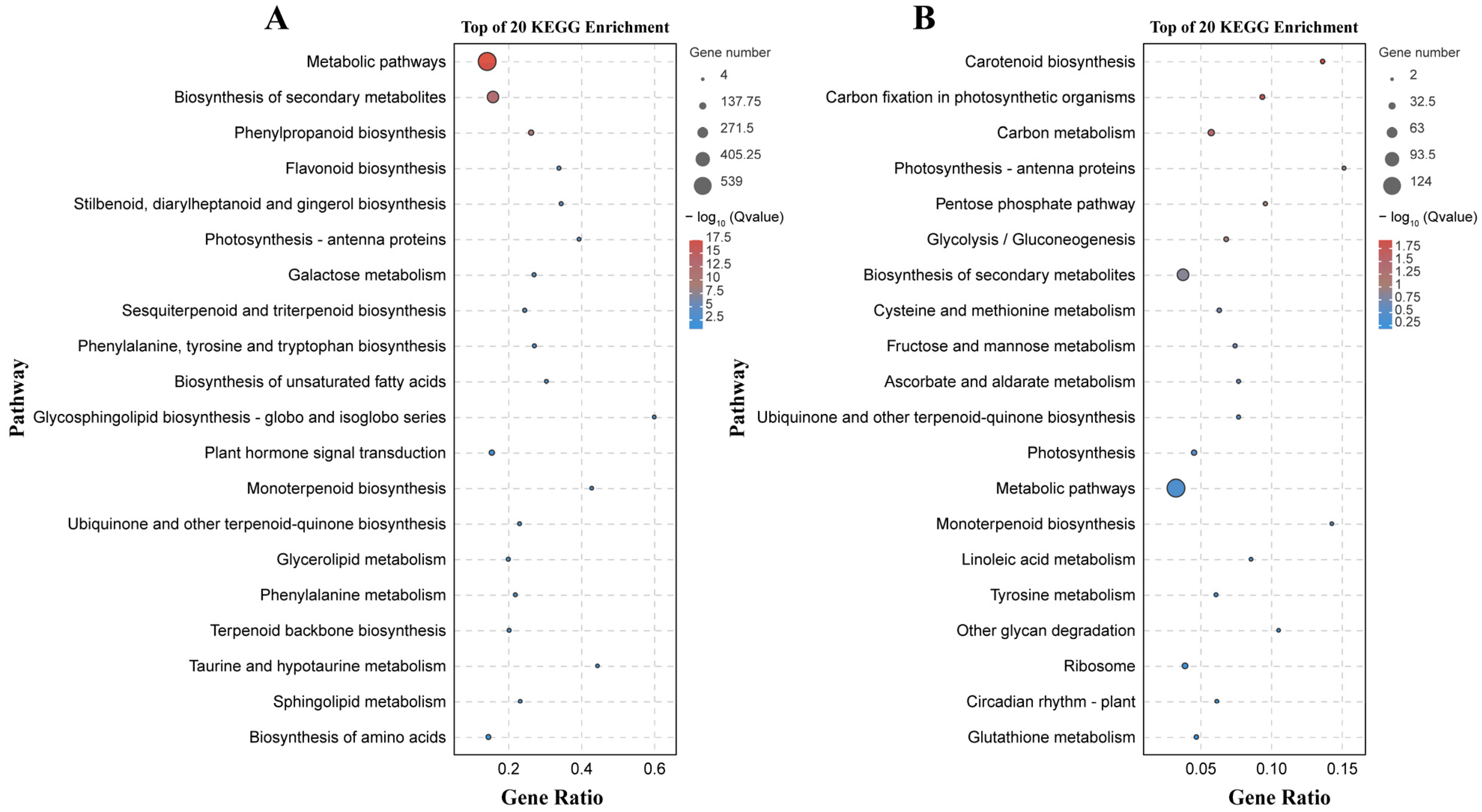
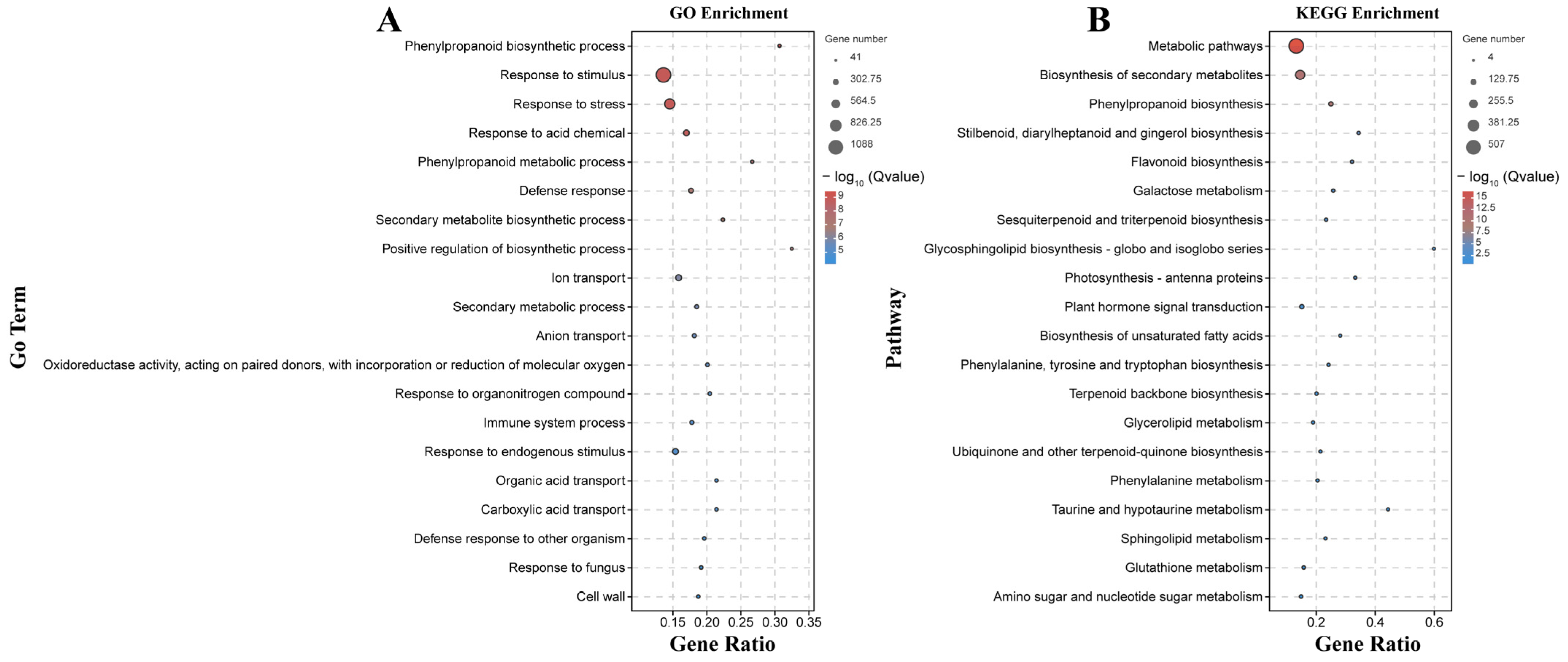
2.4. Functional Characterization according to GO Terms and KEGG Pathways
2.5. Analysis of the DEGs Exclusive to B158
2.6. Analysis of the TFs
3. Discussion
3.1. Response of the Resistant Variety B158 to Anthracnose
3.2. Involvement of TFs in Biological Stress Networks
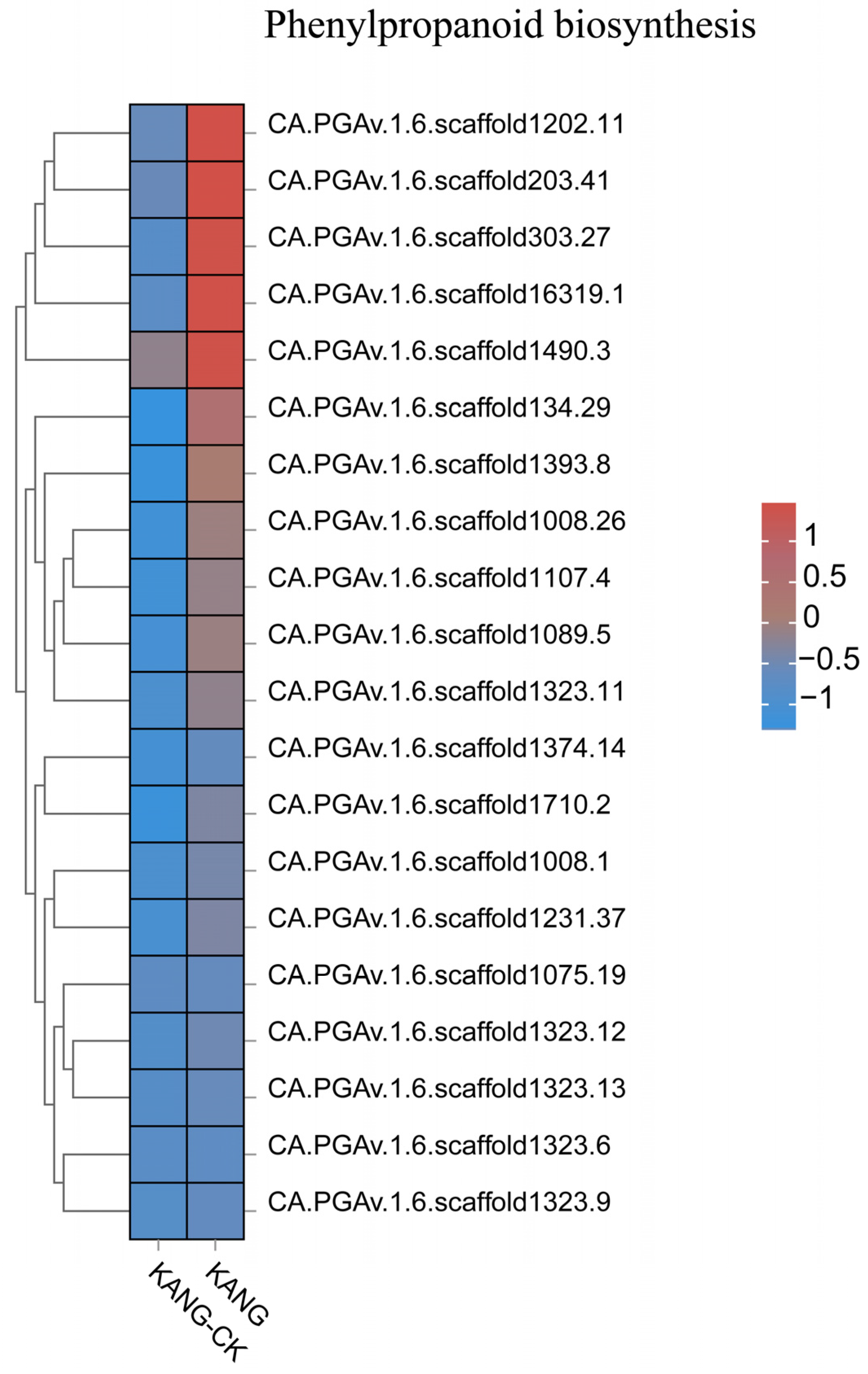
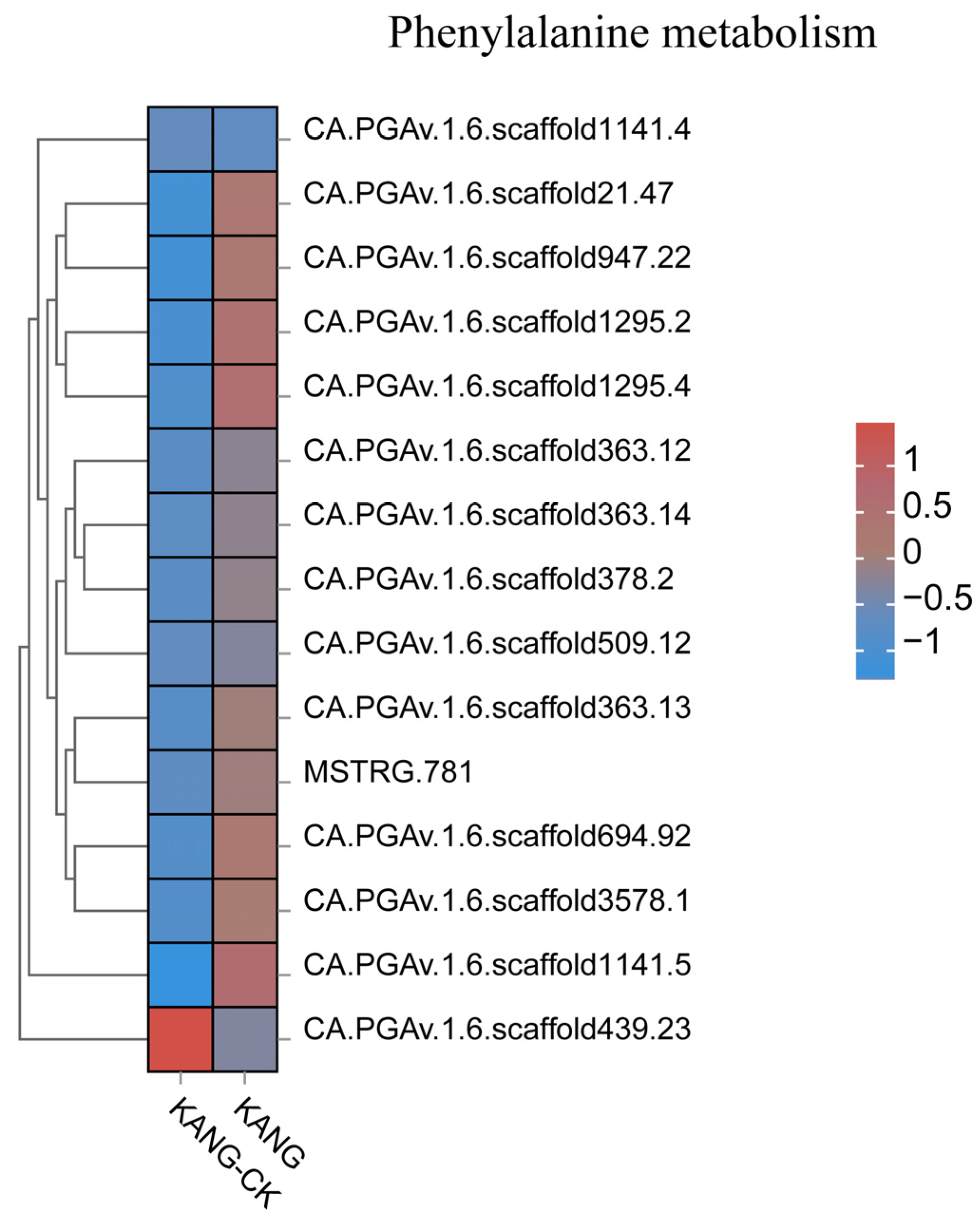
3.3. Analysis of Disease Resistance-Related Pathways
4. Materials and Methods
4.1. Colletotrichum capsici Culture
4.2. Statistical Analysis of Pepper Disease Spot Data
4.3. RNA Extraction and Sequencing Library Construction
4.4. Transcriptomics Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, L.; Zhang, L.; Wang, Y.; Zhan, H.; Yang, J.; Peng, L.; Yang, L.; He, B.; Lu, B.; Wang, Y.; et al. Distribution, identification and characterization of Colletotrichum lineola and C. panacicola causing anthracnose on ginseng in northeast China. Crop. Prot. 2020, 137, 105265. [Google Scholar] [CrossRef]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum—current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; An, C.G.; Park, J.S.; Lim, Y.P.; Kim, S. Carotenoid profiling from 27 types of paprika (Capsicum annuum L.) with different colors, shapes, and cultivation methods. Food Chem. 2016, 201, 64–71. [Google Scholar] [CrossRef]
- Bosland, P.W.; Votava, E.J. Peppers: Vegetable and Spice Capsicums. N. Z. J. Crop Hortic. Sci. 2013, 41, 102–103. [Google Scholar] [CrossRef]
- Sydow, H. Beitrage zur kenntnis der PilzHora des Siidlichen Ostindiens-I. Ann. Mycol. 1913, 11, 329–330. [Google Scholar]
- Johnston, P.R.; Jones, D. Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia 1997, 89, 420–430. [Google Scholar] [CrossRef]
