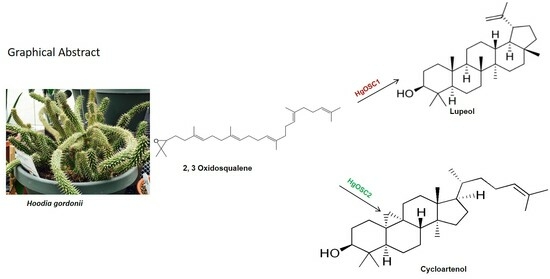
Identification and Functional Characterization of Oxidosqualene Cyclases from Medicinal Plant Hoodia gordonii
Abstract
1. Introduction
2. Results
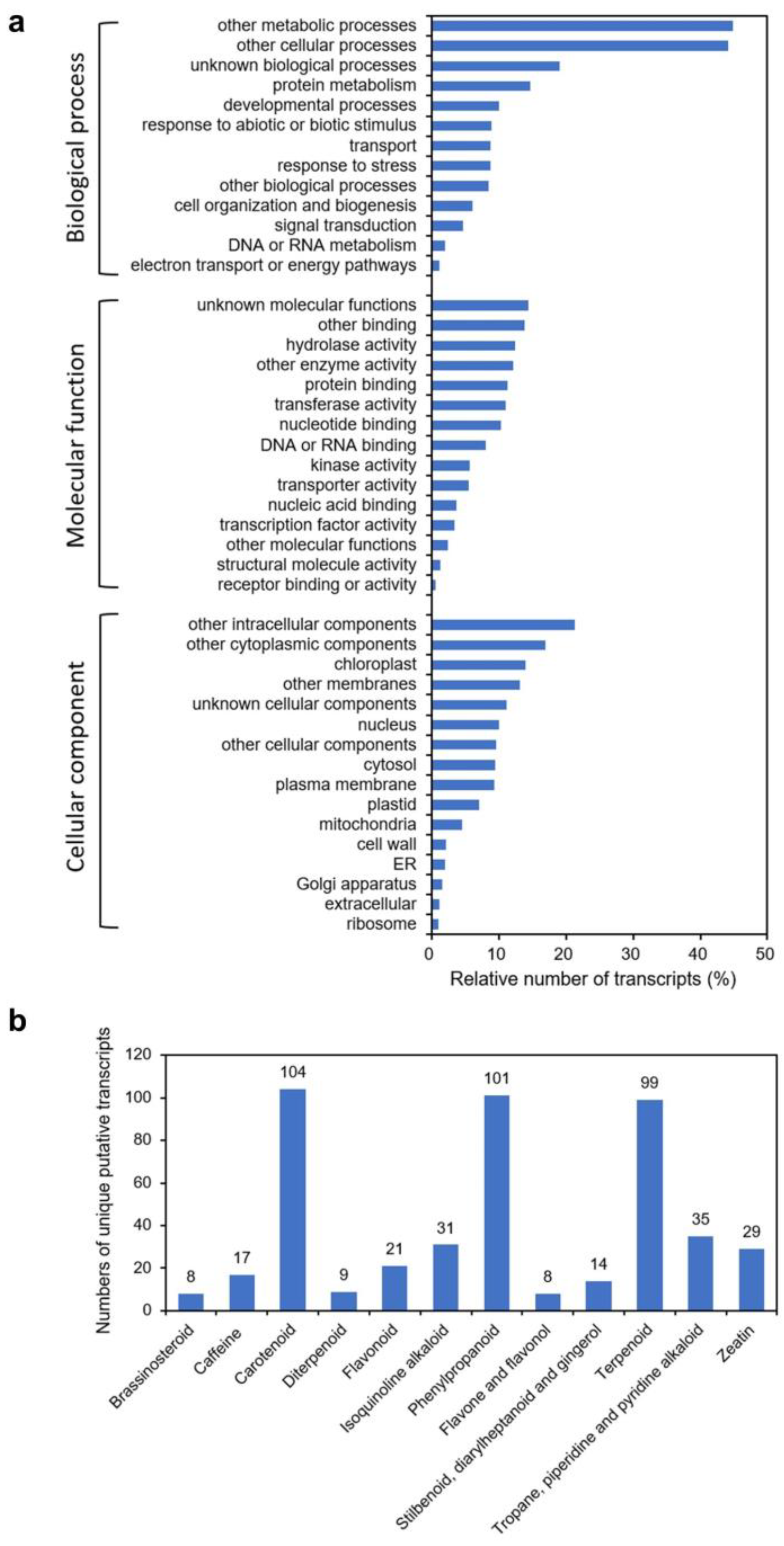
2.1. Isolation and Identification of cDNA Clones for Triterpene Synthases
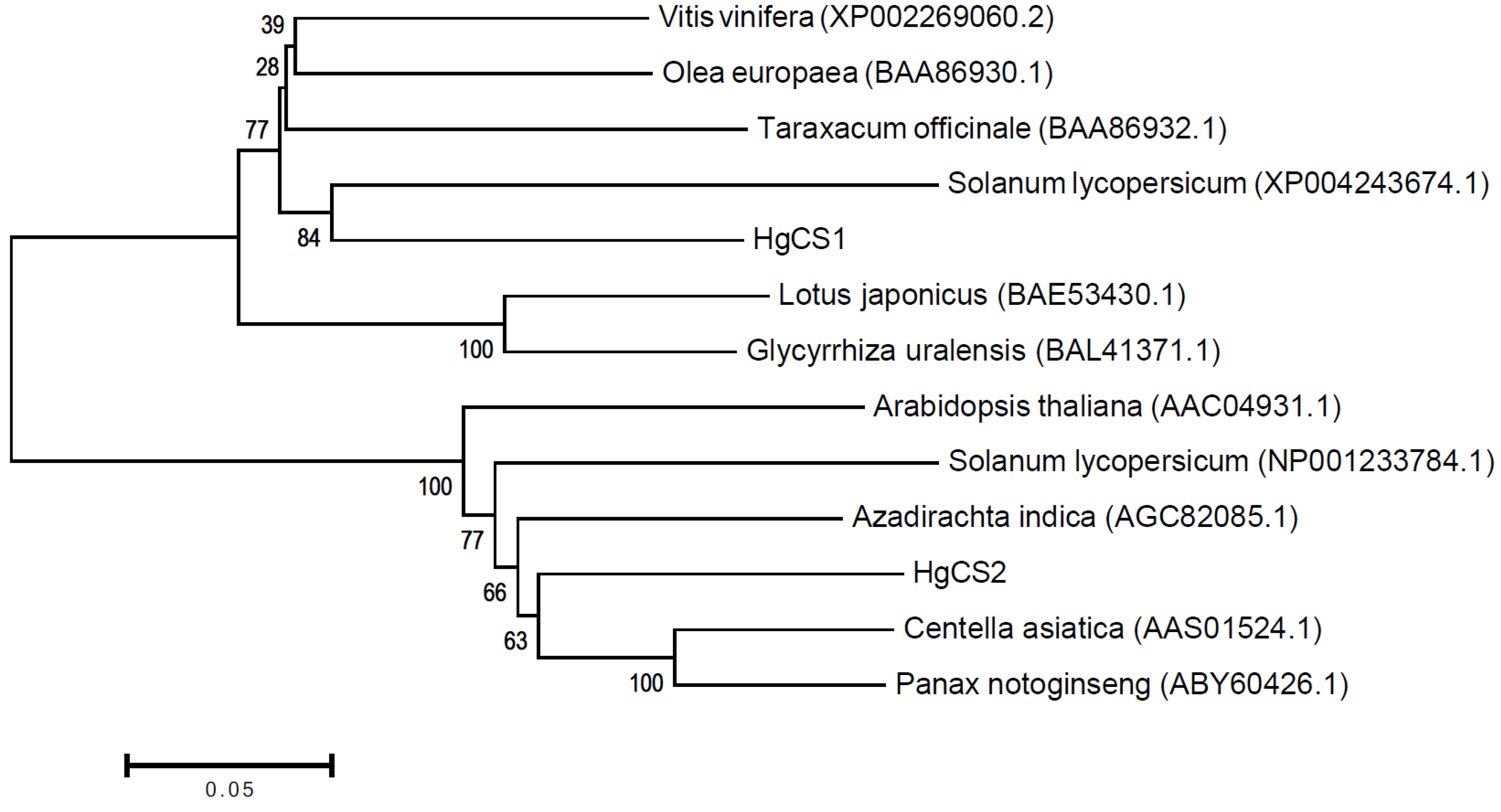
2.2. In Silico Characterization and Phylogenetic Analysis
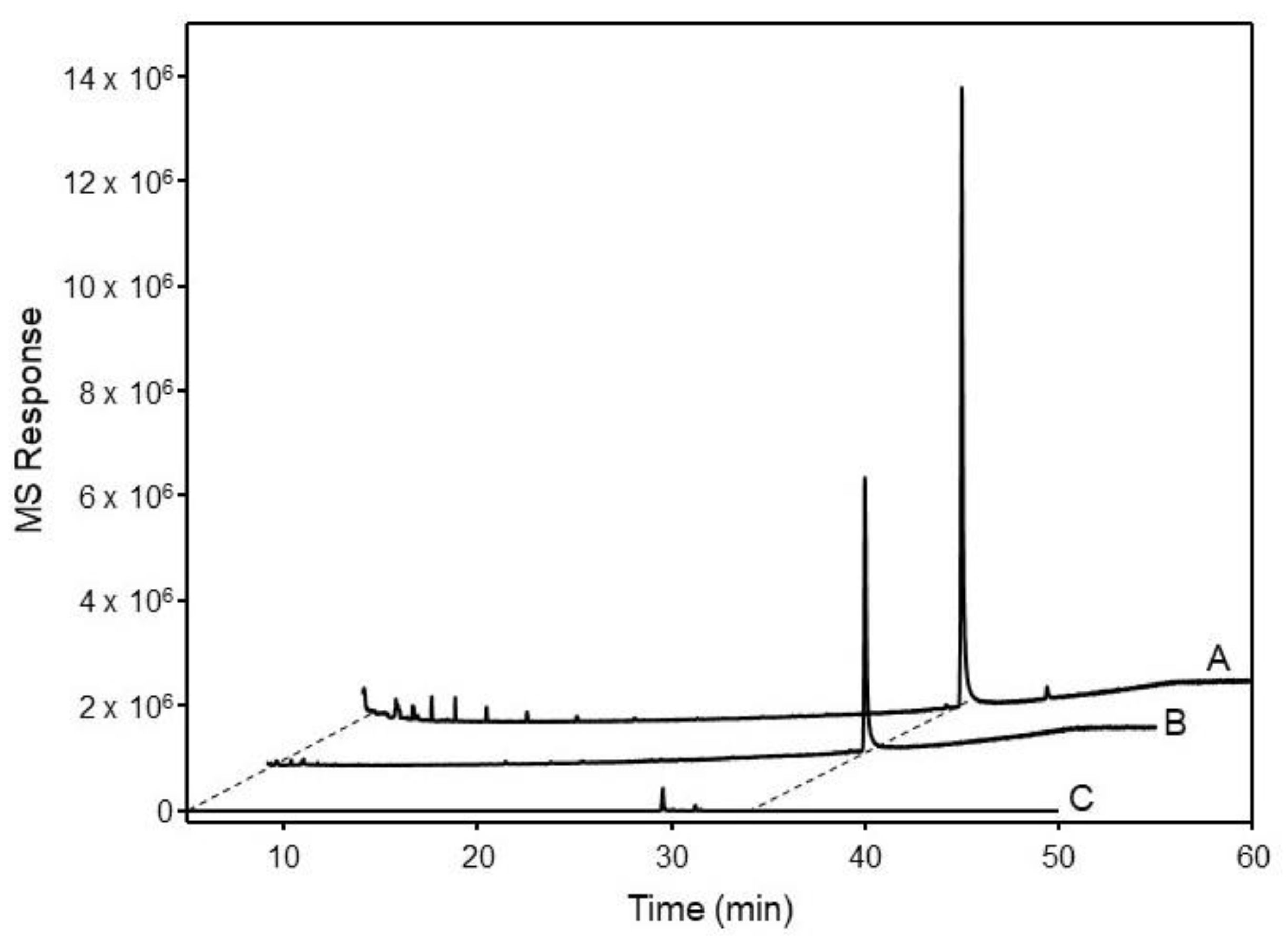
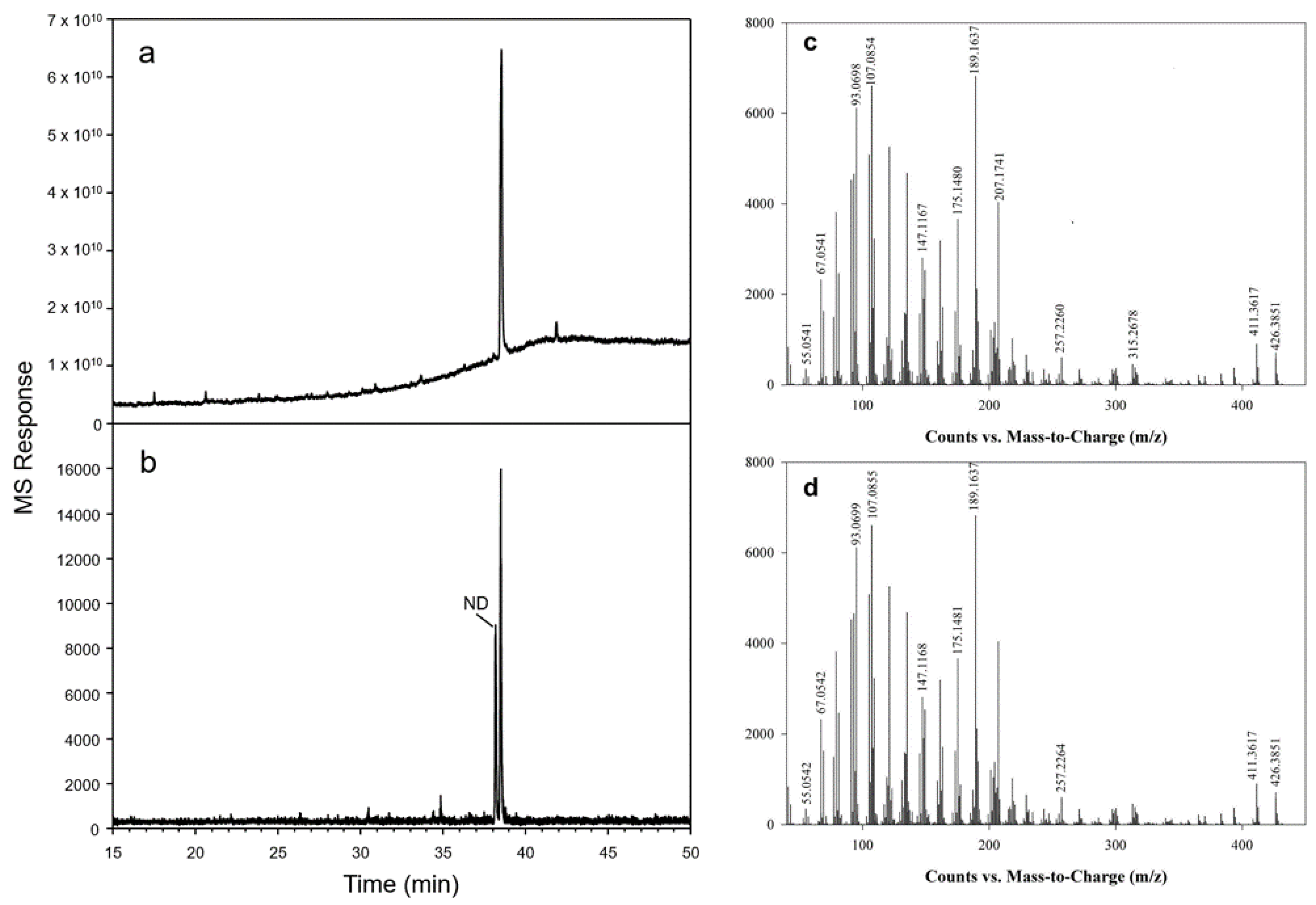
2.3. Functional Characterization of OSCs in Yeast
2.4. Yeast Spotting Assay
2.5. Functional Expression of HgOSC1 and HgOSC2 in Nicotiana benthamiana
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Isolation of RNA
4.3. Functional Annotation of the Transcriptome Dataset and Pathway Analyses
4.4. Isolation of cDNA Clones for Terpene Synthases
4.5. Phylogenetic Analysis
4.6. Expression of OSCs in Saccharomyces cerevisiae
4.7. GC-MS Analysis
4.8. TLC and NMR Analysis of HgOSC1 Products
4.9. Yeast Spotting Assay
4.10. Transient Expression of Hoodia OSCs in Nicotiana benthamiana
