When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development
Abstract
1. Introduction
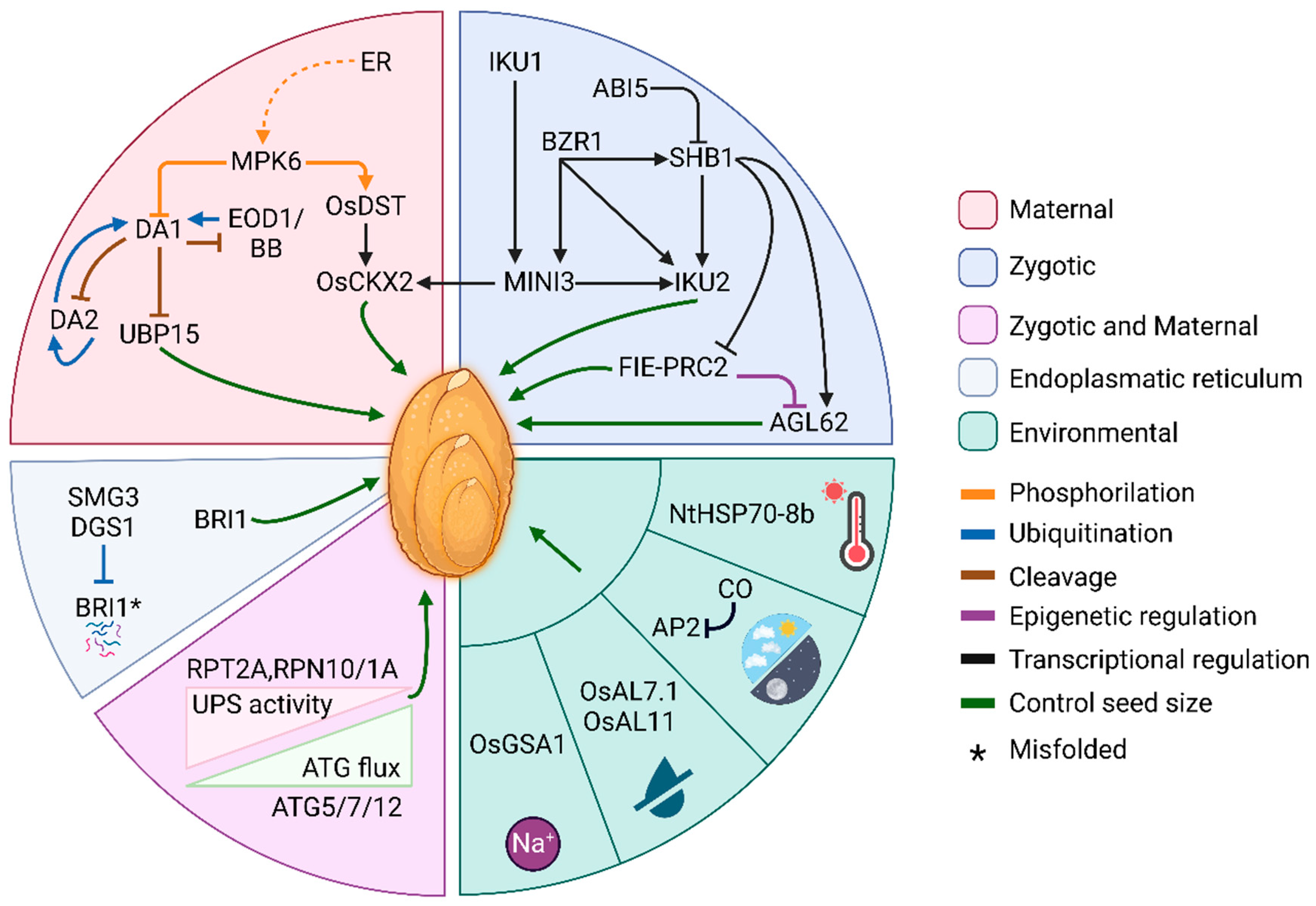
2. Developmental Control of Seed Size
2.1. Maternal and Zygotic Control of Seed Size
2.2. Does Cell Wall Constrain Seed Growth?
3. Seed Size as Determinant of Plant Success
3.1. Is Seed Size Related to Tolerance to Environmental Stresses?
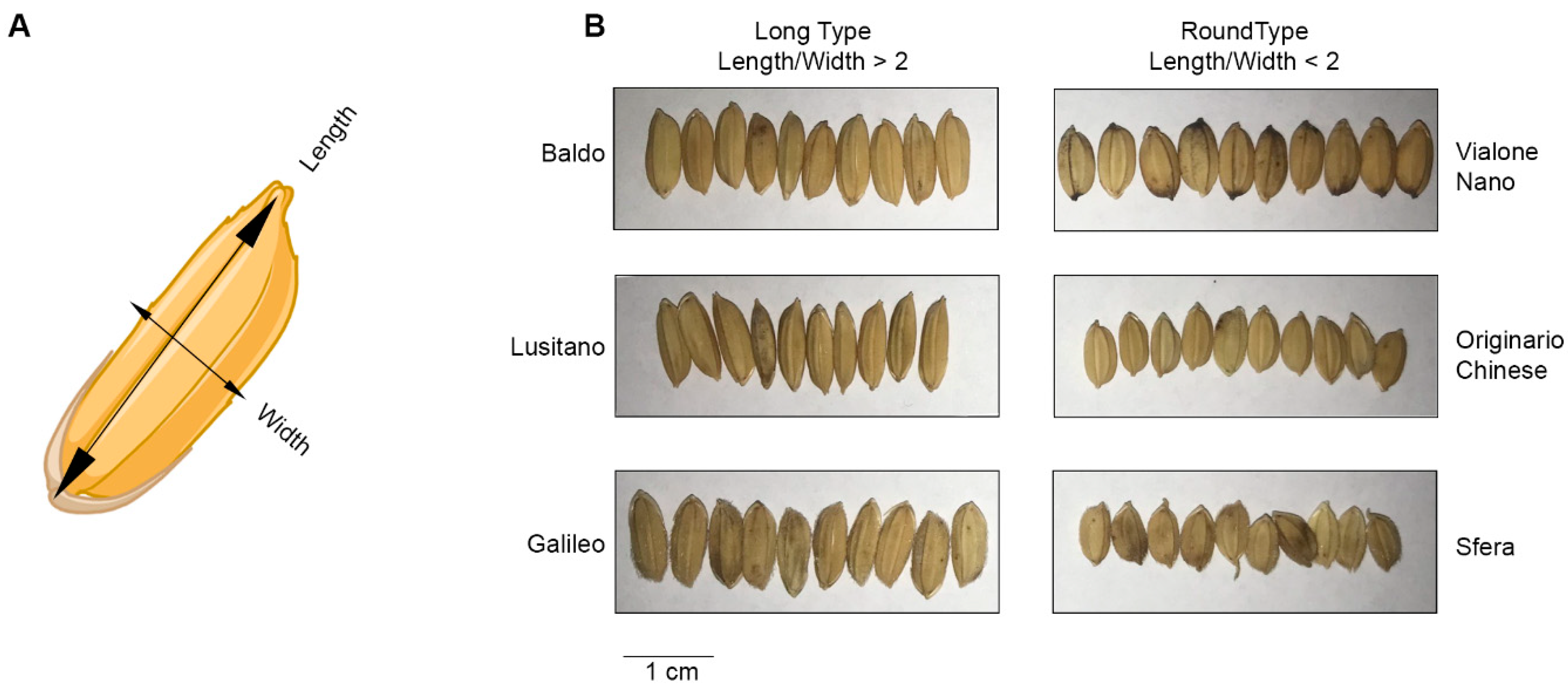
3.2. Seed Size Is a Quantitative and Qualitative Agronomic Trait for Cereals
3.3. Does Seed Size Influence the Success of Seed Germination?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Black, M. Darwin and Seeds. Seed Sci. Res. 2009, 19, 193–199. [Google Scholar] [CrossRef]
- Friedman, W.E. Developmental and Evolutionary Hypotheses for the Origin of Double Fertilization and Endosperm. C. R. Acad. Sci. III 2001, 324, 559–567. [Google Scholar] [CrossRef]
- Haughn, G.; Chaudhury, A. Genetic Analysis of Seed Coat Development in Arabidopsis. Trends Plant Sci. 2005, 10, 472–477. [Google Scholar] [CrossRef]
- Lafon-Placette, C.; Köhler, C. Embryo and Endosperm, Partners in Seed Development. Curr. Opin. Plant Biol. 2014, 17, 64–69. [Google Scholar] [CrossRef]
- Beeckman, T.; De Rycke, R.; Viane, R.; Inzé, D. Histological Study of Seed Coat Development in Arabidopsis Thaliana. J. Plant Res. 2000, 113, 139–148. [Google Scholar] [CrossRef]
- Chaudhury, A.M.; Koltunow, A.; Payne, T.; Luo, M.; Tucker, M.R.; Dennis, E.S.; Peacock, W.J. Control of Early Seed Development. Annu. Rev. Cell Dev. Biol. 2001, 17, 677–699. [Google Scholar] [CrossRef]
- Garcia, D.; Fitz Gerald, J.N.; Berger, F. Maternal Control of Integument Cell Elongation and Zygotic Control of Endosperm Growth Are Coordinated to Determine Seed Size in Arabidopsis. Plant Cell 2005, 17, 52–60. [Google Scholar] [CrossRef]
- Sundaresan, V. Control of Seed Size in Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 17887–17888. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Tsukaya, H. Mechanism of Leaf-Shape Determination. Annu. Rev. Plant Biol. 2006, 57, 477–496. [Google Scholar] [CrossRef]
- Adamski, N.M.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local Maternal Control of Seed Size by KLUH/CYP78A5-Dependent Growth Signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar] [CrossRef]
- Fang, W.; Wang, Z.; Cui, R.; Li, J.; Li, Y. Maternal Control of Seed Size by EOD3/CYP78A6 in Arabidopsis Thaliana. Plant J. 2012, 70, 929–939. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The Ubiquitin Receptor DA1 Interacts with the E3 Ubiquitin Ligase DA2 to Regulate Seed and Organ Size in Arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of Final Seed and Organ Size by the DA1 Gene Family in Arabidopsis Thaliana. Genes. Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking Differential Domain Functions of the GS3 Protein to Natural Variation of Grain Size in Rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Ubiquitin-Mediated Control of Seed Size in Plants. Front. Plant Sci. 2014, 5, 332. [Google Scholar] [CrossRef]
- Dong, H.; Dumenil, J.; Lu, F.-H.; Na, L.; Vanhaeren, H.; Naumann, C.; Klecker, M.; Prior, R.; Smith, C.; McKenzie, N.; et al. Ubiquitylation Activates a Peptidase That Promotes Cleavage and Destabilization of Its Activating E3 Ligases and Diverse Growth Regulatory Proteins to Limit Cell Proliferation in Arabidopsis. Genes. Dev. 2017, 31, 197–208. [Google Scholar] [CrossRef]
- Huang, K.; Wang, D.; Duan, P.; Zhang, B.; Xu, R.; Li, N.; Li, Y. Wide and Thick Grain 1, Which Encodes an Otubain-like Protease with Deubiquitination Activity, Influences Grain Size and Shape in Rice. Plant J. 2017, 91, 849–860. [Google Scholar] [CrossRef]
- Xu, R.; Duan, P.; Yu, H.; Zhou, Z.; Zhang, B.; Wang, R.; Li, J.; Zhang, G.; Zhuang, S.; Lyu, J.; et al. Control of Grain Size and Weight by the OsMKKK10-OsMKK4-OsMAPK6 Signaling Pathway in Rice. Mol. Plant 2018, 11, 860–873. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Liu, X.; Pai, Q.; Wang, Y.; Wu, X. Molecular Network for Regulation of Seed Size in Plants. Int. J. Mol. Sci. 2023, 24, 10666. [Google Scholar] [CrossRef]
- Jiang, S.; Jin, X.; Liu, Z.; Xu, R.; Hou, C.; Zhang, F.; Fan, C.; Wu, H.; Chen, T.; Shi, J.; et al. Natural Variation in SSW1 Coordinates Seed Growth and Nitrogen Use Efficiency in Arabidopsis. Cell Rep. 2024, 43, 114150. [Google Scholar] [CrossRef]
- Santiago, J.P.; Tegeder, M. Connecting Source with Sink: The Role of Arabidopsis AAP8 in Phloem Loading of Amino Acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef]
- Schmidt, R.; Stransky, H.; Koch, W. The Amino Acid Permease AAP8 Is Important for Early Seed Development in Arabidopsis Thaliana. Planta 2007, 226, 805–813. [Google Scholar] [CrossRef]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.-G. GASA4, One of the 14-Member Arabidopsis GASA Family of Small Polypeptides, Regulates Flowering and Seed Development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Trapalis, M.; Li, S.F.; Parish, R.W. The Arabidopsis GASA10 Gene Encodes a Cell Wall Protein Strongly Expressed in Developing Anthers and Seeds. Plant Sci. 2017, 260, 71–79. [Google Scholar] [CrossRef]
- Koorneef, M.; Elgersma, A.; Hanhart, C.J.; van Loenen-Martinet, E.P.; van Rijn, L.; Zeevaart, J.A.D. A Gibberellin Insensitive Mutant of Arabidopsis Thaliana. Physiol. Plant. 1985, 65, 33–39. [Google Scholar] [CrossRef]