- Kim, K.D.; Oh, A.B.J.; Yang, J. Differential Interactions of a Colletotrichum gloeosporioides Isolate with Green and Red Pepper Fruits. Phytoparasitica 1999, 27, 97–106. [Google Scholar] [CrossRef]
- Pakdeevaraporn, P.; Wasee, S.; Taylor, P.W.J.; Mongkolporn, O. Inheritance of resistance to anthracnose caused by Colletotrichum capsici in Capsicum. Plant Breed. 2005, 124, 206–208. [Google Scholar] [CrossRef]
- Sharma, P.N.; Kaur, M.; Sharma, O.P.; Sharma, P.; Pathania, A. Morphological, Pathological and Molecular Variability in Colletotrichum capsici, the Cause of Fruit Rot of Chillies in the Subtropical Region of North-western India. J. Phytopathol. 2005, 153, 232–237. [Google Scholar] [CrossRef]
- Simmonds, J. A study of the species of Colletotrichum causing ripe fruit rots in Queensland. Qld. J. Agric. Anim. Sci. 1966, 22, 437–459. [Google Scholar]
- Than, P.; Jeewon, R.; Hyde, K.; Pongsupasamit, S.; Mongkolporn, O.; Taylor, P. Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathol. 2008, 57, 562–572. [Google Scholar] [CrossRef]
- Voorrips, R.E.; Finkers, R.; Sanjaya, L.; Groenwold, R. QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annuum and C. chinense. Theor. Appl. Genet. 2004, 109, 1275–1282. [Google Scholar] [CrossRef]
- Saxena, A.; Raghuwanshi, R.; Gupta, V.K.; Singh, H.B. Chilli Anthracnose: The Epidemiology and Management. Front. Microbiol. 2016, 7, 1527. [Google Scholar] [CrossRef]
- Zhao, H.; Zhao, H.; Wang, J.; Wang, B.; Wang, Y. Stress stimulation induced resistance of plant. Colloids Surf. B Biointerfaces 2005, 43, 174–178. [Google Scholar] [CrossRef]
