An Optimized and Cost-Effective RNA Extraction Method for Secondary Metabolite-Enriched Tissues of Norway Spruce (Picea abies)
Abstract
1. Introduction
2. Results and Discussion
3. Material and Methods
3.1. Plant Material and Sampling
3.2. Total RNA Extraction
3.2.1. RNeasy® Plant Mini Kit Protocol
3.2.2. Spectrum Plant Total RNA Kit Protocol
3.2.3. TRIzol Protocol
3.2.4. TRIzol-Column Hybrid Protocol
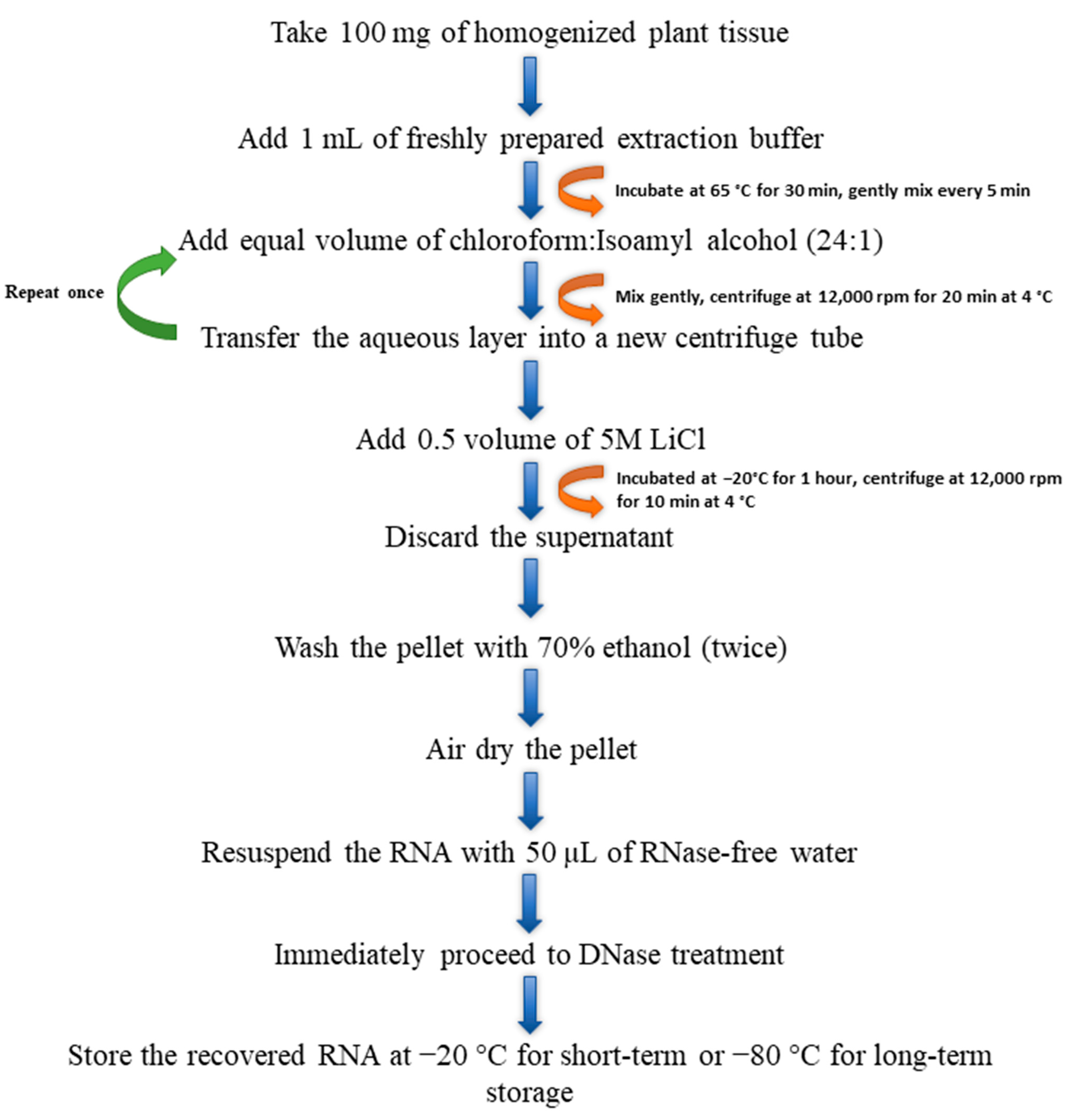
3.2.5. Modified CTAB Method
3.3. DNase Treatment and RNA Analysis
3.4. RNA Sequencing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caudullo, G.; Tinner, W.; De Rigo, D. Picea abies in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Publication Office of the European Union: Luxembourg, 2016; pp. 114–116. [Google Scholar]
- Jactel, H.; Nicoll, B.C.; Branco, M.; Gonzalez-Olabarria, J.R.; Grodzki, W.; Långström, B.; Moreira, F.; Netherer, S.; Orazio, C.; Piou, D.; et al. The influences of forest stand management on biotic and abiotic risks of damage. Ann. For. Sci. 2009, 66, 701. [Google Scholar] [CrossRef]
- Trubin, A.; Kozhoridze, G.; Zabihi, K.; Modlinger, R.; Singh, V.V.; Surový, P.; Jakuš, R. Detection of susceptible Norway spruce to bark beetle attack using PlanetScope multispectral imagery. Front. For. Glob. Chang. 2023, 6, 1130721. [Google Scholar] [CrossRef]
- Zabihi, K.; Singh, V.V.; Trubin, A.; Tomášková, I.; Blaženec, M.; Surový, P.; Jakuš, R. Sap flow as a function of variables within nested scales: Ordinary least squares vs. spatial regression models. Environ. Res. Ecol. 2023, 2, 025002. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Sun, H.; Han, X.; Peng, Y.; Liu, J.; Liu, K.; Ding, Y.; Wang, C.; Du, B. Transcriptome profiles reveal the growth-promoting mechanism of Paenibacillus polymyxa YC0136 on tobacco (Nicotiana tabacum L.). Front. Microbiol. 2020, 11, 584174. [Google Scholar] [CrossRef]