- Dill, A.; Sun, T. Synergistic Derepression of Gibberellin Signaling by Removing RGA and GAI Function in Arabidopsis Thaliana. Genetics 2001, 159, 777–785. [Google Scholar] [CrossRef]
- Gomez, M.D.; Ventimilla, D.; Sacristan, R.; Perez-Amador, M.A. Gibberellins Regulate Ovule Integument Development by Interfering with the Transcription Factor ATS. Plant Physiol. 2016, 172, 2403–2415. [Google Scholar] [CrossRef]
- Gómez, M.D.; Fuster-Almunia, C.; Ocaña-Cuesta, J.; Alonso, J.M.; Pérez-Amador, M.A. RGL2 Controls Flower Development, Ovule Number and Fertility in Arabidopsis. Plant Sci. 2019, 281, 82–92. [Google Scholar] [CrossRef]
- Gómez Jiménez, M.D.; Cored, I.; Barro-Trastoy, D.; Sanchez-Matilla, J.; Tornero, P.; Pérez-Amador, M.Á. DELLA Proteins Positively Regulate Seed Size in Arabidopsis. Development 2023, 150, dev201853. [Google Scholar] [CrossRef]
- Klucher, K.M.; Chow, H.; Reiser, L.; Fischer, R.L. The AINTEGUMENTA Gene of Arabidopsis Required for Ovule and Female Gametophyte Development Is Related to the Floral Homeotic Gene APETALA2. Plant Cell 1996, 8, 137–153. [Google Scholar] [CrossRef]
- Wing, I.S.; De Cian, E.; Mistry, M.N. Global Vulnerability of Crop Yields to Climate Change. J. Environ. Econ. Manag. 2021, 109, 102462. [Google Scholar] [CrossRef]
- Gasparis, S.; Miłoszewski, M.M. Genetic Basis of Grain Size and Weight in Rice, Wheat, and Barley. Int. J. Mol. Sci. 2023, 24, 16921. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, X.; Xu, G.; Zhang, X. Mechanisms Controlling Seed Size by Early Endosperm Development. Seed Biol. 2023, 2, 1–11. [Google Scholar] [CrossRef]
- Disch, S.; Anastasiou, E.; Sharma, V.K.; Laux, T.; Fletcher, J.C.; Lenhard, M. The E3 Ubiquitin Ligase BIG BROTHER Controls Arabidopsis Organ Size in a Dosage-Dependent Manner. Curr. Biol. 2006, 16, 272–279. [Google Scholar] [CrossRef]
- Chen, Y.; Vermeersch, M.; Van Leene, J.; De Jaeger, G.; Li, Y.; Vanhaeren, H. A Dynamic Ubiquitination Balance of Cell Proliferation and Endoreduplication Regulators Determines Plant Organ Size. Sci. Adv. 2024, 10, eadj2570. [Google Scholar] [CrossRef]
- Wu, X.; Cai, X.; Zhang, B.; Wu, S.; Wang, R.; Li, N.; Li, Y.; Sun, Y.; Tang, W. ERECTA Regulates Seed Size Independently of Its Intracellular Domain via MAPK-DA1-UBP15 Signaling. Plant Cell 2022, 34, 3773–3789. [Google Scholar] [CrossRef]
- Guo, T.; Lu, Z.-Q.; Shan, J.-X.; Ye, W.-W.; Dong, N.-Q.; Lin, H.-X. ERECTA1 Acts Upstream of the OsMKKK10-OsMKK4-OsMPK6 Cascade to Control Spikelet Number by Regulating Cytokinin Metabolism in Rice. Plant Cell 2020, 32, 2763–2779. [Google Scholar] [CrossRef]
- Shpak, E.D.; Lakeman, M.B.; Torii, K.U. Dominant-Negative Receptor Uncovers Redundancy in the Arabidopsis ERECTA Leucine-Rich Repeat Receptor–Like Kinase Signaling Pathway That Regulates Organ Shape. Plant Cell 2003, 15, 1095–1110. [Google Scholar] [CrossRef]
- Shpak, E.D.; Berthiaume, C.T.; Hill, E.J.; Torii, K.U. Synergistic Interaction of Three ERECTA-Family Receptor-like Kinases Controls Arabidopsis Organ Growth and Flower Development by Promoting Cell Proliferation. Development 2004, 131, 1491–1501. [Google Scholar] [CrossRef]
- Wang, H.; Ngwenyama, N.; Liu, Y.; Walker, J.C.; Zhang, S. Stomatal Development and Patterning Are Regulated by Environmentally Responsive Mitogen-Activated Protein Kinases in Arabidopsis. Plant Cell 2007, 19, 63–73. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-Activated Protein Kinase Cascades in Plant Signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, H.; He, Y.; Liu, Y.; Walker, J.C.; Torii, K.U.; Zhang, S. A MAPK Cascade Downstream of ERECTA Receptor-Like Protein Kinase Regulates Arabidopsis Inflorescence Architecture by Promoting Localized Cell Proliferation. Plant Cell 2012, 24, 4948–4960. [Google Scholar] [CrossRef]
- Wang, J.-L.; Tang, M.-Q.; Chen, S.; Zheng, X.-F.; Mo, H.-X.; Li, S.-J.; Wang, Z.; Zhu, K.-M.; Ding, L.-N.; Liu, S.-Y.; et al. Down-Regulation of BnDA1, Whose Gene Locus Is Associated with the Seeds Weight, Improves the Seeds Weight and Organ Size in Brassica Napus. Plant Biotechnol. J. 2017, 15, 1024–1033. [Google Scholar] [CrossRef]
- Xie, G.; Li, Z.; Ran, Q.; Wang, H.; Zhang, J. Over-Expression of Mutated ZmDA1 or ZmDAR1 Gene Improves Maize Kernel Yield by Enhancing Starch Synthesis. Plant Biotechnol. J. 2018, 16, 234–244. [Google Scholar] [CrossRef]
- Shi, C.; Ren, Y.; Liu, L.; Wang, F.; Zhang, H.; Tian, P.; Pan, T.; Wang, Y.; Jing, R.; Liu, T.; et al. Ubiquitin Specific Protease 15 Has an Important Role in Regulating Grain Width and Size in Rice. Plant Physiol. 2019, 180, 381–391. [Google Scholar] [CrossRef]
- Strasser, R. Protein Quality Control in the Endoplasmic Reticulum of Plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef]
- Vembar, S.S.; Brodsky, J.L. One Step at a Time: Endoplasmic Reticulum-Associated Degradation. Nat. Rev. Mol. Cell Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef]
- Li, J.; Zhang, B.; Duan, P.; Yan, L.; Yu, H.; Zhang, L.; Li, N.; Zheng, L.; Chai, T.; Xu, R.; et al. An Endoplasmic Reticulum-Associated Degradation-Related E2-E3 Enzyme Pair Controls Grain Size and Weight through the Brassinosteroid Signaling Pathway in Rice. Plant Cell 2023, 35, 1076–1091. [Google Scholar] [CrossRef]