- Sharma, R.; Mahanty, B.; Mishra, R.; Joshi, R.K. Genome wide identification and expression analysis of pepper C2H2 zinc finger transcription factors in response to anthracnose pathogen Colletotrichum truncatum. 3 Biotech. 2021, 11, 118. [Google Scholar] [CrossRef]
- Mishra, R.; Mohanty, J.N.; Mahanty, B.; Joshi, R.K. A single transcript CRISPR/Cas9 mediated mutagenesis of CaERF28 confers anthracnose resistance in chilli pepper (Capsicum annuum L.). Planta 2021, 254, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yan, C.; Ning, Q.; Ruyi, J.Y.M.; Yuhua, Y. Comparative Transcriptome Analysis of Phytophthora blight Resistance of Pepper at Seedling Stage. Acta Agric. Boreali-Sin. 2019, 34, 194–202. [Google Scholar]
- Yao, W.; Zhao, K.; Cheng, Z.; Li, X.; Zhou, B.; Jiang, T. Transcriptome Analysis of Poplar Under Salt Stress and Over-Expression of Transcription Factor NAC57 Gene Confers Salt Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1121. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Shi, S.; Nasir, F.; Chang, C.; Li, W.; Tran, L.P.; Tian, C. Comparative analysis of the root transcriptomes of cultivated and wild rice varieties in response to Magnaporthe oryzae infection revealed both common and species-specific pathogen responses. Rice 2018, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Shi, J.; Li, X.; Liu, J.; Geng, Q.; Shi, H.; Ke, Y.; Sun, Q. Transcriptome analysis reveals the molecular mechanisms of the defense response to gray leaf spot disease in maize. BMC Genom. 2018, 19, 742. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, J.; Bocobza, S.; Panda, S.; Sonawane, P.; Cardenas, P.D.; Lashbrooke, J.; Kamble, A.; Shahaf, N.; Meir, S.; Bovy, A.; et al. Analysis of wild tomato introgression lines elucidates the genetic basis of transcriptome and metabolome variation underlying fruit traits and pathogen response. Nat. Genet. 2020, 52, s1111–s1121. [Google Scholar] [CrossRef]
