Exploring Genomics and Microbial Ecology: Analysis of Bidens pilosa L. Genetic Structure and Soil Microbiome Diversity by RAD-Seq and Metabarcoding
Abstract
1. Introduction
2. Results
2.1. Genomic Data and SNP Calling
2.2. Genetic Diversity Analysis
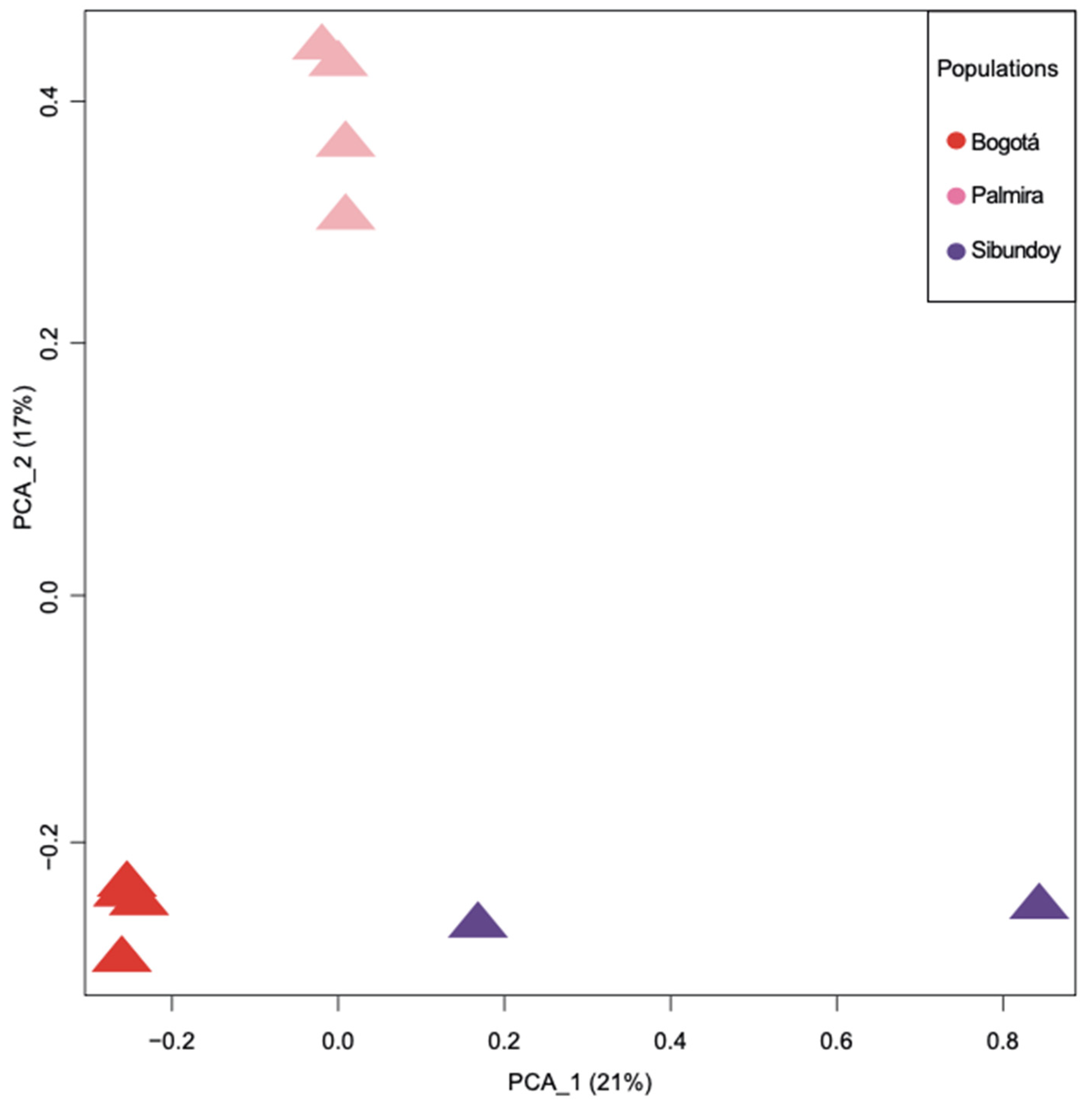
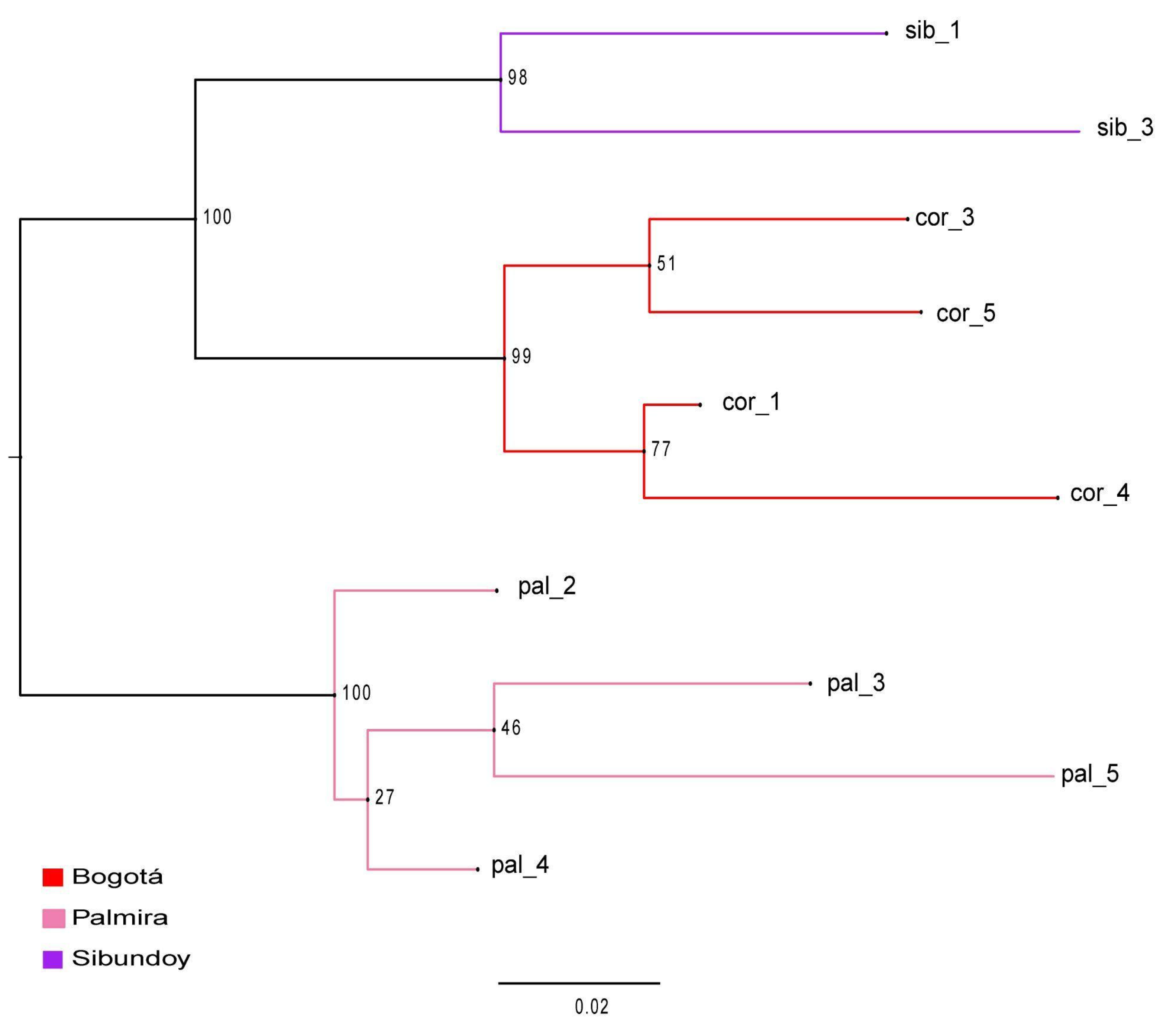
2.3. Population Structure
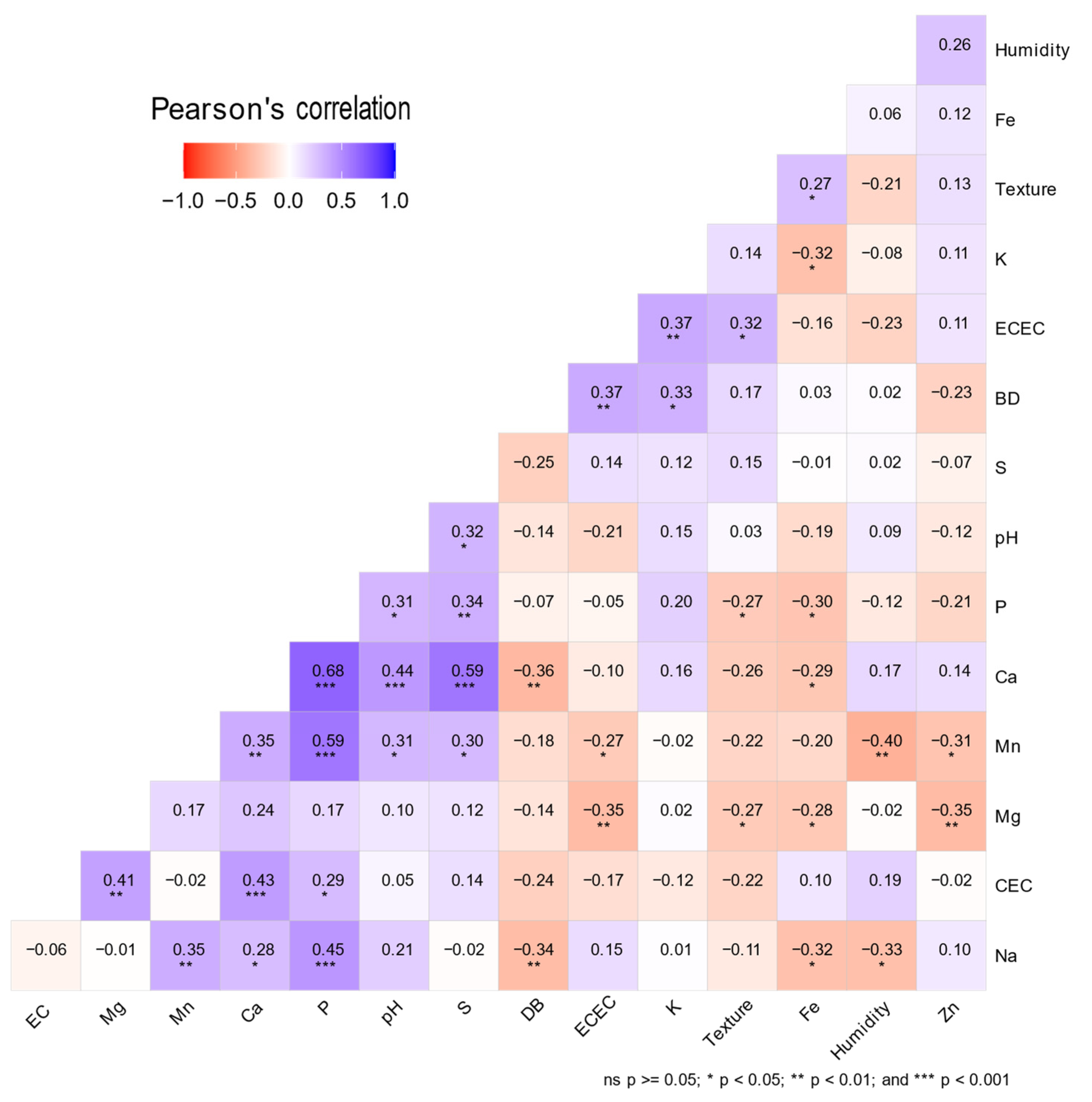
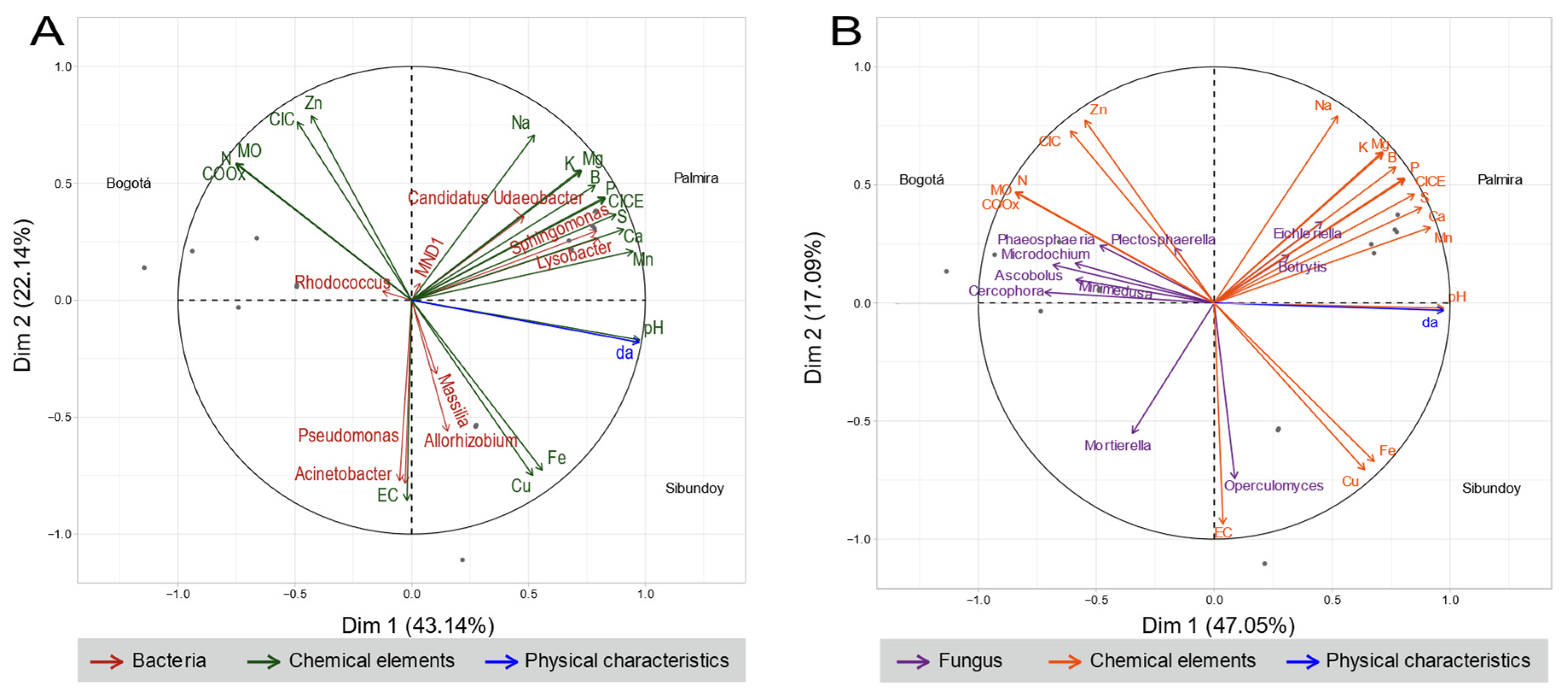
2.4. Physicochemical Analysis of Soils
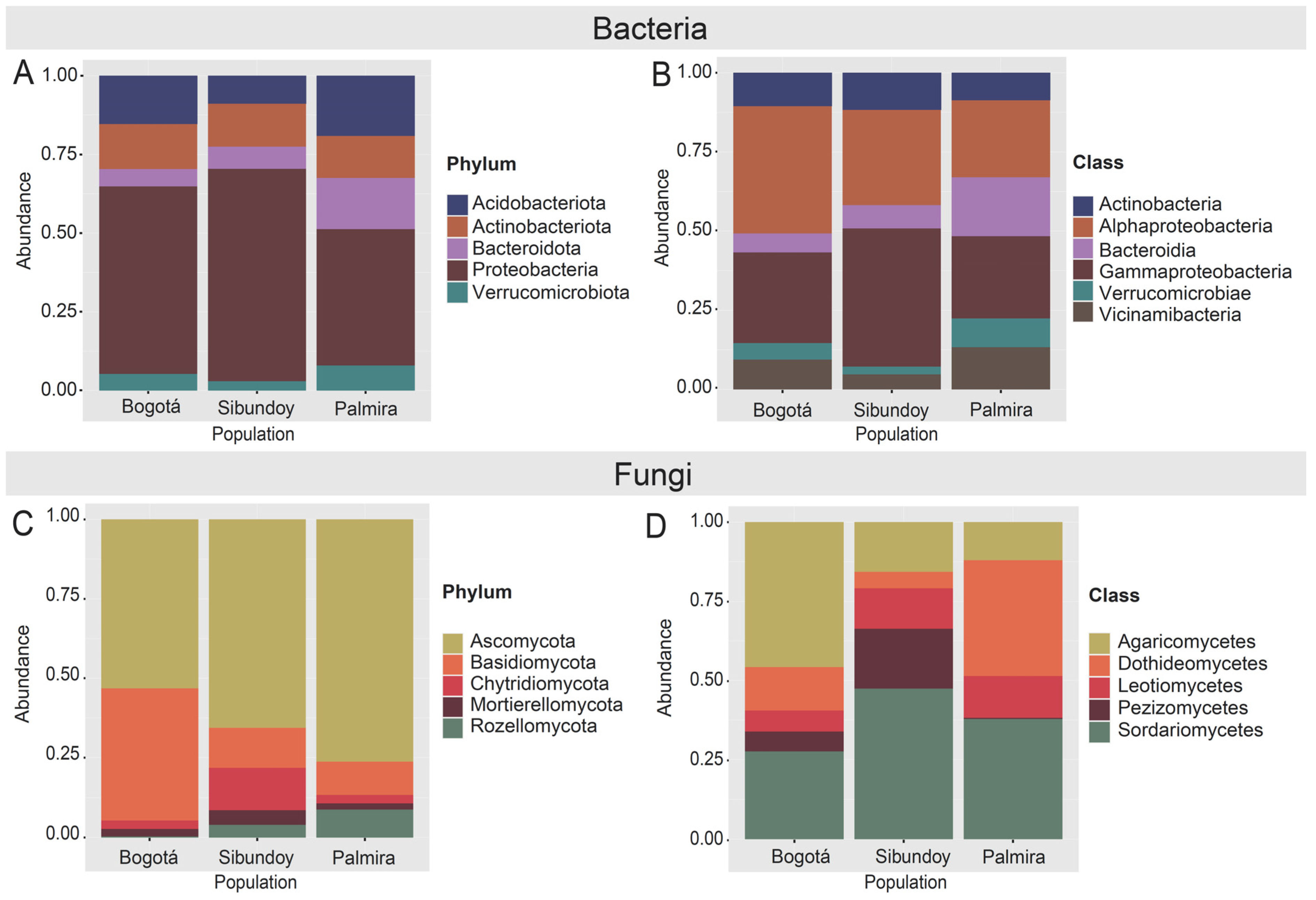
2.5. Microbiome Composition Analysis
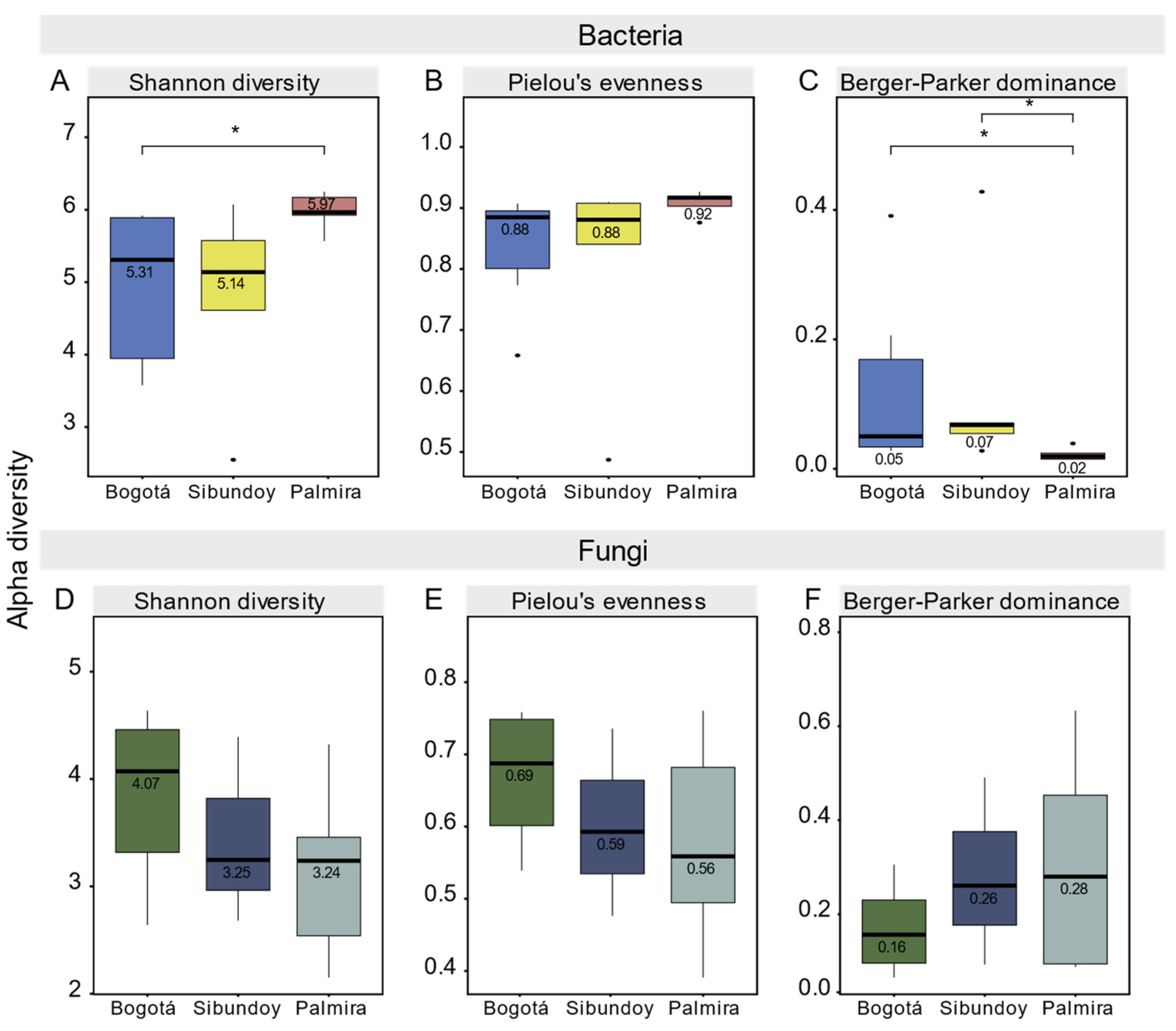
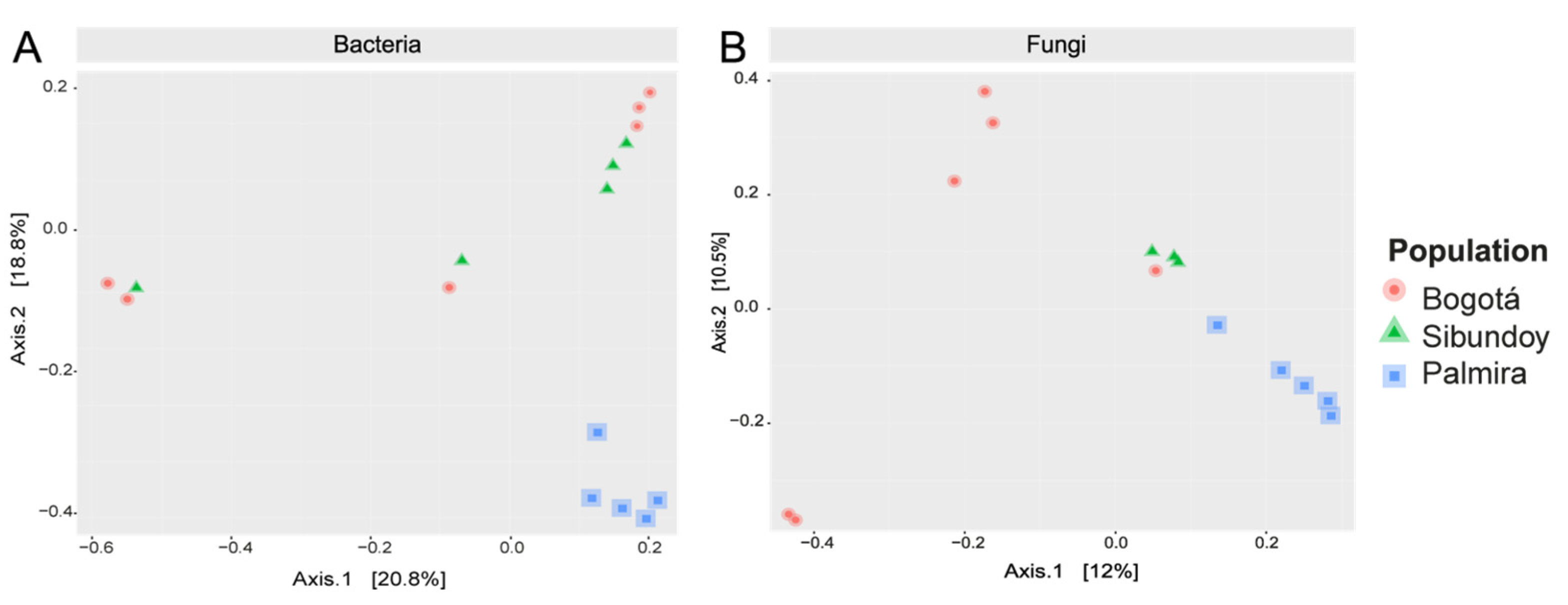
2.6. Alpha and Beta Diversity of Bacteria and Fungi
3. Discussion
4. Materials and Methods
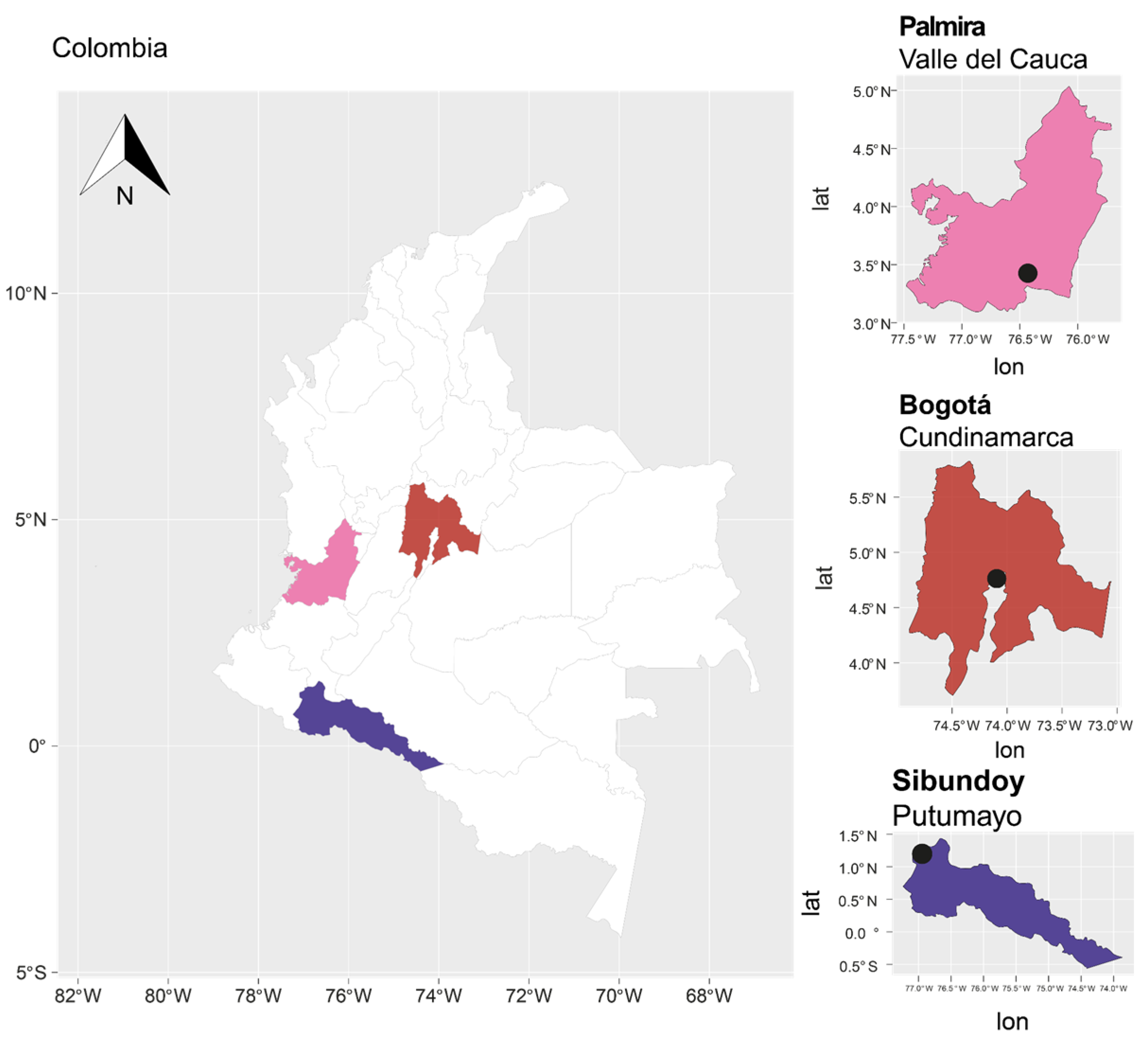
4.1. Collection of Material
4.2. DNA Extraction and Sequencing
4.3. Genetic Diversity and Population Structure of B. pilosa
4.4. Physicochemical and Microbial Sequence Analysis in Soils
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartolome, A.P.; Villaseñor, I.M.; Yang, W.-C. Bidens pilosa L. (Asteraceae): Botanical Properties, Traditional Uses, Phytochemistry, and Pharmacology. Evid. Based Complement. Alternat. Med. 2013, 2013, 340215. [Google Scholar] [CrossRef]