- Kurepa, J.; Wang, S.; Li, Y.; Zaitlin, D.; Pierce, A.J.; Smalle, J.A. Loss of 26S Proteasome Function Leads to Increased Cell Size and Decreased Cell Number in Arabidopsis Shoot Organs. Plant Physiol. 2009, 150, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Hua, Z. The Ubiquitin–26S Proteasome System and Autophagy Relay Proteome Homeostasis Regulation during Silique Development. Plant J. 2022, 111, 1324–1339. [Google Scholar] [CrossRef]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jürgens, G.; Berger, F. Arabidopsis Haiku Mutants Reveal New Controls of Seed Size by Endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ Motif Protein IKU1 Regulates Endosperm Growth and Seed Size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY Family Gene, and HAIKU2 (IKU2), a Leucine-Rich Repeat (LRR) KINASE Gene, Are Regulators of Seed Size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Kang, X.; Li, W.; Zhou, Y.; Ni, M. A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement. PLOS Genet. 2013, 9, e1003347. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Kang, X.; Zhao, X.; Zhang, X.; Ni, M. SHORT HYPOCOTYL UNDER BLUE1 Associates with MINISEED3 and HAIKU2 Promoters in Vivo to Regulate Arabidopsis Seed Development. Plant Cell 2009, 21, 106–117. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Zhao, X.Y.; Shao, X.X.; Wang, F.; Zhou, C.; Liu, Y.G.; Zhang, Y.; Zhang, X.S. Abscisic Acid Regulates Early Seed Development in Arabidopsis by ABI5-Mediated Transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 2014, 26, 1053–1068. [Google Scholar] [CrossRef]
- Jiang, W.-B.; Huang, H.-Y.; Hu, Y.-W.; Zhu, S.-W.; Wang, Z.-Y.; Lin, W.-H. Brassinosteroid Regulates Seed Size and Shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis Histidine Phosphotransfer Proteins Are Redundant Positive Regulators of Cytokinin Signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis Cytokinin Receptor Mutants Reveal Functions in Shoot Growth, Leaf Senescence, Seed Size, Germination, Root Development, and Cytokinin Metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin Oxidase Regulates Rice Grain Production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield in Arabidopsis Thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nie, X.; Tan, J.L.H.; Berger, F. Integration of Epigenetic and Genetic Controls of Seed Size by Cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15479–15484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cheng, F.; Xiao, Y.; Kang, X.; Wang, X.; Kuang, R.; Ni, M. Global Analysis of Canola Genes Targeted by SHORT HYPOCOTYL UNDER BLUE 1 during Endosperm and Embryo Development. Plant J. 2017, 91, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Köhler, C.; Hennig, L. Keeping the Gate Closed: Functions of the Polycomb Repressive Complex PRC2 in Development. Plant J. 2015, 83, 121–132. [Google Scholar] [CrossRef]
- Wu, D.; Wei, Y.; Zhao, X.; Li, B.; Zhang, H.; Xu, G.; Lv, J.; Zhang, D.; Zhang, X.; Ni, M. Ancestral Function but Divergent Epigenetic Regulation of HAIKU2 Reveals Routes of Seed Developmental Evolution. Mol. Plant 2022, 15, 1575–1589. [Google Scholar] [CrossRef]
- Aegerter-Wilmsen, T.; Heimlicher, M.B.; Smith, A.C.; de Reuille, P.B.; Smith, R.S.; Aegerter, C.M.; Basler, K. Integrating Force-Sensing and Signaling Pathways in a Model for the Regulation of Wing Imaginal Disc Size. Development 2012, 139, 3221–3231. [Google Scholar] [CrossRef]
- Shraiman, B.I. Mechanical Feedback as a Possible Regulator of Tissue Growth. Proc. Natl. Acad. Sci. USA 2005, 102, 3318–3323. [Google Scholar] [CrossRef]
- Nakayama, N.; Smith, R.S.; Mandel, T.; Robinson, S.; Kimura, S.; Boudaoud, A.; Kuhlemeier, C. Mechanical Regulation of Auxin-Mediated Growth. Curr. Biol. 2012, 22, 1468–1476. [Google Scholar] [CrossRef]
- Uyttewaal, M.; Burian, A.; Alim, K.; Landrein, B.; Borowska-Wykręt, D.; Dedieu, A.; Peaucelle, A.; Ludynia, M.; Traas, J.; Boudaoud, A.; et al. Mechanical Stress Acts via Katanin to Amplify Differences in Growth Rate between Adjacent Cells in Arabidopsis. Cell 2012, 149, 439–451. [Google Scholar] [CrossRef]
- Fal, K.; Korsbo, N.; Alonso-Serra, J.; Teles, J.; Liu, M.; Refahi, Y.; Chabouté, M.-E.; Jönsson, H.; Hamant, O. Tissue Folding at the Organ–Meristem Boundary Results in Nuclear Compression and Chromatin Compaction. Proc. Natl. Acad. Sci. USA 2021, 118, e2017859118. [Google Scholar] [CrossRef] [PubMed]
- Landrein, B.; Kiss, A.; Sassi, M.; Chauvet, A.; Das, P.; Cortizo, M.; Laufs, P.; Takeda, S.; Aida, M.; Traas, J.; et al. Mechanical Stress Contributes to the Expression of the STM Homeobox Gene in Arabidopsis Shoot Meristems. eLife 2015, 4, e07811. [Google Scholar] [CrossRef] [PubMed]
- Beauzamy, L.; Nakayama, N.; Boudaoud, A. Flowers under Pressure: Ins and Outs of Turgor Regulation in Development. Ann. Bot. 2014, 114, 1517–1533. [Google Scholar] [CrossRef]
- Ali, O.; Cheddadi, I.; Landrein, B.; Long, Y. Revisiting the Relationship between Turgor Pressure and Plant Cell Growth. New Phytol. 2023, 238, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Beauzamy, L.; Fourquin, C.; Dubrulle, N.; Boursiac, Y.; Boudaoud, A.; Ingram, G. Endosperm Turgor Pressure Decreases during Early Arabidopsis Seed Development. Development 2016, 143, 3295–3299. [Google Scholar] [CrossRef] [PubMed]