- Yan, W.J.; Pendi, F.H.; Hussain, H. Improved CTAB method for RNA extraction of thick waxy leaf tissues from sago palm (Metroxylon sagu Rottb.). Chem. Biol. Technol. Agric. 2022, 9, 63. [Google Scholar] [CrossRef]
- Ghawana, S.; Paul, A.; Kumar, H.; Kumar, A.; Singh, H.; Bhardwaj, P.K.; Rani, A.; Singh, R.S.; Raizada, J.; Singh, K.; et al. An RNA isolation system for plant tissues rich in secondary metabolites. BMC Res. Notes 2011, 4, 85. [Google Scholar] [CrossRef] [PubMed]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Agarwal, R.; Furtado, A.; Henry, R.J.; Mitter, N. A comprehensive high-quality DNA and RNA extraction protocol for a range of cultivars and tissue types of the woody crop avocado. Plants 2022, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Gudenschwager, O.; González-Agüero, M.; Defilippi, B.G. A general method for high-quality RNA isolation from metabolite-rich fruits. S. Afr. J. Bot. 2012, 83, 186–192. [Google Scholar] [CrossRef]
- Huang, H.H.; Xu, L.L.; Tong, Z.K.; Lin, E.P.; Liu, Q.P.; Cheng, L.J.; Zhu, M.Y. De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genom. 2012, 13, 648. [Google Scholar] [CrossRef] [PubMed]
- Morante-Carriel, J.; Sellés-Marchart, S.; Martínez-Márquez, A.; Martínez-Esteso, M.J.; Luque, I.; Bru-Martínez, R. RNA isolation from loquat and other recalcitrant woody plants with high quality and yield. Anal. Biochem. 2014, 452, 46–53. [Google Scholar] [CrossRef]
- Wang, S.X.; Hunter, W.; Plant, A. Isolation and purification of functional total RNA from woody branches and needles of Sitka and white spruce. Biotechniques 2000, 28, 292–296. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Sasi, S.; Krishnan, S.; Kodackattumannil, P.; Shamisi, A.A.; Aldarmaki, M.; Lekshmi, G.; Kottackal, M.; Amiri, K.M. DNA-free high-quality RNA extraction from 39 difficult-to-extract plant species (representing seasonal tissues and tissue types) of 32 families, and its validation for downstream molecular applications. Plant Methods 2023, 19, 84. [Google Scholar] [CrossRef]