- Wahjudi, M.; Meira, G.; Santoso, H.; Irwansyah, A.Z. Bidens pilosa Linn.: Beautiful Weed for the Healthy Mouth—A Mini Review. E3S Web Conf. 2023, 374, 00023. [Google Scholar] [CrossRef]
- Wu, L.; Chen, H.; Chen, D.; Wang, S.; Wu, Y.; Wang, B.; Liu, S.; Yue, L.; Yu, J.; Bai, Y. Soil Biota Diversity and Plant Diversity Both Contributed to Ecosystem Stability in Grasslands. Ecol. Lett. 2023, 26, 858–868. [Google Scholar] [CrossRef]
- Xuan, T.D.; Khanh, T.D. Chemistry and Pharmacology of Bidens pilosa: An Overview. J. Pharm. Investig. 2016, 46, 91–132. [Google Scholar] [CrossRef]
- Li, Q.; Guo, J.; Zhang, H.; Zhao, M. The competition between Bidens pilosa and Setaria viridis alters soil microbial composition and soil ecological function. J. Integr. Agric. 2023; in press. [Google Scholar] [CrossRef]
- Dv, M.; As, R. A Review on the Potential of Underutilized Blackjack (Biden pilosa) Naturally Occurring in Sub-Saharan Africa. Heliyon 2022, 8, e09586. [Google Scholar] [CrossRef]
- Rodríguez-Mesa, X.M.; Contreras Bolaños, L.A.; Mejía, A.; Pombo, L.M.; Modesti Costa, G.; Santander González, S.P. Immunomodulatory Properties of Natural Extracts and Compounds Derived from Bidens pilosa L.: Literature Review. Pharmaceutics 2023, 15, 1491. [Google Scholar] [CrossRef]
- Yan, Z.; Chen, Z.; Zhang, L.; Wang, X.; Zhang, Y.; Tian, Z. Bioactive Polyacetylenes from Bidens pilosa L. and Their Anti-Inflammatory Activity. Nat. Prod. Res. 2022, 36, 6353–6358. [Google Scholar] [CrossRef]