- Creff, A.; Ali, O.; Bied, C.; Bayle, V.; Ingram, G.; Landrein, B. Evidence That Endosperm Turgor Pressure Both Promotes and Restricts Seed Growth and Size. Nat. Commun. 2023, 14, 67. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Liu, M.; Yan, P.; Niu, F.; Ma, F.; Hu, J.; He, S.; Cui, J.; Yuan, X.; et al. OsEXPA7 Encoding an Expansin Affects Grain Size and Quality Traits in Rice (Oryza sativa L.). Rice 2024, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Creff, A.; Brocard, L.; Ingram, G. A Mechanically Sensitive Cell Layer Regulates the Physical Properties of the Arabidopsis Seed Coat. Nat. Commun. 2015, 6, 6382. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Lapierre, C.; Alvarado, C.; Yoshinaga, A.; Barron, C.; Bouchet, B.; Bakan, B.; Saulnier, L.; Devaux, M.-F.; Girousse, C.; et al. Ferulate and Lignin Cross-Links Increase in Cell Walls of Wheat Grain Outer Layers during Late Development. Plant Sci. 2018, 276, 199–207. [Google Scholar] [CrossRef]
- Schopfer, P. Biomechanics of Plant Growth. Am. J. Bot. 2006, 93, 1415–1425. [Google Scholar] [CrossRef]
- Kierzkowski, D.; Nakayama, N.; Routier-Kierzkowska, A.-L.; Weber, A.; Bayer, E.; Schorderet, M.; Reinhardt, D.; Kuhlemeier, C.; Smith, R.S. Elastic Domains Regulate Growth and Organogenesis in the Plant Shoot Apical Meristem. Science 2012, 335, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Kutschera, U. Cessation of Cell Elongation in Rye Coleoptiles Is Accompanied by a Loss of Cell-Wall Plasticity. J. Exp. Bot. 1996, 47, 1387–1394. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Nanoscale Structure, Mechanics and Growth of Epidermal Cell Walls. Curr. Opin. Plant Biol. 2018, 46, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Levesque-Tremblay, G.; Pelloux, J.; Braybrook, S.A.; Müller, K. Tuning of Pectin Methylesterification: Consequences for Cell Wall Biomechanics and Development. Planta 2015, 242, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.A.; Le Guillou, L.; Bron, E.; Kuhlemeier, C.; Höfte, H. Pectin-Induced Changes in Cell Wall Mechanics Underlie Organ Initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef] [PubMed]
- Parre, E.; Geitmann, A. Pectin and the Role of the Physical Properties of the Cell Wall in Pollen Tube Growth of Solanum Chacoense. Planta 2005, 220, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Levesque-Tremblay, G.; Bartels, S.; Weitbrecht, K.; Wormit, A.; Usadel, B.; Haughn, G.; Kermode, A.R. Demethylesterification of Cell Wall Pectins in Arabidopsis Plays a Role in Seed Germination. Plant Physiol. 2013, 161, 305–316. [Google Scholar] [CrossRef]
- Di Marzo, M.; Babolin, N.; Viana, V.E.; de Oliveira, A.C.; Gugi, B.; Caporali, E.; Herrera-Ubaldo, H.; Martínez-Estrada, E.; Driouich, A.; de Folter, S.; et al. The Genetic Control of SEEDSTICK and LEUNIG-HOMOLOG in Seed and Fruit Development: New Insights into Cell Wall Control. Plants 2022, 11, 3146. [Google Scholar] [CrossRef]
- Vogel, J. Unique Aspects of the Grass Cell Wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef]
- Yang, X.; Wilkinson, L.G.; Aubert, M.K.; Houston, K.; Shirley, N.J.; Tucker, M.R. Ovule Cell Wall Composition Is a Maternal Determinant of Grain Size in Barley. New Phytol. 2023, 237, 2136–2147. [Google Scholar] [CrossRef]
- Stork, J.; Harris, D.; Griffiths, J.; Williams, B.; Beisson, F.; Li-Beisson, Y.; Mendu, V.; Haughn, G.; DeBolt, S. CELLULOSE SYNTHASE9 Serves a Nonredundant Role in Secondary Cell Wall Synthesis in Arabidopsis Epidermal Testa Cells. Plant Physiol. 2010, 153, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Mendu, V.; Griffiths, J.S.; Persson, S.; Stork, J.; Downie, A.B.; Voiniciuc, C.; Haughn, G.W.; DeBolt, S. Subfunctionalization of Cellulose Synthases in Seed Coat Epidermal Cells Mediates Secondary Radial Wall Synthesis and Mucilage Attachment. Plant Physiol. 2011, 157, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Hématy, K.; Höfte, H. Growth Control and Cell Wall Signaling in Plants. Annu. Rev. Plant Biol. 2012, 63, 381–407. [Google Scholar] [CrossRef] [PubMed]
- McQueen-Mason, S.J.; Cosgrove, D.J. Expansin Mode of Action on Cell Walls (Analysis of Wall Hydrolysis, Stress Relaxation, and Binding). Plant Physiol. 1995, 107, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Yennawar, N.H.; Li, L.-C.; Dudzinski, D.M.; Tabuchi, A.; Cosgrove, D.J. Crystal Structure and Activities of EXPB1 (Zea m 1), a β-Expansin and Group-1 Pollen Allergen from Maize. Proc. Natl. Acad. Sci. USA 2006, 103, 14664–14671. [Google Scholar] [CrossRef]
- Sun, Q.; Li, Y.; Gong, D.; Hu, A.; Zhong, W.; Zhao, H.; Ning, Q.; Tan, Z.; Liang, K.; Mu, L.; et al. A NAC-EXPANSIN Module Enhances Maize Kernel Size by Controlling Nucellus Elimination. Nat. Commun. 2022, 13, 5708. [Google Scholar] [CrossRef] [PubMed]
- Calderini, D.F.; Castillo, F.M.; Arenas-M, A.; Molero, G.; Reynolds, M.P.; Craze, M.; Bowden, S.; Milner, M.J.; Wallington, E.J.; Dowle, A.; et al. Overcoming the Trade-off between Grain Weight and Number in Wheat by the Ectopic Expression of Expansin in Developing Seeds Leads to Increased Yield Potential. New Phytol. 2021, 230, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 Mediates the Balance between Growth and Resistance to Biotic Stress in Rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Luo, Y.; Wang, G.; Feng, C.; Li, H. Genome-Wide Identification of Expansin Genes in Brachypodium Distachyon and Functional Characterization of BdEXPA27. Plant Sci. 2020, 296, 110490. [Google Scholar] [CrossRef]