- Johnson, M.T.J.; Carpenter, E.J.; Tian, Z.; Bruskiewich, R.; Burris, J.N.; Carrigan, C.T.; Chase, M.W.; Clarke, N.D.; Covshoff, S.; Depamphilis, C.W.; et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE 2012, 7, e50226. [Google Scholar] [CrossRef]
- Lee, S.; Moon, J.S.; Ko, T.S.; Petros, D.; Goldsbrough, P.B.; Korban, S.S. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 2003, 131, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.G.; Honda, C.; Kita, M.; Zhang, Z.; Tsuda, T.; Moriguchi, T. A simple protocol for RNA isolation from fruit trees containing high levels of polysaccharides and polyphenol compounds. Plant Mol. Biol. Rep. 2002, 20, 69a–69g. [Google Scholar] [CrossRef]
- Kolosova, N.; Miller, B.; Ralph, S.; Ellis, B.E.; Douglas, C.; Ritland, K.; Bohlmann, J. Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechniques 2004, 36, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Meisel, L.; Fonseca, B.; González, S.; Baeza-Yates, R.; Cambiazo, V.; Campos, R.; Gonzalez, M.; Orellana, A.; Retamales, J.; Silva, H. A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biol. Res. 2005, 38, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.; Trivedi, P.; Solomos, T.; Tucker, M. Isolation of high-quality RNA from apple (Malus domestica) fruit. J. Agric. Food Chem. 2006, 54, 5227–5229. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A. “RNAprep-Trizol combined with Columns” Untergasser’s Lab. Winter 2008. Available online: https://www.untergasser.de/lab/protocols/rna_prep_comb_trizol_v1_0.htm (accessed on 1 August 2023).
- Vennapusa, A.R.; Somayandal, C.J.; Jagadish, S.V.K. A universal method for high quality RNA extraction from plant tissues rich in starch, proteins and fiber. Sci. Rep. 2020, 10, 16887. [Google Scholar] [CrossRef]