- Dofuor, A.K.; Djameh, G.I.; Amoa-Bosompem, M.; Kwain, S.; Osei, E.; Tetevi, G.M.; Ayertey, F.; Bolah, P.; Okine, L.K.; Kyeremeh, K.; et al. In Vitro Effects and Mechanisms of Action of Bidens pilosa in Trypanosoma brucei. J. Tradit. Complement. Med. 2022, 12, 260–268. [Google Scholar] [CrossRef]
- Xie, J.; Lou, X.; Lu, Y.; Huang, H.; Yang, Q.; Zhang, Z.; Zhao, W.; Li, Z.; Liu, H.; Du, S.; et al. Suitable light combinations enhance cadmium accumulation in Bidens pilosa L. by regulating the soil microbial communities. Environ. Exp. Bot. 2023, 205, 105128. [Google Scholar] [CrossRef]
- Wei, H.; Yan, W.; Quan, G.; Zhang, J.; Liang, K. Soil Microbial Carbon Utilization, Enzyme Activities and Nutrient Availability Responses to Bidens pilosa and a Non-Invasive Congener under Different Irradiances. Sci. Rep. 2017, 7, 11309. [Google Scholar] [CrossRef]
- Lastra Valdés, H.A.; Ponce de León Rego, H. Bidens pilosa Linné. Rev. Cuba. De Plantas Med. 2001, 6, 28–33. [Google Scholar]
- Fuhrmann, J.J.; Zuberer, D.A. 13—Carbon Transformations and Soil Organic Matter Formation. In Principles and Applications of Soil Microbiology, 3rd ed.; Gentry, T.J., Fuhrmann, J.J., Zuberer, D.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 327–361. ISBN 978-0-12-820202-9. [Google Scholar]