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Papadopoulou, K.; Melton, R.E.; Leggett, M.; Daniels, M.J.; Osbourn, A.E. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 1999, 96, 12923–12928. [Google Scholar] [CrossRef] [PubMed]
- Gas-Pascual, E.; Berna, A.; Bach, T.J.; Schaller, H. Plant oxidosqualene metabolism: Cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana. PLoS ONE 2014, 9, e109156. [Google Scholar] [CrossRef] [PubMed]
- Abe, I. The oxidosqualene cyclases: One substrate, diverse products. In Natural Products: Discourse, Diversity and Design; Osbourn, A., Goss, R.J., Carter, G.T., Eds.; Wiley Blackwell: Oxford, UK, 2014; pp. 297–316. [Google Scholar]
- Sandeep Misra, R.C.; Chanotiya, C.S.; Mukhopadhyay, P.; Ghosh, S. Oxidosqualene cyclase and CYP716 enzymes contribute to triterpene structural diversity in the medicinal tree banaba. New Phytol. 2019, 222, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.-Y.; Yin, Y.; Lei, T.; Shi, Y.-H.; Gao, W.; Zhang, X.-N.; Li, J. A cycloartenol synthase from the steroidal saponin biosynthesis pathway of Paris. polyphylla. J. Asian Nat. Prod. Res. 2021, 23, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Pang, Y.; Chen, Q.; Li, C.; Lu, B. Oxidosqualene cyclases in triterpenoids biosynthesis: A review. Chin. J. Biotechnol. 2022, 38, 443–459. [Google Scholar]