- Bae, J.M.; Kwak, M.S.; Noh, S.A.; Oh, M.-J.; Kim, Y.-S.; Shin, J.S. Overexpression of Sweetpotato Expansin cDNA (IbEXP1) Increases Seed Yield in Arabidopsis. Transgenic Res. 2014, 23, 657–667. [Google Scholar] [CrossRef]
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell Wall Integrity Maintenance during Plant Development and Interaction with the Environment. Nat. Plant. 2019, 5, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Bacete, L.; Hamann, T. The Role of Mechanoperception in Plant Cell Wall Integrity Maintenance. Plants 2020, 9, 574. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, L.; Ye, H.; Yu, X.; Algreen, A.; Yin, Y. Three Related Receptor-like Kinases Are Required for Optimal Cell Elongation in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.-M. FERONIA Receptor-like Kinase Regulates RHO GTPase Signaling of Root Hair Development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [PubMed]
- Nibau, C.; Wu, H.; Cheung, A.Y. RAC/ROP GTPases: “hubs” for Signal Integration and Diversification in Plants. Trends Plant Sci. 2006, 11, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, J.; Huang, Y.; Liu, L.; Li, D.; Chen, L.; Luan, S. FERONIA Receptor Kinase Controls Seed Size in Arabidopsis Thaliana. Mol. Plant 2014, 7, 920–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, D.; Yang, Z.; Jiang, S.; Qu, J.; He, W.; Liu, Z.; Xing, J.; Ma, Y.; Lin, Q.; et al. Roles of FERONIA-like Receptor Genes in Regulating Grain Size and Quality in Rice. Sci. China Life Sci. 2021, 64, 294–310. [Google Scholar] [CrossRef] [PubMed]
- Engelsdorf, T.; Gigli-Bisceglia, N.; Veerabagu, M.; McKenna, J.F.; Vaahtera, L.; Augstein, F.; Van der Does, D.; Zipfel, C.; Hamann, T. The Plant Cell Wall Integrity Maintenance and Immune Signaling Systems Cooperate to Control Stress Responses in Arabidopsis Thaliana. Sci. Signal. 2018, 11, eaao3070. [Google Scholar] [CrossRef]
- Mielke, S.; Zimmer, M.; Meena, M.K.; Dreos, R.; Stellmach, H.; Hause, B.; Voiniciuc, C.; Gasperini, D. Jasmonate Biosynthesis Arising from Altered Cell Walls Is Prompted by Turgor-Driven Mechanical Compression. Sci. Adv. 2021, 7, eabf0356. [Google Scholar] [CrossRef]
- Lorrai, R.; Erguvan, Ö.; Raggi, S.; Jonsson, K.; Široká, J.; Tarkowská, D.; Novák, O.; Verger, S.; Robert, S.; Ferrari, S. Cell Wall Integrity Modulates a PHYTOCHROME-INTERACTING FACTOR (PIF)—HOOKLESS1 (HLS1) Signalling Module Controlling Apical Hook Formation in Arabidopsis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 Protein Is a Jasmonate Receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef]
- Yan, J.; Yao, R.; Chen, L.; Li, S.; Gu, M.; Nan, F.; Xie, D. Dynamic Perception of Jasmonates by the F-Box Protein COI1. Mol. Plant 2018, 11, 1237–1247. [Google Scholar] [CrossRef]
- Hu, S.; Yang, H.; Gao, H.; Yan, J.; Xie, D. Control of Seed Size by Jasmonate. Sci. China Life Sci. 2021, 64, 1215–1226. [Google Scholar] [CrossRef]
- Mehra, P.; Pandey, B.K.; Verma, L.; Prusty, A.; Singh, A.P.; Sharma, S.; Malik, N.; Bennett, M.J.; Parida, S.K.; Giri, J.; et al. OsJAZ11 Regulates Spikelet and Seed Development in Rice. Plant Direct 2022, 6, e401. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, Y.; Wu, T.-T.; Zhang, G.-L.; Yin, H.; Chen, W.; Wang, S.; Chang, F.; Gou, J.-Y. Cloning of Wheat Keto-Acyl Thiolase 2B Reveals a Role of Jasmonic Acid in Grain Weight Determination. Nat. Commun. 2020, 11, 6266. [Google Scholar] [CrossRef]
- Yang, L.; Song, W.; Xu, C.; Sapey, E.; Jiang, D.; Wu, C. Effects of High Night Temperature on Soybean Yield and Compositions. Front. Plant Sci. 2023, 14, 1065604. [Google Scholar] [CrossRef]
- Kino, R.I.; Pellny, T.K.; Mitchell, R.A.C.; Gonzalez-Uriarte, A.; Tosi, P. High Post-Anthesis Temperature Effects on Bread Wheat (Triticum aestivum L.) Grain Transcriptome during Early Grain-Filling. BMC Plant Biol. 2020, 20, 170. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Khungar, L.; Shimphrui, R.; Tiwari, L.D.; Tripathi, G.; Sarkar, N.K.; Agarwal, S.-K.; Agarwal, M.; Grover, A. AtHsp101 Research Sets Course of Action for the Genetic Improvement of Crops against Heat Stress. J. Plant Biochem. Biotechnol. 2020, 29, 715–732. [Google Scholar] [CrossRef]
- Tiwari, L.D.; Kumar, R.; Sharma, V.; Sahu, A.K.; Sahu, B.; Naithani, S.C.; Grover, A. Stress and Development Phenotyping of Hsp101 and Diverse Other Hsp Mutants of Arabidopsis Thaliana. J. Plant Biochem. Biotechnol. 2021, 30, 889–905. [Google Scholar] [CrossRef]
- Singh, G.; Sarkar, N.K.; Grover, A. Hsp70, sHsps and Ubiquitin Proteins Modulate HsfA6a-Mediated Hsp101 Transcript Expression in Rice (Oryza sativa L.). Physiol. Plant. 2021, 173, 2055–2067. [Google Scholar] [CrossRef]
- Song, Z.; Li, Y.; Jia, Y.; Lian, W.; Jia, H. An Endoplasmic Reticulum-Localized NtHSP70-8 Confers Drought Tolerance in Tobacco by Regulating Water Loss and Antioxidant Capacity. Environ. Exp. Bot. 2021, 188, 104519. [Google Scholar] [CrossRef]