- Ahmed, M.; Sarwar, M.B.; Ashfaq, R.; Ahmed, A.; Yanag, X.; ud Din, S.; Sajid, M.; Syed, Q.; Abidi, S.H.; Wang, X. Low-cost and highly efficient: A method for high-quality nucleic acid isolation from cotton fibres. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kiefer, E.; Heller, W.; Ernst, D. A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol. Biol. Rep. 2000, 18, 33–39. [Google Scholar] [CrossRef]
- Yockteng, R.; Almeida, A.M.; Yee, S.; Andre, T.; Hill, C.; Specht, C.D. A method for extracting high-quality RNA from diverse plants for next-generation sequencing and gene expression analyses. Appl. Plant Sci. 2013, 1, 1300070. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 11, 951–969. [Google Scholar] [CrossRef]
- Mitchell, N.; McAssey, E.V.; Hodel, R.G.J. Emerging methods in botanical DNA/RNA extraction. Appl. Plant Sci. 2023, 11, e11530. [Google Scholar] [CrossRef]
- Bhardwaj, K.; Silva, A.S.; Atanassova, M.; Sharma, R.; Nepovimova, E.; Musilek, K.; Sharma, R.; Alghuthaymi, M.A.; Dhanjal, D.S.; Nicoletti, M.; et al. Conifers Phytochemicals: A Valuable Forest with Therapeutic Potential. Molecules 2021, 26, 3005. [Google Scholar] [CrossRef] [PubMed]
- Seçgin, Z.; Gökdemir, G.; Atabay, E.S.; Kurt Kızıldoğan, A.; Kavas, M. Development of new total RNA isolation method for tissues with rich phenolic compounds. Turk. J. Biochem. 2020, 45, 343–350. [Google Scholar] [CrossRef]
- Carpinetti, P.D.A.; Fioresi, V.S.; Ignez da Cruz, T.; de Almeida, F.A.N.; Canal, D.; Ferreira, A.; Ferreira, M.F.D.S. Efficient method for isolation of high-quality RNA from Psidium guajava L. tissues. PLoS ONE 2021, 16, e0255245. [Google Scholar] [CrossRef]
- Xiao, H.; Kim, W.S.; Meng, B. A highly effective and versatile technology for the isolation of RNAs from grapevines and other woody perennials for use in virus diagnostics. Virol. J. 2015, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, E.A.; Ergul, A.; AlKayal, F.; DeLuc, L.; Cushman, J.C.; Cramer, G.R. Comparison of methods for isolating high-quality RNA from leaves of grapevine. Am. J. Enol. Vitic. 2005, 56, 400–406. [Google Scholar] [CrossRef]