- Singh, G.; Passsari, A.K.; Singh, P.; Leo, V.V.; Subbarayan, S.; Kumar, B.; Singh, B.P.; Lalhlenmawia, H.; Kumar, N.S. Pharmacological Potential of Bidens pilosa L. and Determination of Bioactive Compounds Using UHPLC-QqQLIT-MS/MS and GC/MS. BMC Complement. Altern. Med. 2017, 17, 492. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X.; Tan, W.; Ling, Q.; Pei, F.; Luo, S.; Qin, P.; Yuan, H.; Huang, L.; Yu, F. Metagenomics Combined with Metabolomics Reveals the Effect of Enterobacter sp. Inoculation on the Rhizosphere Microenvironment of Bidens pilosa L. in Heavy Metal Contaminated Soil. J. Hazard. Mater. 2023, 458, 132033. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.; Li, C.; Wu, C.; Chen, F.; Chen, X.; Zhang, F. Nitrate Nitrogen and pH Correlate with Changes in Rhizosphere Microbial Community Assemblages during Invasion of Ambrosia artemisiifolia and Bidens pilosa. Microbiol. Spectr. 2023, 11, e0364922. [Google Scholar] [CrossRef]
- Hartman, K.; Tringe, S.G. Interactions between Plants and Soil Shaping the Root Microbiome under Abiotic Stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef]
- Aqeel, M.; Ran, J.; Hu, W.; Irshad, M.K.; Dong, L.; Akram, M.A.; Eldesoky, G.E.; Aljuwayid, A.M.; Chuah, L.F.; Deng, J. Plant-Soil-Microbe Interactions in Maintaining Ecosystem Stability and Coordinated Turnover under Changing Environmental Conditions. Chemosphere 2023, 318, 137924. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends in Plant Science 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Rahman, L.; Shinwari, Z.K.; Iqrar, I.; Rahman, L.; Tanveer, F. An Assessment on the Role of Endophytic Microbes in the Therapeutic Potential of Fagonia Indica. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 53. [Google Scholar] [CrossRef]
- Daba, A.; Berecha, G.; Tadesse, M.; Belay, A. Evaluation of the Herbicidal Potential of Some Fungal Species against Bidens pilosa, the Coffee Farming Weeds. Saudi J. Biol. Sci. 2021, 28, 6408–6416. [Google Scholar] [CrossRef]
- Tang, S.; Xu, Y.; Zeng, K.; Liang, X.; Shi, X.; Liu, K.; Ma, J.; Yu, F.; Li, Y. Comparative Study on Plant Growth-Promoting Bacterial Inoculation by Irrigation and Spraying for Promoting Bidens pilosa L. Phytoremediation of Cadmium-Contaminated Soil. Ecotoxicol. Environ. Saf. 2023, 254, 114764. [Google Scholar] [CrossRef]
- Caujapé Castells, J.; Jaén Molina, R.; Cabrera García, N. El Banco de ADN de la flora canaria: Creación, progresos y líneas futuras de desarrollo. Botánica Macaronésica 2006, 26, 3–18. [Google Scholar]
- Wright, S. The Relation of Livestock Breeding to Theories of Evolution. J. Anim. Sci. 1978, 46, 1192–1200. [Google Scholar] [CrossRef]