- Kushiro, T.; Shibuya, M.; Ebizuka, Y. Beta-amyrin synthase—Cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur. J. Biochem. 1998, 256, 238–244. [Google Scholar] [CrossRef]
- Wang, Z.; Yeats, T.; Han, H.; Jetter, R. Cloning and characterization of oxidosqualene cyclases from Kalanchoe daigremontiana: Enzymes catalyzing up to 10 rearrangement steps yielding friedelin and other triterpenoids. J. Biol. Chem. 2010, 285, 29703–29712. [Google Scholar] [CrossRef]
- Kim, O.T.; Lee, J.W.; Bang, K.H.; Kim, Y.C.; Hyun, D.Y.; Cha, S.W.; Choi, Y.E.; Jin, M.L.; Hwang, B. Characterization of a dammarenediol synthase in Centella asiatica (L.) Urban. Plant Physiol. Biochem. 2009, 47, 998–1002. [Google Scholar] [CrossRef]
- Shibuya, M.; Katsube, Y.; Otsuka, M.; Zhang, H.; Tansakul, P.; Xiang, T.; Ebizuka, Y. Identification of a product specific beta-amyrin synthase from Arabidopsis thaliana. Plant Physiol. Biochem. 2009, 47, 26–30. [Google Scholar] [CrossRef]
- Sawai, S.; Uchiyama, H.; Mizuno, S.; Aoki, T.; Akashi, T.; Ayabe, S.I.; Takahashi, T. Molecular characterization of an oxidosqualene cyclase that yields shionone, a unique tetracyclic triterpene ketone of Aster tataricus. FEBS Lett. 2011, 585, 1031–1036. [Google Scholar] [CrossRef]