- Colquhoun, T.A.; Kim, J.Y.; Wedde, A.E.; Levin, L.A.; Schmitt, K.C.; Schuurink, R.C.; Clark, D.G. PhMYB4 fine-tunes the floral volatile signature of Petunia x hybrida through PhC4H. J. Exp. Bot. 2011, 62, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Yin, X.R.; Zeng, J.K.; Ge, H.; Song, M.; Xu, C.J.; Li, X.; Ferguson, I.B.; Chen, K.S. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 2014, 65, 4349–4359. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lu, Y.H.W.; Cao, C.W.X.; Guo, X. GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 2012, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Kanth, B.K.; Ahn, J.Y.; Kim, J.H.; Lee, G.J. Genome-Wide Transcriptomic Identification and Functional Insight of Lily WRKY Genes Responding to Botrytis Fungal Disease. Plants 2021, 10, 776. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D.; et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 2014, 150, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Le Hénanff, G.; Profizi, C.; Courteaux, B.; Rabenoelina, F.; Clément, G.; Baillieul, F.; Baillieul, F.; Cordelier, S.; Dhondt-Cordelier, S. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013, 64, 4877–4893. [Google Scholar] [CrossRef]
- Liu, J.; Shen, Y.; Cao, H.; He, K.; Chu, Z.; Li, N. OsbHLH057 targets the AATCA cis-element to regulate disease resistance and drought tolerance in rice. Plant Cell Rep. 2022, 41, 1285–1299. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X.; et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018, 98, 289–302. [Google Scholar] [CrossRef]