- Gomez-Cadenas, A.; Vives, V.; Zandalinas, S.I.; Manzi, M.; Sanchez-Perez, A.M.; Perez-Clemente, R.M.; Arbona, V. Abscisic Acid: A Versatile Phytohormone in Plant Signaling and Beyond. Curr. Protein Pept. Sci. 2015, 16, 413–434. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, S.I.; Tripathy, D.; Luo, Y.; Dion, E.; Fukui, G.; Kumazaki, A.; et al. ABA Is Required for Plant Acclimation to a Combination of Salt and Heat Stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, J.; Li, M.; Zhang, S.; Song, S.; Wang, W.; Wang, S.; Chang, J.; Xia, Z.; Zhang, S.; et al. NtHSP70-8b Positively Regulates Heat Tolerance and Seed Size in Nicotiana Tabacum. Plant Physiol. Biochem. 2023, 201, 107901. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, X.; Xia, H.; Wang, L.; Chen, S.; Xu, K.; Yang, F.; Zou, Y.; Wang, Y.; Zhu, J.; et al. Natural Variation of Alfin-like Family Affects Seed Size and Drought Tolerance in Rice. Plant J. 2022, 112, 1176–1193. [Google Scholar] [CrossRef] [PubMed]
- Lampayan, R.M.; Rejesus, R.M.; Singleton, G.R.; Bouman, B.A. Adoption and Economics of Alternate Wetting and Drying Water Management for Irrigated Lowland Rice. Field Crop. Res. 2015, 170, 95–108. [Google Scholar] [CrossRef]
- Dong, N.-Q.; Sun, Y.; Guo, T.; Shi, C.-L.; Zhang, Y.-M.; Kan, Y.; Xiang, Y.-H.; Zhang, H.; Yang, Y.-B.; Li, Y.-C.; et al. UDP-Glucosyltransferase Regulates Grain Size and Abiotic Stress Tolerance Associated with Metabolic Flux Redirection in Rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tong, H.; Xiao, Y.; Che, R.; Xu, F.; Hu, B.; Liang, C.; Chu, J.; Li, J.; Chu, C. Activation of Big Grain1 Significantly Improves Grain Size by Regulating Auxin Transport in Rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11102–11107. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.-Q.; Ye, W.-W.; Shan, J.-X.; Lin, H.-X. Tillering and Small Grain 1 Dominates the Tryptophan Aminotransferase Family Required for Local Auxin Biosynthesis in Rice. J. Integr. Plant Biol. 2020, 62, 581–600. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Johansson, M.; Staiger, D. Time to Flower: Interplay between Photoperiod and the Circadian Clock. J. Exp. Bot. 2015, 66, 719–730. [Google Scholar] [CrossRef]
- Yu, B.; He, X.; Tang, Y.; Chen, Z.; Zhou, L.; Li, X.; Zhang, C.; Huang, X.; Yang, Y.; Zhang, W.; et al. Photoperiod Controls Plant Seed Size in a CONSTANS-Dependent Manner. Nat. Plants 2023, 9, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Fan, J.; Jiang, Z.; Basso, B.; Sala, F.; Spada, A.; Grassi, F.; Lu, B.-R. The Puzzle of Italian Rice Origin and Evolution: Determining Genetic Divergence and Affinity of Rice Germplasm from Italy and Asia. PLoS ONE 2013, 8, e80351. [Google Scholar] [CrossRef]
- Chen, R.; Xiao, N.; Lu, Y.; Tao, T.; Huang, Q.; Wang, S.; Wang, Z.; Chuan, M.; Bu, Q.; Lu, Z.; et al. A de Novo Evolved Gene Contributes to Rice Grain Shape Difference between Indica and Japonica. Nat. Commun. 2023, 14, 5906. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Hou, Q.; Si, L.; Huang, X.; Luo, J.; Lu, D.; Zhu, J.; Shangguan, Y.; Miao, J.; Xie, Y.; et al. The PLATZ Transcription Factor GL6 Affects Grain Length and Number in Rice. Plant Physiol. 2019, 180, 2077–2090. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy Number Variation at the GL7 Locus Contributes to Grain Size Diversity in Rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef]
- Qi, P.; Lin, Y.-S.; Song, X.-J.; Shen, J.-B.; Huang, W.; Shan, J.-X.; Zhu, M.-Z.; Jiang, L.; Gao, J.-P.; Lin, H.-X. The Novel Quantitative Trait Locus GL3.1 Controls Rice Grain Size and Yield by Regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Yu, H.-X.; Ye, W.-W.; Shan, J.-X.; Dong, N.-Q.; Guo, T.; Kan, Y.; Xiang, Y.-H.; Zhang, H.; Yang, Y.-B.; et al. A Rice QTL GS3.1 Regulates Grain Size through Metabolic-Flux Distribution between Flavonoid and Lignin Metabolons without Affecting Stress Tolerance. Commun. Biol. 2021, 4, 1171. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-L.; Dong, N.-Q.; Guo, T.; Ye, W.-W.; Shan, J.-X.; Lin, H.-X. A Quantitative Trait Locus GW6 Controls Rice Grain Size and Yield through the Gibberellin Pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a Major QTL for Grain Length and Weight and Minor QTL for Grain Width and Thickness in Rice, Encodes a Putative Transmembrane Protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and Initial Characterization of GW5, a Major QTL Associated with Rice Grain Width and Weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 Acts in the Brassinosteroid Signalling Pathway to Regulate Grain Width and Weight in Rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Song, X.-J.; Huang, W.; Shi, M.; Zhu, M.-Z.; Lin, H.-X. A QTL for Rice Grain Width and Weight Encodes a Previously Unknown RING-Type E3 Ubiquitin Ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of Grain Size and Rice Yield by GL2-Mediated Brassinosteroid Responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Hou, Q.; Liu, K.; Gu, Z.; Dai, B.; Wang, A.; Feng, Q.; Zhao, Y.; Zhou, C.; Zhu, J.; et al. Armadillo Repeat Only Protein GS10 Negatively Regulates Brassinosteroid Signaling to Control Rice Grain Size. Plant Physiol. 2023, 192, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Abbas, W.; Shalmani, A.; Zhang, J.; Sun, Q.; Zhang, C.; Li, W.; Cui, Y.; Xiong, M.; Li, Y. The GW5-WRKY53-SGW5 Module Regulates Grain Size Variation in Rice. New Phytol. 2024, 242, 2011–2025. [Google Scholar] [CrossRef] [PubMed]
