A Method for Electroporation of Cre Recombinase Protein into Intact Nicotiana tabacum Cells
Abstract
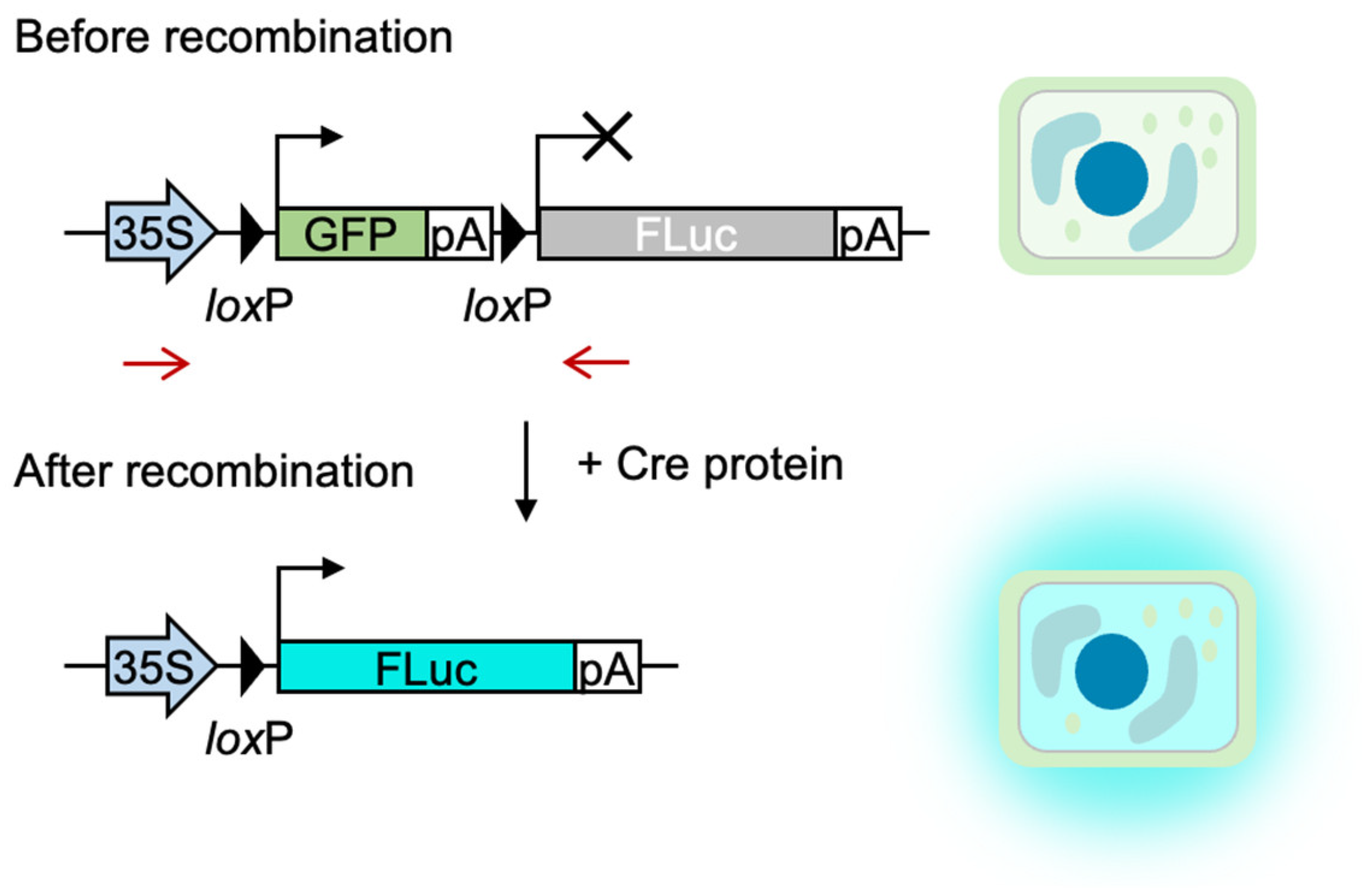
1. Introduction
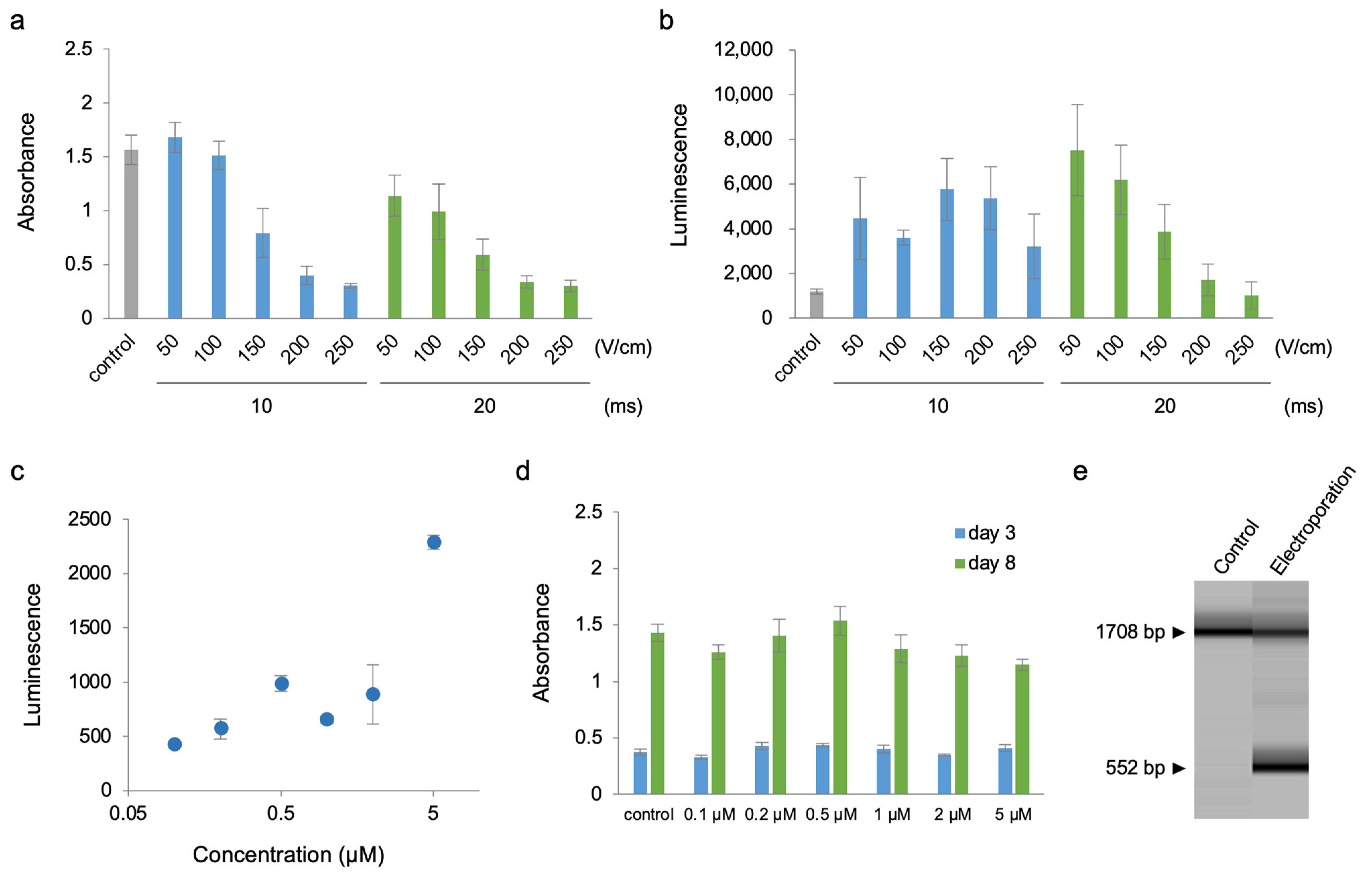
2. Results
3. Discussion
4. Methods
4.1. Plasmid Construct Preparation
4.2. Preparation of Cre Protein
4.3. BY-2 Cell Line and Agrobacterium Tumefaciens (Rhizobium radiobacter) Culture
4.4. Agrobacterium-Mediated Transformation of BY-2 Cells
4.5. Electroporation
4.6. Luminescence Quantification in Cells
4.7. Cell Counting Kit-8 (CCK8) Assay
4.8. Genomic PCR of Cre-Responsive Reporter Cassette
4.9. Quantification and Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Branda, C.S.; Dymecki, S.M. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 2004, 6, 7–28. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A. Cre recombinase: The universal reagent for genome tailoring. Genesis 2000, 26, 99–109. [Google Scholar] [CrossRef]