- Lewinsohn, E.; Steele, C.L.; Croteau, R. Simple isolation of functional RNA from woody stems of gymnosperms. Plant Mol. Biol. Rep. 1994, 12, 20–25. [Google Scholar] [CrossRef]
- Nizam, A.; Kalath, H.; Kumar, A. A modified method for efficient RNA isolation from mangrove root tissues rich in secondary metabolites. BioTechniques 2023, 74, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Masoomi-Aladizgeh, F.; Jabbari, L.; Khayam Nekouei, R.; Aalami, A.; Atwell, B.J.; Haynes, P.A. A universal protocol for high-quality DNA and RNA isolation from diverse plant species. PLoS ONE 2023, 14, e0295852. [Google Scholar] [CrossRef]
- Schultz, D.J.; Craig, R.; Cox-Foster, D.L.; Mumma, R.O.; Medford, J.I. RNA isolation from recalcitrant plant tissue. Plant Mol. Biol. Rep. 1994, 12, 310–316. [Google Scholar] [CrossRef]
- Azevedo, H.; Lino-Neto, T.; Tavares, R.M. An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol. Biol. Rep. 2003, 21, 333–338. [Google Scholar] [CrossRef]
- Gautam, A. Lithium Chloride-Based Isolation of RNA. In DNA and RNA Isolation Techniques for Non-Experts; Techniques in Life Science and Biomedicine for the Non-Expert; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
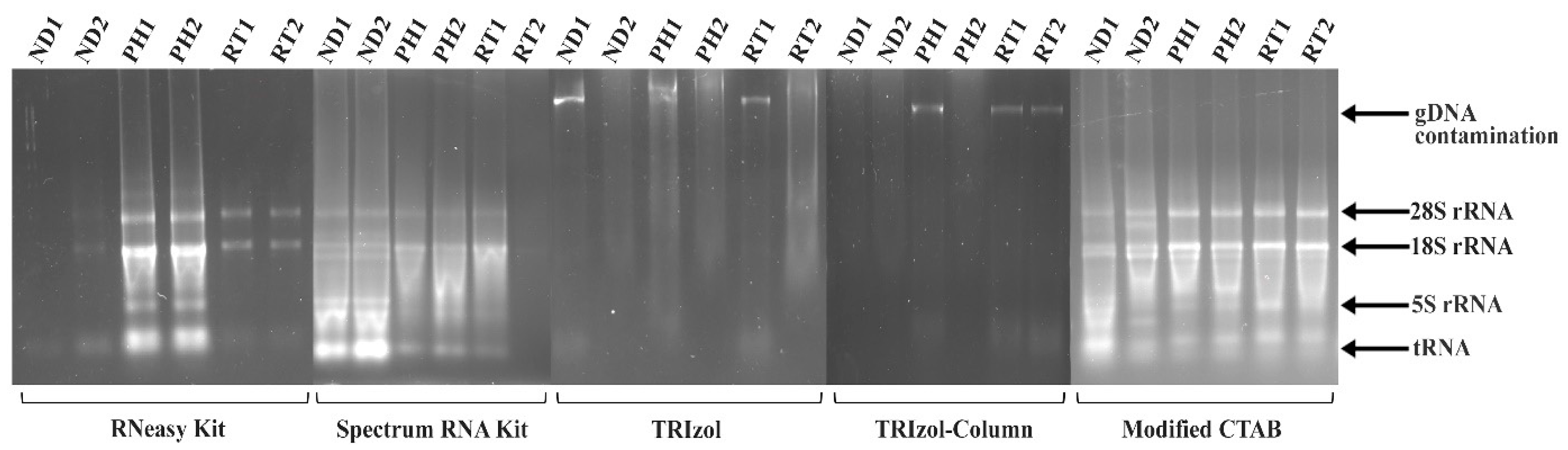
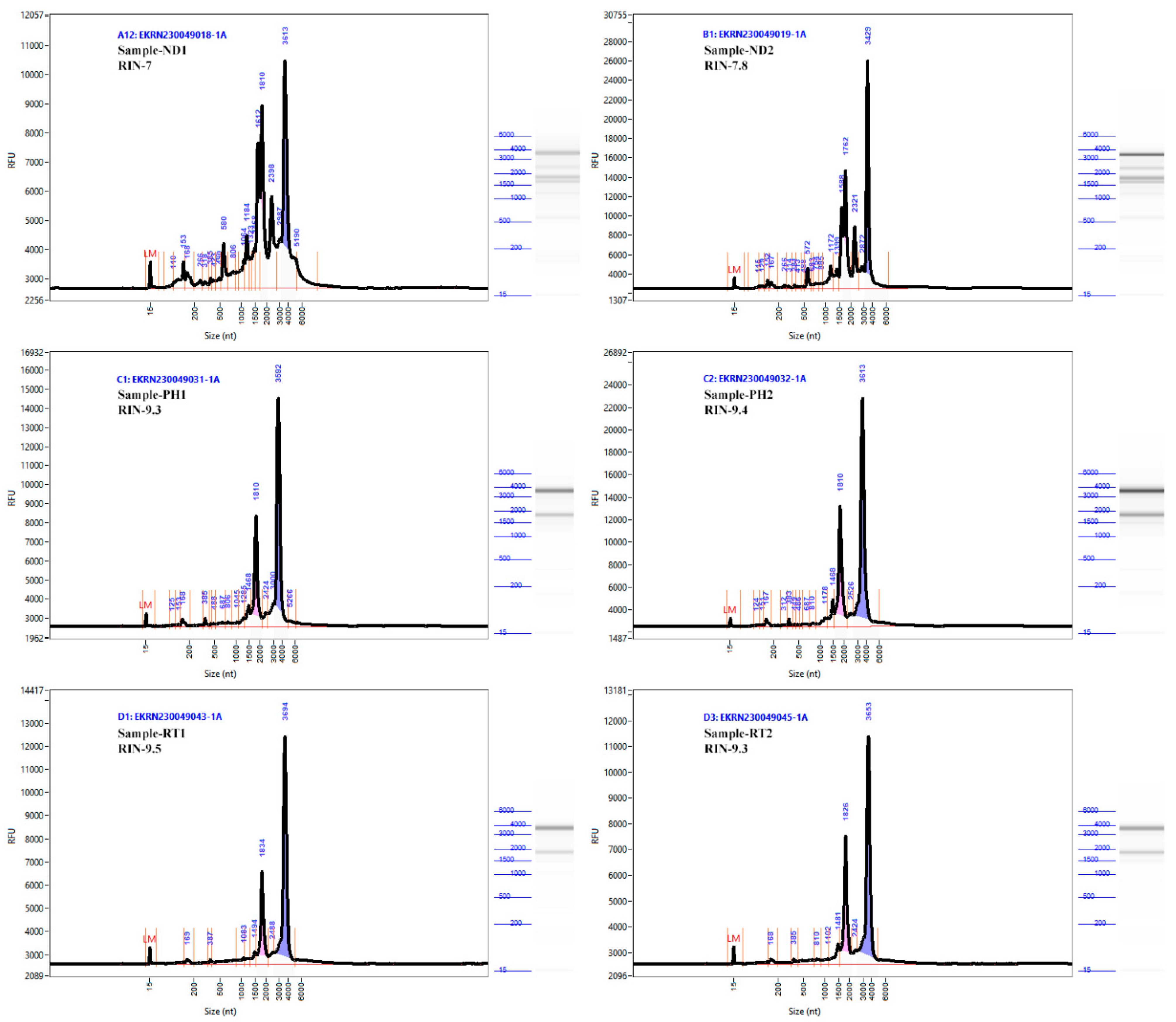
| Absorbance Ratio | ||||
|---|---|---|---|---|
| Method | Replicate | 260/280 | 260/230 | RNA Concentration (ng/µL) |
| RNeasy® Plant Mini Kit | ND1 | 1.69 | 0.02 | 4.2 |
| ND2 | 1.68 | 0.09 | 5.3 | |
| PH1 | 1.87 | 0.58 | 49.1 | |
| PH2 | 1.81 | 0.63 | 51.0 | |
| RT1 | 1.36 | 0.16 | 7.9 | |
| RT2 | 1.34 | 0.08 | 5.7 | |
| Spectrum Plant Total RNA Kit | ND1 | 1.90 | 0.94 | 154.5 |
| ND2 | 1.92 | 0.98 | 175.6 | |
| PH1 | 1.81 | 0.86 | 62.6 | |
| PH2 | 1.97 | 1.34 | 68.2 | |
| RT1 | 1.79 | 0.82 | 27.5 | |
| RT2 | 1.09 | 0.15 | 2.7 | |
| TRIzol | ND1 | 1.29 | 0.19 | 59.9 |
| ND2 | 1.13 | 0.16 | 83.4 | |
| PH1 | 0.74 | 0.28 | 173.0 | |
| PH2 | 0.92 | 0.20 | 146.4 | |
| RT1 | 1.29 | 0.32 | 395.2 | |
| RT2 | 1.26 | 0.24 | 253.8 | |