- Vyšniauskienė, R.; Rančelienė, V.; Naugžemys, D.; Patamsytė, J.; Sinkevičienė, Z.; Butkuvienė, J.; Žvingila, D. Genetic Diversity of Populations of Bidens Genera Invasive and Native Species in Lithuania. Zemdirbyste-Agriculture 2018, 105, 183–190. [Google Scholar] [CrossRef]
- Calderón, J.; Alán, E.; Barrantes, U. Estructura, dimensiones y producción de propágulos sexuales de malezas del trópico húmedo. Agron. Mesoam. 2000, 11, 31–39. [Google Scholar] [CrossRef]
- Zhong, Z.; Wang, X.; Zhang, X.; Zhang, W.; Xu, Y.; Ren, C.; Han, X.; Yang, G. Edaphic Factors but Not Plant Characteristics Mainly Alter Soil Microbial Properties along a Restoration Chronosequence of Pinus tabulaeformis Stands on Mt. Ziwuling, China. For. Ecol. Manag. 2019, 453, 117625. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.; Régnière, J.; Vasseur, L.; Lin, J.; Huang, S.; Ke, F.; Chen, S.; Li, J.; Huang, J.; et al. Large-Scale Genome-Wide Study Reveals Climate Adaptive Variability in a Cosmopolitan Pest. Nat. Commun. 2021, 12, 7206. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Wu, X.; Jiang, M.; Liu, M.; Wang, B.; Yu, Q.; Chen, H.; Wang, L.; Liu, C. Analysis of the Complete Plastomes of Bidens pilosa L. 1753 (Asteraceae, Coreopsideae) from Beijing, China Reveals High Genetic Diversity and Possible Misidentifications. Mitochondrial DNA B Resour. 2023, 8, 612–618. [Google Scholar] [CrossRef]