- Perochon, A.; Kahla, A.; Vranic, M.; Jia, J.; Malla, K.B.; Craze, M.; Wallington, E.; Doohan, F.M. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol. J. 2019, 17, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Yang, S.; Wu, R.; Cao, J.; Shen, L.; Guan, D.; Shuilin, H. Pepper NAC-type transcription factor NAC2c balances the trade-off between growth and defense responses. Plant Physiol. 2021, 186, 2169–2189. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhou, J.; Sun, Y.; Qiao, B.; Wan, T.; Guo, R.; Zhang, J.; Shan, D.; Cai, Y. Comparative transcriptome and metabolome analyses of cherry leaves spot disease caused by Alternaria alternata. Front Plant Sci. 2023, 14, 1129515. [Google Scholar] [CrossRef]
- Tronchet, M.; Balague, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef]
- Zhou, X.H.; Erlin, G.; Yujing, W.; Yanlong, L.; Daojun, Y.; Longfu, Z. GhROP6 involved in cotton resistance to Verticillium wilt through regulating jasmonic acid synthesis and lignin metabolism. Cotton Sci. 2022, 34, 79–92. [Google Scholar] [CrossRef]
- Sharma, N.K.; Yadav, S.; Gupta, S.K.; Irulappan, V.; Francis, A.; Senthil-Kumar, M.; Chattopadhyay, D. MicroRNA397 regulates tolerance to drought and fungal infection by regulating lignin deposition in chickpea root. Plant Cell Environ. 2023, 11, 3501–3517. [Google Scholar] [CrossRef]
- Mishra, J.; Srivastava, R.; Trivedi, P.K.; Verma, P.C. Effect of virus infection on the secondary metabolite production and phytohormone biosynthesis in plants. 3 Biotech. 2020, 10, 547. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, X.; Liu, H.; Wang, Y.; Nocker, S.; Tu, M.; Fang, J.; Guo, J.; Li, Z.; Wang, X. Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic. Res. 2022, 9, uhab064. [Google Scholar] [CrossRef]