- Ankrum, J.A.; Dastidar, R.G.; Ong, J.F.; Levy, O.; Karp, J.M. Performance-enhanced mesenchymal stem cells via intracellular delivery of steroids. Sci. Rep. 2014, 4, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Livet, J.; Weissman, T.A.; Kang, H.; Draft, R.W.; Lu, J.; Bennis, R.A.; Sanes, J.R.; Lichtman, J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Furuhata, Y.; Sakai, A.; Murakami, T.; Morikawa, M.; Nakamura, C.; Yoshizumi, T.; Fujikura, U.; Nishida, K.; Kato, Y. A method using electroporation for the protein delivery of Cre recombinase into cultured Arabidopsis cells with an intact cell wall. Sci. Rep. 2019, 9, 2163. [Google Scholar] [CrossRef] [PubMed]
- Furuhata, Y.; Sakai, A.; Kato, Y. Protein electroporation of Cre recombinase into cultured Arabidopsis cells with an intact cell wall. Protoc. Exch. 2019, 1–9. [Google Scholar] [CrossRef]
- Doran, P.M. Therapeutically important proteins from in vitro plant tissue culture systems. Curr. Med. Chem. 2013, 20, 1047–1055. [Google Scholar]
- Leifert, J.A.; Harkins, S.; Whitton, J.L. Full-length proteins attached to the HIV tat protein transduction domain are neither transduced between cells, nor exhibit enhanced immunogenicity. Gene Ther. 2002, 9, 1422–1428. [Google Scholar] [CrossRef]
- Cedeño, C.; Pauwels, K.; Tompa, P. Protein delivery into plant cells: Toward in vivo structural biology. Front. Plant Sci. 2017, 8, 519. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furuhata, Y.; Egi, E.; Murakami, T.; Kato, Y. A Method for Electroporation of Cre Recombinase Protein into Intact Nicotiana tabacum Cells. Plants 2023, 12, 1631. https://doi.org/10.3390/plants12081631
Furuhata Y, Egi E, Murakami T, Kato Y. A Method for Electroporation of Cre Recombinase Protein into Intact Nicotiana tabacum Cells. Plants. 2023; 12(8):1631. https://doi.org/10.3390/plants12081631
Chicago/Turabian StyleFuruhata, Yuichi, Emiko Egi, Tomi Murakami, and Yoshio Kato. 2023. "A Method for Electroporation of Cre Recombinase Protein into Intact Nicotiana tabacum Cells" Plants 12, no. 8: 1631. https://doi.org/10.3390/plants12081631
APA StyleFuruhata, Y., Egi, E., Murakami, T., & Kato, Y. (2023). A Method for Electroporation of Cre Recombinase Protein into Intact Nicotiana tabacum Cells. Plants, 12(8), 1631. https://doi.org/10.3390/plants12081631