- Miranda, J.M.C.; Mendoza, B.A.; Díaz-Soto, M.T. Actividad farmacológica y composición fitoquímica de Bidens pilosa L. Rev. Cuba. De Farm. 2021, 54, 4. [Google Scholar]
- Naylor, D.; McClure, R.; Jansson, J. Trends in Microbial Community Composition and Function by Soil Depth. Microorganisms 2022, 10, 540. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S rRNA and 16S rRNA Genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Nobarinezhad, M.H.; Wallace, L.E. Fine-Scale Genetic Structure in Rhizosphere Microbial Communities Associated with Chamaecrista fasciculata (Fabaceae). Ecol. Evol. 2023, 13, e10570. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative Metagenomic, Phylogenetic and Physiological Analyses of Soil Microbial Communities across Nitrogen Gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.; Lara-Gutiérrez, J.; Stocker, R. Environmental Fluctuations and Their Effects on Microbial Communities, Populations and Individuals. FEMS Microbiol. Rev. 2021, 45, fuaa068. [Google Scholar] [CrossRef] [PubMed]
- Vasco Palacios, A.M.; Franco Molano, A.E. Diversidad de Macrohongos Colombianos (Ascomycota—Basidiomycota). 2021. Available online: https://ipt.biodiversidad.co/sib/resource?r=udea_mhongos_literatura_001 (accessed on 30 November 2023).