| TRIzol-column hybrid | ND1 | 1.34 | 0.35 | 9.8 |
| ND2 | 1.33 | 0.40 | 7.9 | |
| PH1 | 1.01 | 0.21 | 10.9 | |
| PH2 | 1.15 | 0.29 | 13.8 | |
| RT1 | 1.27 | 0.43 | 13.7 | |
| RT2 | 1.26 | 0.45 | 13.1 | |
| Modified CTAB | ND1 | 2.05 | 1.98 | 476.5 |
| ND2 | 2.06 | 2.16 | 578.7 | |
| PH1 | 2.05 | 2.04 | 418.9 | |
| PH2 | 2.05 | 1.99 | 685.1 | |
| RT1 | 2.03 | 1.83 | 308.8 | |
| RT2 | 2.05 | 1.88 | 347.8 | |
| Sample | Raw Reads (Gb) | Effective (%) | Error (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| ND1 | 146,415,518 | 98.85 | 0.02 | 98.41 | 95.27 | 46.39 |
| ND2 | 122,608,254 | 99.22 | 0.02 | 98.14 | 94.49 | 45.92 |
| PH1 | 142,090,562 | 98.45 | 0.02 | 98.26 | 94.90 | 45.36 |
| PH2 | 147,244,540 | 98.34 | 0.02 | 98.35 | 95.15 | 45.12 |
| RT1 | 141,041,240 | 98.49 | 0.02 | 98.35 | 95.06 | 45.24 |
| RT2 | 149,003,516 | 99.09 | 0.02 | 98.35 | 95.13 | 45.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, V.V.; Naseer, A.; Sellamuthu, G.; Jakuš, R. An Optimized and Cost-Effective RNA Extraction Method for Secondary Metabolite-Enriched Tissues of Norway Spruce (Picea abies). Plants 2024, 13, 389. https://doi.org/10.3390/plants13030389
Singh VV, Naseer A, Sellamuthu G, Jakuš R. An Optimized and Cost-Effective RNA Extraction Method for Secondary Metabolite-Enriched Tissues of Norway Spruce (Picea abies). Plants. 2024; 13(3):389. https://doi.org/10.3390/plants13030389
Chicago/Turabian StyleSingh, Vivek Vikram, Aisha Naseer, Gothandapani Sellamuthu, and Rastislav Jakuš. 2024. "An Optimized and Cost-Effective RNA Extraction Method for Secondary Metabolite-Enriched Tissues of Norway Spruce (Picea abies)" Plants 13, no. 3: 389. https://doi.org/10.3390/plants13030389
APA StyleSingh, V. V., Naseer, A., Sellamuthu, G., & Jakuš, R. (2024). An Optimized and Cost-Effective RNA Extraction Method for Secondary Metabolite-Enriched Tissues of Norway Spruce (Picea abies). Plants, 13(3), 389. https://doi.org/10.3390/plants13030389