- Botella, J.R. Can Heterotrimeric G Proteins Help to Feed the World? Trends Plant Sci. 2012, 17, 563–568. [Google Scholar] [CrossRef]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-Protein Pathway Determines Grain Size in Rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wu, K.; Wang, B.; Liu, H.; Guo, S.; Guo, X.; Luo, W.; Sun, S.; Ouyang, Y.; Fu, X.; et al. The RING E3 Ligase CLG1 Targets GS3 for Degradation via the Endosome Pathway to Determine Grain Size in Rice. Mol. Plant 2021, 14, 1699–1713. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a Gene Associated with Grain Size Increased Yields during Rice Domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Wan, X.; Weng, J.; Zhai, H.; Wang, J.; Lei, C.; Liu, X.; Guo, T.; Jiang, L.; Su, N.; Wan, J. Quantitative Trait Loci (QTL) Analysis For Rice Grain Width and Fine Mapping of an Identified QTL Allele Gw-5 in a Recombination Hotspot Region on Chromosome 5. Genetics 2008, 179, 2239–2252. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S.; et al. Natural Variation in the Promoter of GSE5 Contributes to Grain Size Diversity in Rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, X.; Jiang, J.; Wang, X. Functions and Mechanisms of Brassinosteroids in Regulating Crop Agronomic Traits. Plant Cell Physiol. 2024, pcae044. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; He, M.; Mei, E.; Zhang, B.; Tang, J.; Xu, M.; Liu, J.; Li, X.; Wang, Z.; Tang, W.; et al. WRKY53 Integrates Classic Brassinosteroid Signaling and the Mitogen-Activated Protein Kinase Pathway to Regulate Rice Architecture and Seed Size. Plant Cell 2021, 33, 2753–2775. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, X.; Li, Q.; Xiao, Y.; Zhang, G.; Yin, W.; Niu, M.; Meng, W.; Dong, N.; Liu, J.; et al. The U-Box Ubiquitin Ligase TUD1 Promotes Brassinosteroid-Induced GSK2 Degradation in Rice. Plant Commun. 2022, 4, 100450. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, Z.; Lin, J.; Guan, B.; Chen, J.; Zhang, Z.; Chen, F.; Jiang, L.; Zheng, J.; Wang, T.; et al. Gw2.1, a New Allele of GW2, Improves Grain Weight and Grain Yield in Rice. Plant Sci. 2022, 325, 111495. [Google Scholar] [CrossRef]
- Choi, B.S.; Kim, Y.J.; Markkandan, K.; Koo, Y.J.; Song, J.T.; Seo, H.S. GW2 Functions as an E3 Ubiquitin Ligase for Rice Expansin-Like 1. Int. J. Mol. Sci. 2018, 19, 1904. [Google Scholar] [CrossRef]
- Hao, J.; Wang, D.; Wu, Y.; Huang, K.; Duan, P.; Li, N.; Xu, R.; Zeng, D.; Dong, G.; Zhang, B.; et al. The GW2-WG1-OsbZIP47 Pathway Controls Grain Size and Weight in Rice. Mol. Plant 2021, 14, 1266–1280. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, L.; He, Z. Understanding the Regulation of Cereal Grain Filling: The Way Forward. J. Integr. Plant Biol. 2023, 65, 526–547. [Google Scholar] [CrossRef]
- Achary, V.M.M.; Reddy, M.K. CRISPR-Cas9 Mediated Mutation in GRAIN WIDTH and WEIGHT2 (GW2) Locus Improves Aleurone Layer and Grain Nutritional Quality in Rice. Sci. Rep. 2021, 11, 21941. [Google Scholar] [CrossRef]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular Networks Regulating Arabidopsis Seed Maturation, After-ripening, Dormancy and Germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gao, M.; Liang, X.; Xu, M.; Liu, X.; Zhang, Y.; Liu, X.; Liu, J.; Gao, Y.; Qu, S.; et al. Quantitative Trait Loci for Seed Size Variation in Cucurbits—A Review. Front. Plant Sci. 2020, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Wang, D.; He, R.; Liu, S.; Zhu, J.-K. Mutations in MIR396e and MIR396f Increase Grain Size and Modulate Shoot Architecture in Rice. Plant Biotechnol. J. 2020, 18, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, Q.; Tian, B.; He, F.; Chen, Y.; Bai, G.; Akhunova, A.; Trick, H.N.; Akhunov, E. Gene Editing of the Wheat Homologs of TONNEAU 1-recruiting Motif Encoding Gene Affects Grain Shape and Weight in Wheat. Plant J. 2019, 100, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cadenas, A.; Zentella, R.; Walker-Simmons, M.K.; Ho, T.-H.D. Gibberellin/Abscisic Acid Antagonism in Barley Aleurone Cells: Site of Action of the Protein Kinase PKABA1 in Relation to Gibberellin Signaling Molecules. Plant Cell 2001, 13, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Asatsuma, S.; Sawada, C.; Itoh, K.; Okito, M.; Kitajima, A.; Mitsui, T. Involvement of Alpha-Amylase I-1 in Starch Degradation in Rice Chloroplasts. Plant Cell Physiol. 2005, 46, 858–869. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, D.D.; Köhler, C. Signalling Events Regulating Seed Coat Development. Biochem. Soc. Trans. 2014, 42, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.B.; Østergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit Development and Ripening. Annu. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of Glucosidase via Stress-Induced Polymerization Rapidly Increases Active Pools of Abscisic Acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; McGregor, C.; Zhang, Y.; Gong, G.; Zhang, H.; Guo, S.; Sun, H.; Cai, W.; Zhang, J.; Xu, Y. An Integrated Genetic Map Based on Four Mapping Populations and Quantitative Trait Loci Associated with Economically Important Traits in Watermelon (Citrullus lanatus). BMC Plant Biol. 2014, 14, 33. [Google Scholar] [CrossRef]