- Gillman, G.P.; Sumpter, E.A. Modification to the Compulsive Exchange Method for Measuring Exchange Characteristics of Soils. Soil Res. 1986, 24, 61–66. [Google Scholar] [CrossRef]
- Cerón Rincón, L.E.; Ancízar Aristizábal Gutiérrez, F. Dinámica del ciclo del nitrógeno y fósforo en suelos. Rev. Colomb. Biotecnol. 2012, 14, 285–295. [Google Scholar]
- Braga, R.M.; Dourado, M.N.; Araújo, W.L. Microbial Interactions: Ecology in a Molecular Perspective. Braz. J. Microbiol. 2016, 47, 86–98. [Google Scholar] [CrossRef]
- Monteiro LC, P.; de Matos CD, C.; de Oliveira Cândido, A.; de Oliveira Mendes, T.A.; Santana, M.F.; Costa, M.D. Changes in Rhizosphere Microbial Diversity and Composition Due to NaCl Addition to the Soil Modify the Outcome of Maize-Weed Interactions. Appl. Soil Ecol. 2021, 159, 103818. [Google Scholar] [CrossRef]
- Capote, S.; Orta, R.; Pérez, E. Estrategia de Reproducción de Una Mala Hierba: Bidens pilosa L. Rev. Jardín Botánico Nac. 1986, 7, 73–79. [Google Scholar]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil Origin and Plant Genotype Structure Distinct Microbiome Compartments in the Model Legume Medicago Truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. Isolation of Plant DNA from Faesh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Rochette, N.C.; Catchen, J.M. Deriving Genotypes from RAD-Seq Short-Read Data Using Stacks. Nat. Protoc. 2017, 12, 2640–2659. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An Analysis Tool Set for Population Genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and Genotyping Loci de Novo from Short-Read Sequences. G3 2011, 1, 171–182. [Google Scholar] [CrossRef]
- Rochette, N.C.; Rivera-Colón, A.G.; Catchen, J.M. Stacks 2: Analytical Methods for Paired-End Sequencing Improve RADseq-Based Population Genomics. Mol. Ecol. 2019, 28, 4737–4754. [Google Scholar] [CrossRef]
- Maruki, T.; Lynch, M. Genotype Calling from Population-Genomic Sequencing Data. G3 2017, 7, 1393–1404. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Liu, J.-X. StructureSelector: A Web-Based Software to Select and Visualize the Optimal Number of Clusters Using Multiple Methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z.; Gong, Z.; Liang, Y.; Ai, X.; Sang, Z.; Guo, J.; Li, X.; Zheng, J. Analysis of the Genetic Structure and Diversity of Upland Cotton Groups in Different Planting Areas Based on SNP Markers. Gene 2022, 809, 146042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cartabia, A.; Garcés-Ruiz, M.; Herent, M.-F.; Quetin-Leclercq, J.; Ortiz, S.; Declerck, S.; Lalaymia, I. Arbuscular Mycorrhizal Fungi Impact the Production of Alkannin/Shikonin and Their Derivatives in Alkanna Tinctoria Tausch. Grown in Semi-Hydroponic and Pot Cultivation Systems. Front. Microbiol. 2023, 14, 1216029. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Du, W.; Tang, R.; Liu, Y.; Zou, X.; Yuan, D.; Wang, Z.; Liu, J.; Guo, J.; Yang, X.; et al. Genomic History and Forensic Characteristics of Sherpa Highlanders on the Tibetan Plateau Inferred from High-Resolution InDel Panel and Genome-Wide SNPs. Forensic. Sci. Int. Genet. 2022, 56, 102633. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 30 November 2023).
- Wickham, H. Ggplot2; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]

| Population | Variant | Polymorphic Sites | Private Alleles |
|---|---|---|---|
| Bogotá | 181,987 | 121,310 | 45,318 |
| Sibundoy | 115,553 | 54,323 | 30,557 |
| Palmira | 220,679 | 184,661 | 39,856 |
| Pop | Usable Loci | Ts | Tv | Polymorphic Locus | Na | SNPs | Nucleotide Composition % | π | Ho | He | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T | A | G | ||||||||||
| Bogotá | 2407 | 722 | 389 | 121,310 | 45,318 | 1111 | 25.33 | 25.14 | 25.13 | 24.40 | 0.361 +/− 0.236 | 0.674 | 0.574 |
| Sibundoy | 1837 | 632 | 336 | 54,323 | 30,557 | 968 | 25.94 | 25.22 | 24.77 | 24.07 | 0.422 +/− 0.276 | 0.551 | 0.561 |
| Palmira | 1597 | 655 | 391 | 184,661 | 39,856 | 1406 | 25.83 | 25.28 | 24.47 | 24.42 | 0.420 +/− 0.229 | 0.512 | 0.444 |
| Source of Variation | df. | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices |
|---|---|---|---|---|---|
| Among populations | 2 | 1200.527 | 50.537 | 12 | FST = 0.115 (p < 0.001) |
| Within individuals | 7 | 4363.083 | 388.927 | 88 | FIT = 0.013 (p > 0.05) |
| Bogotá | Sibundoy | Palmira | |
|---|---|---|---|
| Bogotá | 0 | 0.007 | 0.056 |
| Sibundoy | 0.329 ± 0.011 | 0 | 0.087 |
| Palmira | 0.067 ± 0.008 | 0.061 ± 0.008 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes-Ardila, W.L.; Rugeles-Silva, P.A.; Duque-Zapata, J.D.; Vélez-Martínez, G.A.; Tarazona Pulido, L.; Cardona Tobar, K.M.; Díaz Gallo, S.A.; Muñoz Flórez, J.E.; Díaz-Ariza, L.A.; López-Alvarez, D. Exploring Genomics and Microbial Ecology: Analysis of Bidens pilosa L. Genetic Structure and Soil Microbiome Diversity by RAD-Seq and Metabarcoding. Plants 2024, 13, 221. https://doi.org/10.3390/plants13020221
Reyes-Ardila WL, Rugeles-Silva PA, Duque-Zapata JD, Vélez-Martínez GA, Tarazona Pulido L, Cardona Tobar KM, Díaz Gallo SA, Muñoz Flórez JE, Díaz-Ariza LA, López-Alvarez D. Exploring Genomics and Microbial Ecology: Analysis of Bidens pilosa L. Genetic Structure and Soil Microbiome Diversity by RAD-Seq and Metabarcoding. Plants. 2024; 13(2):221. https://doi.org/10.3390/plants13020221
Chicago/Turabian StyleReyes-Ardila, Wendy Lorena, Paula Andrea Rugeles-Silva, Juan Diego Duque-Zapata, Glever Alexander Vélez-Martínez, Lina Tarazona Pulido, Karen Melissa Cardona Tobar, Sergio Alberto Díaz Gallo, Jaime Eduardo Muñoz Flórez, Lucia Ana Díaz-Ariza, and Diana López-Alvarez. 2024. "Exploring Genomics and Microbial Ecology: Analysis of Bidens pilosa L. Genetic Structure and Soil Microbiome Diversity by RAD-Seq and Metabarcoding" Plants 13, no. 2: 221. https://doi.org/10.3390/plants13020221
APA StyleReyes-Ardila, W. L., Rugeles-Silva, P. A., Duque-Zapata, J. D., Vélez-Martínez, G. A., Tarazona Pulido, L., Cardona Tobar, K. M., Díaz Gallo, S. A., Muñoz Flórez, J. E., Díaz-Ariza, L. A., & López-Alvarez, D. (2024). Exploring Genomics and Microbial Ecology: Analysis of Bidens pilosa L. Genetic Structure and Soil Microbiome Diversity by RAD-Seq and Metabarcoding. Plants, 13(2), 221. https://doi.org/10.3390/plants13020221