- Shan, D.; Wang, C.; Zheng, X.; Hu, Z.; Zhu, Y.; Zhao, Y.; Jiang, A.; Zhang, H.; Shi, K.; Bai, Y. MKK4-MPK3-WRKY17-mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 2021, 186, 1202–1219. [Google Scholar] [CrossRef]
- Deng, B.; Wang, W.; Ruan, C.; Deng, L.; Yao, S.; Zeng, K. Involvement of CsWRKY70 in salicylic acid-induced citrus fruit resistance against Penicillium digitatum. Hortic. Res. 2020, 7, 157. [Google Scholar] [CrossRef]
- Jiang, Y.; Zheng, W.; Li, J.; Liu, P.; Zhong, K.; Jin, P.; Xu, M.; Yang, J.; Chen, J. NbWRKY40 Positively Regulates the Response of Nicotiana benthamiana to Tomato Mosaic Virus via Salicylic Acid Signaling. Front. Plant Sci. 2020, 11, 603518. [Google Scholar] [CrossRef]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Shi, L.P.; Yang, S.; Qiu, S.S.; Ma, X.L.; Cai, J.S.; Guan, D.Y.; Wang, Z.H.; He, S.L. A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol. Plant Pathol. 2021, 22, 3–18. [Google Scholar] [CrossRef]
- Yang, S.; Cai, W.; Shen, L.; Cao, J.; Liu, C.; Hu, J.; Guan, D.; He, S. A CaCDPK29-CaWRKY27b module promotes CaWRKY40-mediated thermotolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 2022, 233, 1843–1863. [Google Scholar] [CrossRef]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef]
- Hussain, A.; Li, X.; Weng, Y.; Liu, Z.; Ashraf, M.F.; Noman, A.; Yang, S.; Ifnan, M.; Qiu, S.; Yang, Y. CaWRKY22 acts as a positive regulator in pepper response to Ralstonia solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int. J. Mol. Sci. 2018, 19, 1426. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Lin, J.; Chen, Y.; Li, G.X.; Guan, D.; Zheng, S.J.; He, S. A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J. Exp. Bot. 2019, 70, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Khan, M.I.; Albaqami, M.; Mahpara, S.; Noorka, I.R.; Ahmed, M.A.A.; Aljuaid, B.S.; El-Shehawi, A.M.; Liu, Z.; Farooq, S.; et al. CaWRKY30 Positively Regulates Pepper Immunity by Targeting CaWRKY40 against Ralstonia solanacearum Inoculation through Modulating Defense-Related Genes. Int. J. Mol. Sci. 2021, 22, 12091. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, Y.; Cai, W.; Liu, C.; Hu, J.; Shen, L.; Huang, X.; Guan, D.; He, S. CaWRKY28 Cys249 is Required for Interaction with CaWRKY40 in the Regulation of Pepper Immunity to Ralstonia solanacearum. Mol. Plant Microbe Interact. 2021, 34, 733–745. [Google Scholar] [CrossRef]
- Wang, Y.; Dang, F.; Liu, Z.; Wang, X.; Eulgem, T.; Lai, Y.; Yu, L.; She, J.; Shi, Y.; Lin, J.; et al. CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 2013, 14, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Ifnan Khan, M.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; Yang, I.K.M.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; et al. CaWRKY40b in Pepper Acts as a Negative Regulator in Response to Ralstonia solanacearum by Directly Modulating Defense Genes Including CaWRKY40. Int. J. Mol. Sci. 2018, 19, 1403. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, X.; Xiao, L.-D.; Yu, Y.-N.; Yan, H.-L.; Gong, Z.-H. CaWRKY50 Acts as a Negative Regulator in Response to Colletotrichum scovillei Infection in Pepper. Plants 2023, 12, 1962. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Clean Reads | Q30 (%) | GC (%) |
|---|---|---|---|---|
| B158-T-1 | 65,218,986 | 64,650,424 | 94.85 | 43.51 |
| B158-T-2 | 94,914,578 | 93,939,304 | 94.65 | 44.93 |
| B158-T-3 | 63,687,242 | 63,081,338 | 94.63 | 43.27 |
| B158-CK-1 | 65,847,442 | 65,195,778 | 94.59 | 42.28 |
| B158-CK-2 | 65,128,704 | 64,532,454 | 94.66 | 42.40 |