- Wang, Z.; Guhling, O.; Yao, R.; Li, F.; Yeats, T.H.; Rose, J.K.; Jetter, R. Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol. 2011, 155, 540–552. [Google Scholar] [CrossRef]
- Dhar, N.; Rana, S.; Razdan, S.; Bhat, W.W.; Hussain, A.; Dhar, R.S.; Vaishnavi, S.; Hamid, A.; Vishwakarma, R.; Lattoo, S.K. Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) dunal. J. Biol. Chem. 2014, 289, 17249–17267. [Google Scholar] [CrossRef]
- Russell, P.J.; Swindells, C. Chemical characterisation of Hoodia gordonii extract. Food Chem. Toxicol. 2012, 50, S6–S13. [Google Scholar] [CrossRef] [PubMed]
- Poralla, K.; Hewelt, A.; Prestwich, G.D.; Abe, I.; Reipen, I.; Sprenger, G. A specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem. Sci. 2012, 19, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, S.; Tseng, Y.; Lee, Y.; Chu, F. Cloning and characterization of a 2,3-oxidosqualene cyclase from Eleutherococcus trifoliatus. Holzforschung 2012, 67, 463–471. [Google Scholar] [CrossRef]
- Ladhari, A.; Chappell, J. Unravelling triterpene biosynthesis through functional characterization of an oxidosqualene cyclase (OSC) from Cleome arabica L. Plant Physiol. Biochem. 2019, 144, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Shibuya, M.; Lee, M.S.; Sankawa, U.; Ebizuka, Y. Molecular cloning of pea cDNA encoding cycloartenol synthase and its functional expression in yeast. Biol. Pharm. Bull. 1997, 20, 770–775. [Google Scholar] [CrossRef]
- Zhu, Z.; Liu, W.; Ge, L.; Yu, Q.; Zhao, Z.; Yang, J.; Wan, H. Molecular cloning and characterization of a cDNA encoding cycloartenol synthase from Fritillaria thunbergii Miq. Afr. J. Biotechnol. 2012, 11, 6896–6903. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [PubMed]
- Pan, Z.; Bajsa-Hirschel, J.; Vaughn, J.N.; Rimando, A.M.; Baerson, S.R.; Duke, S.O. In vivo assembly of the sorgoleone biosynthetic pathway and its impact on agroinfiltrated leaves of Nicotiana benthamiana. New Phytol. 2021, 230, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, L.; Toepfer, R. Development of plasmid vectors. In Bioengineering of Custom-Tailored Rape Varieties; Brauer, D., Roebbelen, G., Toepfer, R., Eds.; Gesellschaft fuer Pflanzenzuechtung: Goettingen, Germany, 1999; pp. 155–171. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parveen, I.; Wang, M.; Lee, J.; Zhao, J.; Zhu, Y.; Chittiboyina, A.G.; Khan, I.A.; Pan, Z. Identification and Functional Characterization of Oxidosqualene Cyclases from Medicinal Plant Hoodia gordonii. Plants 2024, 13, 231. https://doi.org/10.3390/plants13020231
Parveen I, Wang M, Lee J, Zhao J, Zhu Y, Chittiboyina AG, Khan IA, Pan Z. Identification and Functional Characterization of Oxidosqualene Cyclases from Medicinal Plant Hoodia gordonii. Plants. 2024; 13(2):231. https://doi.org/10.3390/plants13020231
Chicago/Turabian StyleParveen, Iffat, Mei Wang, Joseph Lee, Jianping Zhao, Yingjie Zhu, Amar G. Chittiboyina, Ikhlas A. Khan, and Zhiqiang Pan. 2024. "Identification and Functional Characterization of Oxidosqualene Cyclases from Medicinal Plant Hoodia gordonii" Plants 13, no. 2: 231. https://doi.org/10.3390/plants13020231
APA StyleParveen, I., Wang, M., Lee, J., Zhao, J., Zhu, Y., Chittiboyina, A. G., Khan, I. A., & Pan, Z. (2024). Identification and Functional Characterization of Oxidosqualene Cyclases from Medicinal Plant Hoodia gordonii. Plants, 13(2), 231. https://doi.org/10.3390/plants13020231