- Prothro, J.; Sandlin, K.; Abdel-Haleem, H.; Bachlava, E.; White, V.; Knapp, S.; McGregor, C. Main and Epistatic Quantitative Trait Loci Associated with Seed Size in Watermelon. J. Am. Soc. Hortic. Sci. 2012, 137, 452–457. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Guo, S.; Tian, S.; Zhang, J.; Ren, Y.; Li, M.; Gong, G.; Zhang, H.; Xu, Y. CRISPR/Cas9-Mediated Mutagenesis of ClBG1 Decreased Seed Size and Promoted Seed Germination in Watermelon. Hortic. Res. 2021, 8, 70. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S.; Huang, Y.; Ma, X.; Tan, L.; Liu, F.; Lv, Q.; Zhu, Z.; Hu, M.; Fu, Y.; et al. OsMADS17 Simultaneously Increases Grain Number and Grain Weight in Rice. Nat. Commun. 2023, 14, 3098. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Fujisawa, Y.; Kobayashi, M.; Ashikari, M.; Iwasaki, Y.; Kitano, H.; Matsuoka, M. Rice Dwarf Mutant D1, Which Is Defective in the α Subunit of the Heterotrimeric G Protein, Affects Gibberellin Signal Transduction. Proc. Natl. Acad. Sci. USA 2000, 97, 11638–11643. [Google Scholar] [CrossRef]
- Yang, X.; Lu, J.; Shi, W.-J.; Chen, Y.-H.; Yu, J.-W.; Chen, S.-H.; Zhao, D.-S.; Huang, L.-C.; Fan, X.-L.; Zhang, C.-Q.; et al. RGA1 Regulates Grain Size, Rice Quality and Seed Germination in the Small and Round Grain Mutant Srg5. BMC Plant Biol. 2024, 24, 167. [Google Scholar] [CrossRef]
- He, Y.; Wang, M.; Wen, S.; Zhang, Y.; Ma, T.; Du, G. Seed Size Effect on Seedling Growth under Different Light Conditions in the Clonal Herb Ligularia Virgaurea in Qinghai-Tibet Plateau. Acta Ecol. Sin. 2007, 27, 3091–3108. [Google Scholar] [CrossRef]
- Herrera, L.P.; Laterra, P. Do Seed and Microsite Limitation Interact with Seed Size in Determining Invasion Patterns in Flooding Pampa Grasslands? In Herbaceous Plant Ecology; Van Der Valk, A.G., Ed.; Springer: Dordrecht, The Netherlands, 2008; ISBN 978-90-481-2797-9. [Google Scholar]
- Souza, M.L.; Fagundes, M. Seed Size as Key Factor in Germination and Seedling Development of Copaifera Langsdorffii (Fabaceae). Am. J. Plant Sci. 2014, 2014, 48412. [Google Scholar] [CrossRef]
- Phartyal, S.S.; Rosbakh, S.; Ritz, C.; Poschlod, P. Ready for Change: Seed Traits Contribute to the High Adaptability of Mudflat Species to Their Unpredictable Habitat. J. Veg. Sci. 2020, 31, 331–342. [Google Scholar] [CrossRef]
- Vaughton, G.; Ramsey, M. Sources and Consequences of Seed Mass Variation in Banksia marginata (Proteaceae). J. Ecol. 1998, 86, 563–573. [Google Scholar] [CrossRef]
- Gairola, S.; Mahmoud, T.; Shabana, H.A.; AlKetbi, A.; Phartyal, S.S. Effect of Seed Size on Germination in Three Species from Arid Arabian Deserts. Botany 2021, 99, 69–74. [Google Scholar] [CrossRef]
- Fredrick, C.; Chima, U.D.; Jimmy, A.O. Effect of Seed Size on Germination and Early Seedling Growth of Dennettia tripetala (G. Baker). PAT 2020, 16, 94–103. [Google Scholar]
- Chacón, P.; Bustamante, R.; Henríquez, C. The Effect of Seed Size on Germination and Seedling Growth of Cryptocarya alba (Lauraceae) in Chile. Rev. Chil. De. Hist. Nat. 1998, 71, 189–197. [Google Scholar]
- Alam, I.; Batool, K.; Huang, Y.; Liu, J.; Ge, L. Developing Genetic Engineering Techniques for Control of Seed Size and Yield. Int. J. Mol. Sci. 2022, 23, 13256. [Google Scholar] [CrossRef]
| QTL | Locus | Molecular Function | Effect on Seed Size | References |
|---|---|---|---|---|
| GS3 | Os03g0407400 | G-protein | Negative on length | [15,140] |
| GW5 | Os05g0187500 | Regulator of the BR signaling pathway | Negative on width | [141,142] |
| GW2 | Os02g0244100 | E3 Ubiquitin Ligase | Negative on width | [143] |
| GL6 | Os06g0666100 | Transcription factor | Positive on length | [135] |
| GL7 | Os07g0603300 | Regulator of cell cycle (to be confirmed) | Positive on length | [136] |
| GL3.1 | Os03g0646900 | Serine/Threonine phosphatase | Positive on length | [137] |
| GS3.1 | Os03g0229500 | MATE transporter | Negative on length and width | [138] |
| GW6 | Os06g0266800 | GAST family protein | Positive on width | [139] |
| GL2 | Os02g0701300 | Transcription factor | Positive on length | [144] |
| GS10 | Os10g0522601 | Proteins with Armadillo tandem repeats | Negative on width | [145] |
| SGW5 | Os02g0543400 | WD40 repeat-like domain-containing protein | Positive on width | [146] |
| GSA1 | Os03g0757500 | UDP-glucosyltransferase | Positive on width and length | [127] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boccaccini, A.; Cimini, S.; Kazmi, H.; Lepri, A.; Longo, C.; Lorrai, R.; Vittorioso, P. When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development. Plants 2024, 13, 1793. https://doi.org/10.3390/plants13131793
Boccaccini A, Cimini S, Kazmi H, Lepri A, Longo C, Lorrai R, Vittorioso P. When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development. Plants. 2024; 13(13):1793. https://doi.org/10.3390/plants13131793
Chicago/Turabian StyleBoccaccini, Alessandra, Sara Cimini, Hira Kazmi, Andrea Lepri, Chiara Longo, Riccardo Lorrai, and Paola Vittorioso. 2024. "When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development" Plants 13, no. 13: 1793. https://doi.org/10.3390/plants13131793
APA StyleBoccaccini, A., Cimini, S., Kazmi, H., Lepri, A., Longo, C., Lorrai, R., & Vittorioso, P. (2024). When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development. Plants, 13(13), 1793. https://doi.org/10.3390/plants13131793