| B158-CK-3 | 55,746,766 | 55,244,818 | 94.69 | 42.51 |
| B161-T-1 | 103,516,244 | 100,728,566 | 94.81 | 45.81 |
| B161-T-2 | 127,486,314 | 124,524,228 | 94.17 | 49.54 |
| B161-T-3 | 133,622,868 | 132,342,810 | 94.62 | 49.83 |
| B161-CK-1 | 92,814,684 | 91,932,294 | 94.67 | 45.18 |
| B161-CK-2 | 58,120,020 | 57,515,364 | 94.56 | 42.24 |
| B161-CK-3 | 88,593,188 | 87,229,950 | 94.81 | 43.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Chen, B.; Cheng, C.; Fu, B.; Qi, M.; Du, H.; Geng, S.; Zhang, X. Comparative Transcriptomics Analysis Reveals the Differences in Transcription between Resistant and Susceptible Pepper (Capsicum annuum L.) Varieties in Response to Anthracnose. Plants 2024, 13, 527. https://doi.org/10.3390/plants13040527
Wang Y, Chen B, Cheng C, Fu B, Qi M, Du H, Geng S, Zhang X. Comparative Transcriptomics Analysis Reveals the Differences in Transcription between Resistant and Susceptible Pepper (Capsicum annuum L.) Varieties in Response to Anthracnose. Plants. 2024; 13(4):527. https://doi.org/10.3390/plants13040527
Chicago/Turabian StyleWang, Yixin, Bin Chen, Chunyuan Cheng, Bingkun Fu, Meixia Qi, Heshan Du, Sansheng Geng, and Xiaofen Zhang. 2024. "Comparative Transcriptomics Analysis Reveals the Differences in Transcription between Resistant and Susceptible Pepper (Capsicum annuum L.) Varieties in Response to Anthracnose" Plants 13, no. 4: 527. https://doi.org/10.3390/plants13040527
APA StyleWang, Y., Chen, B., Cheng, C., Fu, B., Qi, M., Du, H., Geng, S., & Zhang, X. (2024). Comparative Transcriptomics Analysis Reveals the Differences in Transcription between Resistant and Susceptible Pepper (Capsicum annuum L.) Varieties in Response to Anthracnose. Plants, 13(4), 527. https://doi.org/10.3390/plants13040527






